Combining Fluidic Devices with Microscopy and Flow Cytometry to Study Microbial Transport in Porous Media Across Spatial Scales
Summary
Breakthrough curves (BTCs) are efficient tools to study the transport of bacteria in porous media. Here we introduce tools based on fluidic devices in combination with microscopy and flow cytometric counting to obtain BTCs.
Abstract
Understanding the transport, dispersion and deposition of microorganisms in porous media is a complex scientific task comprising topics as diverse as hydrodynamics, ecology and environmental engineering. Modeling bacterial transport in porous environments at different spatial scales is critical to better predict the consequences of bacterial transport, yet current models often fail to up-scale from laboratory to field conditions. Here, we introduce experimental tools to study bacterial transport in porous media at two spatial scales. The aim of these tools is to obtain macroscopic observables (such as breakthrough curves or deposition profiles) of bacteria injected into transparent porous matrices. At the small scale (10-1000 µm), microfluidic devices are combined with optical video-microscopy and image processing to obtain breakthrough curves and, at the same time, to track individual bacterial cells at the pore scale. At larger scale, flow cytometry is combined with a self-made robotic dispenser to obtain breakthrough curves. We illustrate the utility of these tools to better understand how bacteria are transported in complex porous media such as the hyporheic zone of streams. As these tools provide simultaneous measurements across scales, they pave the way for mechanism-based models, critically important for upscaling. Application of these tools may not only contribute to the development of novel bioremediation applications but also shed new light on the ecological strategies of microorganisms colonizing porous substrates.
Introduction
Studies aiming to understand the transport of microbes through porous media have mainly been driven by concerns of contamination1, the transmission of disease2 and bioremediation3. In this regard, bacteria have mostly been treated as particles in transport models4 and processes such as filtration, straining, gravitational settling or remobilization from biofilms have been identified as drivers of retention or transport of microbes5. However, studying the transport of bacteria through porous landscapes can also inform us on the ecological strategies underpinning their success in these complex environments. Yet, this requires novel experiments and mathematical models operating at the single cell, population or microbial community level.
Natural porous environments, such as those found in the hyporheic zone of streams and rivers, are densely colonized by diverse communities of biofilm-forming microbes6. Biofilms form structures that modify the flow and thus the transport and dispersal of bacteria in the liquid phase7,8. The transport of bacteria at pore scale depends to the constrained space availability in the porous matrix and motility-related dispersal may be an effective way to increase the individual fitness through reduced competition for resources in less densely populated areas. On the other hand, motile bacteria can also reach more isolated regions of the porous matrix and the extended exploration of such areas may provide ecological opportunities to motile populations10. At larger spatial scales, biofilm growth diverts the flow paths also leading to (partial) clogging of pores and, thus, to the establishment of even more channelized and heterogeneous flow conditions11. This has consequences for nutrient supply and dispersal capacity, frequency and distance. Preferential flow, for instance, can generate so-called "fast-tracks" and motile bacteria can attain even higher velocities than the local flow along these tracks12. This is an effective way to increase the exploration of novel habitats.
A variety of tools avail themselves for the study of transport of motile and non-motile bacteria (and particles) in porous media. Numerical models have great predictive capacities important for applications, however are often limited by inherent assumptions4. Laboratory-scale experiments13, 14 combined with breakthrough curve (BTC) modeling have provided important insights in the importance of bacterial cell surface properties for sticking efficiency15. Typically, BTCs (i.e., times series of particle concentration at a fixed location) are obtained via constant-rate releases and measurement of cell numbers at the outflow of the experimental device. In this context, BTCs reflect the advection-dispersion dynamics of bacteria in the porous matrix and can be extended by a sink term accounting for attachment. However, modeling of BTCs alone does not resolve the role of spatial organization of the porous substrate or biofilm for transport processes. Other macroscopic observables like dispersivity or deposition profiles have been proven to provide important information about the spatial distribution or the retained particles or growing communities. Microfluidics is a technology that allows studying transport in porous media by microscopy investigation9,12,16, and except a recent work10, experimental systems are typically constrained to a single length scale of resolution, that is, the pore scale or the entire fluidic device scale.
Here, we introduce a suite of combined methods to study the transport of motile and non-motile bacteria in porous landscapes at different scales. We combine observations of bacterial transport at the pore scale with information at larger scale, by means of BTC analysis. Microfluidic devices built from soft lithography using polydimethylsiloxane (PDMS) are bio-compatible, resistant to a range of chemicals, allow replicability at low costs and provide excellent optical transparency as well as low autofluorescence critical for microscopic observation. Microfluidics based on PDMS has been previously used to study the transport of microbes in simple channels17 or in more complex geometries12. However, typically microfluidics experiments focus on short-term horizons and epi-fluorescence microscopic observation of living cells is commonly restricted to genetically-modified strains (e.g., GFP-tagged strains). Here we present tools to study bacterial transport using PDMS-based microfluidic devices in combination with microscopy and larger devices fabricated from poly(methyl methacrylate) (PMMA, also known as plexiglass) in combination with flow cytometry. PDMS and PMMA differ in gas permeability and surface properties, thus providing complementary opportunities to study bacterial transport. While the microfluidic device provides a more controlled environment, the larger device allows for experiments over extended periods of time or using natural bacterial communities. Microscopy counting at high temporal resolution in a dedicated area is used to obtain BTC in the PDMS-based microfluidic device. To obtain cell counts for BTC modeling from the PMMA-based device, we introduce a self-constructed automated liquid dispenser in combination with flow cytometry. In this setup, cells pass the fluidic device and are consecutively dispensed into 96 well plates. The temporal resolution is restricted by the minimum volume that can be accurately dispensed and thus the medium flow rate through the fluidic device. Fixative in the wells prevents growth and facilitates DNA staining for downstream flow-cytometric enumeration. To prevent bacterial growth during transport experiments we use a minimal medium (termed motility buffer).
Since protocols for the preparation of fluidic devices at different scales are readily available, we only briefly introduce the techniques to produce such devices and rather focus on the experimental procedures to record BTCs. Similarly, various routines exist for the flow cytometric enumeration of microbes and users require expert knowledge to interpret results obtained by flow cytometry. We report the novel use of microfluidic devices in combination with microscopic imaging to record BTCs of fluorescently-tagged cells. At the pore scale, local velocities and trajectories are obtained by means of image processing. Further, we demonstrate the use of a PMMA-based fluidic device in combination with flow-cytometric counting to observe bacterial transport of motile and non-motile cells in porous environments colonized by a native stream biofilm.
Protocol
1. Bacterial culture conditions
- Working under a laminar flow hood, use 100 μL of a glycerol stock of GFP-tagged Pseudomonas putida KT2440 (1 × 107 mL-1, stored at -80 °C) to inoculate 5 mL of Luria-Bertani (LB) medium. Incubate at 30 °C while shaking at 250 rpm overnight.
- The next day, resuspend 100 μL of the overnight culture in 5 mL LB medium and incubate under the same conditions for 5h (exponential phase). Sample a 1 mL aliquot into a 2 mL tube, allow to cool to room temperature (~15 min) and centrifuge (2300 x g for 5 min).
- Remove the supernatant and add 1 mL motility buffer to the pellet. Vortex briefly to homogenize the sample. Dilute to reach the desired cell concentration, e.g., 5 x 105 mL-1.
- For experiments involving natural communities, such as those derived from streams, prepare a non-selective cultivation medium. For instance, use sterile-filtered and autoclaved stream water or an artificial stream water medium amended with a complex carbon source (LB medium).
2. Preparation of a microfluidic device in polydimethylsiloxane (PDMS)
- Design the desired porous geometry by means of computer-aided drafting (CAD) software18, which consists of a matrix of circles (that is the impermeable obstacle to flow), described by radius size and center coordinates.
NOTE: An example of a porous geometry with randomized grain and pore sizes is provided in Figure 1A. An observation channel without obstacles close to the outlet facilitates the acquisition of BTCs. - Based on the chosen geometry, prepare a mold using standard SU-8-photolithography18.
NOTE: Alternatively, molds can also be ordered from dedicated microfabrication facility. In order to obtain heterogeneous fluid flow in the horizontal plane, it is important to design the thickness of the microfluidics chamber of the same order of magnitude as the average pore throat size. However, make sure that the dimensions of the microfluidic device are suitable for observation under the microscope (e.g., work on microscope slides). - Prepare 50 g of PDMS by adding 10% of cross linker (dimethyl, methylhydrogen siloxane copolymer) to 90% of elastomer by weight, using a syringe without needle. Work under clean conditions and avoid dust as much as possible. Mix the two reagents in a clean disposable container and apply vacuum (100 mbar) for 30 min to remove dissolved air and bubbles from the viscous PDMS.
- Place the mold into a Petri dish (100 mm in diameter, 15 mm high). Pour the PDMS onto the mold to the desired height (e.g., 2-5 mm). Cover the petri dish and keep it at 60 °C for 4 h (overnight for thicker layers) to cure.
NOTE: For visualization purpose, light should be able to pass through the PDMS, thus, a thin layer between 2 mm to 5 mm is desirable. Thicker layers (>5 mm) reduce the device transparency and thinner ones are subjected to deformations during application. - Remove the mold from the oven and allow the microfluidic device to cool to room temperature. Once it is cooled, carefully remove the desired portion of PDMS with a scalpel.
NOTE: Strong pressures on the mold result in mold fractures. Do not touch the PDMS with bare hands, as fingerprints will affect optical transparency. - Temporarily seal the bottom of the PDMS channel (where the desired geometry has been engraved) with tape. With a 0.5 mm diameter biopsy puncher, pierce microfluidic channel to create an inlet and an outlet fitting the 0.5 mm (inner diameter) tubing.
NOTE: The soft nature of PDMS will ensure tightness once the tubing will be inserted. Inlet and outlet channels cannot be made after the PDMS has been sealed to the glass. - Seal the microfluidic channel via oxygen plasma bonding using the high frequency generator (plasma bonder, Table of Materials). For this, clean a silicate glass slide (25 mm x 75 mm) with ethanol and let it dry. Remove tape from the PDMS channel and place the channel with the porous side facing up. Treat the silicate glass slide and PDMS surfaces with plasma for about 45 s at room temperature.
- Place the PDMS channel onto the silicate glass slide and heat at 100 °C for 30 min on a hot plate.
- Remove the microfluidic device from the hot plate and cool it to room temperature. Connect the PDMS channel inlet with tubing. Apply vacuum for 30 min to remove air from PDMS, which is almost impermeable to fluids but permeable to gas.
- Prepare 100 mL of motility buffer (10 mM potassium phosphate, 0.1 mM EDTA, supplemented with 1% w/v glucose, pH 7.0) and inject 1 mL of it into the microfluidic device using a syringe pump operated at 10 µL min-1.
NOTE: As the PDMS is under-saturated in gas (due to the previous vacuum step), bubbles will disappear within ~30 min.
3. Preparation of a fluidic device in poly (methyl methacrylate)
- Design the desired geometry with the CAD software. If applicable, make sure that the dimensions are suitable for observation under the microscope (e.g., dimensions of a standard 96 well plate in combination with an appropriate holder). The fluidic device is composed by a base (127 × 127 × 12 mm) and a lid (127 × 127 × 12 mm) of PMMA.
NOTE: Expertise with CAD software is recommended. An example technical drawing is supplied in Figure 1A. - To produce the pore compartment, by means of high precision micromilling (WF31SA, Mikron), remove 0.5 mm from the bottom PMMA layer and mill a groove (1.1 × 1.1 mm) for a rubber O-ring. Drill 12 threaded holes (M5). This will serve as the base of the fluidic device.
NOTE: The dimensions of the fluidic device need to be adjusted to fit the microscope stage and focal distance. A technical drawing is supplied in Figure S1. - Drill two threaded holes (type 1/4-28UNF) for in- and outlet into the top part of the fluidic device, and 12 holes (5.5 mm) for the screws. This will serve as the lid of the fluidic device.
NOTE: Expertise in micro-milling is advisable; the authors use support from a specialized workshop. - In order to clean and sterilize the fluidic device prior to and after each use, soak the base and lid of the fluidic device in HCl 7% and rinse three times with deionized water.
- Screw base and lid together using the 12 threaded holes.
4. Setup of automated dispenser
NOTE: Commercially available liquid dispensers are costly and often do not offer the flexibility to dispense directly from the outflow of the fluidic device. Therefore, assembling a cheap and flexible robotic dispenser system from an XY Plotter Robot (Table of Materials) is recommended.
- In order to dispense the outflow from the fluidic device into 96 well plates, mount the robotic dispenser onto a PMMA plate and mill cavities of 85.8 x 128 mm with a depth of 1 mm to hold several 96 well plates.
- Attach the outflow tube of the fluidic device to the robotic arm of the dispenser.
- To operate the robotic dispenser download bCNC from github: https://github.com/vlachoudis/bCNC and follow the instructions to install the program.
- Download dispenser.py from the supporting material of this article.
NOTE: This python code provides a plugin to bCNC for a simple robotic dispenser layout. - Connect the robotic dispenser to the computer running bCNC and identify the correct COM port.
- In bCNC, click the home button to return the robotic dispenser to the home position.
NOTE: Homing returns the robotic dispenser to a known position (x=0, y=0) and therefore improves the accuracy of the dispenser. - Prior to the experiment, prepare a sufficient number of 96 well plates, with wells containing an appropriate amount of fixative (e.g., final concentration 3.7% formaldehyde).
NOTE: For instance, at a flow rate of 0.2 mL min-1, 100 µL are dispensed into each well every 30 s. Therefore, add 10 µL of 37% formaldehyde to each well to reach a final concentration of Formaldehyde between 2 and 4%. Using eight 96 well plates will allow to operate the experiment for more than 6 h with a total of 768 data points. Moreover, note that GFP-tagged cells may lose their fluorescent signal after fixation using formaldehyde.
5. Analyze bacterial transport using PDMS microfluidic devices
- Place the PDMS microfluidic device previously saturated with the motility buffer on the microscope stage. Use tape to fix the tubing to minimize disturbance of the flow during stage movement.
- Move the microscope stage to the observation channel close to the outlet. Using bright field microscopy or phase contrast, focus to the center of the observation channel and adjust the magnification to visualize individual bacterial cells.
- Switch the light path settings to fluorescence microscopy and adjust the camera exposure time to resolve individual bacterial cells (e.g. 100 ms), or such that fluorescence signals of cells are at least 3x stronger than background noise.
- Next, insert the inlet tubing into a 2 mL tube containing the bacterial suspension. Reverse pump direction and start withdrawing the suspension at a flow rate of 1 µL min-1.
- Scan the cross section of the entire observation channel recording a composite picture every 2 min, over the entire duration of the experiment.
6. Basic image processing
NOTE: The goal of these basic image processing routines is to count the number of bacteria in the recorded images. Optimal processing procedures depend on the technical specifications of the microscope and camera, as well as on the fluorescence properties of the bacterial strain used in the experiment and therefore need to be adjusted.
- First export images in .tiff format.
- Import images to a desired software platform (e.g., MATLAB, ImageJ, R or Python).
- Remove camera noise, which is a random variation of pixel intensity in images and correct for optical aberration. This can be done applying a Gaussian filter to each picture: the size of the filter depends on the quality of the camera sensor (e.g. CCD or CMOS). The optical aberration can be removed by normalizing each picture by a reference image collected in absence of the specimen with the same optical configuration.
- Crop the images to a region of interest.
- Identify a threshold value (pixel intensity), such that values above the threshold include bacterial cells.
NOTE: In case images are unevenly illuminated (because of optical aberration or noise) it may be useful to apply an adaptive threshold, which chooses a threshold value based on local mean intensities. - Subtract from each picture the threshold defined above.
- Binarize the resulting image, such that bacterial cells take a value of 1, whereas background takes a value of 0.
- Remove clusters with an area smaller than the smallest bacterial cell size.
- Sum the binarized image to obtain the total number of pixels of the remaining clusters. Divide the number of pixels by the average size (in pixels) of a bacterial cell: this provides an estimate of the total number of cells.
- Knowing depth of view and area of investigation, transform counts into concentration (particles mL-1).
- It is fundamental to identify the concentration of the injected bacteria suspension. To do this, inject with a syringe 1 mL of bacteria culture suspension into the observation channel of a clean microfluidic device. Record the image and calculate the influent bacterial concentration (C0) as described above (6.1 to 6.10).
- Analyze BTCs by normalizing effluent bacterial concentration (C) with influent bacterial concentration (C0) and plot C/C0 versus time.
7. Analyze bacterial transport at the pore scale
- In order to analyze local velocities and trajectories of bacteria transported through the porous matrix, move the microscope stage to the region of interest and adjust the focus to the center of the microfluidic device.
- Set microscope to bright field or phase contrast.
NOTE: This only in case the fluorescence signal of bacterial cell does not allow recording images at exposure time shorter than the average bacterial time, otherwise use fluorescence microscopy. - Record time-lapse images, at exposure time that captures bacteria displacement (shorter than the average displacement over a number of pixels smaller than the object size), and that optimizes bacterial cell detection (e.g. exposure of 20 ms and images recorded every 50 ms). Record pictures over a sufficient amount of time in order to record enough (to be statistically representative) of the slowest trajectories (e.g. 3 min).
NOTE: Make sure the computer has enough disk space. - To remove background noise, subtract from each image the average of all recorded images. To do that, create a matrix whose result is the sum of intensity of all the images, for each pixel, and divide it by the number of images.
- From the processed image (Im), determine the modulus of the numerical gradient
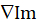 and normalize it by its maximum value (max), as defined below.
and normalize it by its maximum value (max), as defined below.
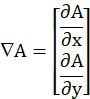
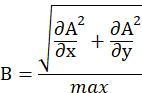
- Binarize the matrix B via intensity thresholding, see steps 6.7-6.8.
- For each time point, record bacteria coordinates (x, y in pixel or mm) and time of image acquisition into a three-column file.
- Finally, apply a particle tracking script to process the recorded data and compute the trajectories of the bacteria. For instance use the established protocol19, and the freely available Matlab Particle Tracking Code: (http://site.physics.georgetown.edu/matlab/).
8. Study bacterial filtration by means of deposition profiles
- To obtain deposition profiles of GFP-tagged P. putida KT2440 cells by fluorescence microscopy, record a composite image of the entire porous channel before, that is the background, and after injection of bacterial suspension through the microfluidic device. Use an exposure time that allows acquiring bacterial fluorescent signal (e.g. 100 ms), without bleaching the signal.
- Export images and import in desired software platform (see 6.1-6.2).
- Remove background from the images recorded after bacterial injection.
- Integrate the total fluorescence signal of retained bacteria along transversal sections of the porous channel.
- In order to compute the deposition profile, plot the integrated florescence signal versus porous channel length.
9. Analyze bacterial transport using PMMA fluidic devices and flow cytometry
- Connect peristaltic pump with the inlet using 50 cm (1 mm inner diameter) tubing, and the outflow with the automated dispenser using the same tubing (50 cm, see section 4).
NOTE: Use the pump to dispense cultivation media, and to inject bacterial cells. Use Luer-lock connectors and three-way valves to shift between medium and bacterial suspension during the constant-rate release. - Pump cultivation medium in the fluidic device. Note the arrival of medium at the outlet tubing fixed to the robotic dispenser.
- Start injecting the bacterial suspension through the PMMA fluidic device at a flow rate of 0.2 mL min-1,
- When bacteria reach the fluidic device inlet turn on the automated dispenser.
- In order to make a pulse injection, inject bacterial suspension for some pore volumes (e.g. 30), and then switch injection to cultivation media until experiment end.
NOTE: pore volume of the fluidic device is approximately 0.2 mL, thus at the prosed flow rate every minute one entire pore volume is exchanged. - Once a 96 well plate is completed, cover the plate to reduce evaporation and store it at 4 °C.
- Analyze bacterial abundance via flow cytometry, following established protocols20.
NOTE: For instance, add 25 µL of the green-fluorescent nucleic acid stain (Table of Materials) at 0.025 mM (in ultrapure water) to each well. It stains bacterial DNA, thus allowing to quantify by flow cytometry the bacterial abundance. Incubate samples for 15 min in the dark and then analyze using a flow cytometer equipped with a 488 nm laser and detectors at 515 nm. - Prior to BTC analysis, consider the dilution of the sample due to the addition of fixative and stain. Correct bacterial abundance by a factor of 1.35 to account for fixative and stain.
Representative Results
To illustrate the functionality of the presented workflow, we performed experiments using genetically modified Pseudomonas putida KT2440, a gram negative motile bacterium important for bioremediation and biotechnology. Genetically modified versions of this strain that express GFP production are commercially available. A non-motile strain of P. putida KT2440 which lacks the relevant structural and regulatory genes for motility is also available. Using both, motile and non-motile GFP tagged P. putida KT2440, we performed sequential experiments in PDMS microfluidic devices with a random array of pillars (Figure 1B) and recorded BTCs (Figure 2A). BTCs have been normalized to the concentration of injected cells (C0). Simultaneously, bacterial trajectories at the pore scale were visualized via image processing and particle tracking as described above (Figure 2B).
Next, we performed experiments with large-scale fluidic devices milled from PMMA (Figure 1A). Motile and non-motile P. putida KT2440 (non-fluorescent) were injected into a regularly spaced porous matrix and BTCs were obtained using the liquid dispenser and flow cytometry counting as described above (Figure 3A). Strikingly, in a porous environment devoid of biofilm, motile and non-motile P. putida KT2440 showed a markedly different transport behavior. In a porous matrix colonized for 48h with a complex stream biofilm community, these differences in BTC between motile and non-motile P. putida KT2440 vanished (Figure 3B.)
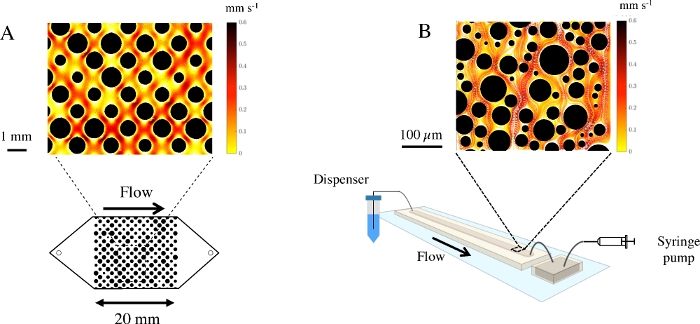
Figure 1: Fluidic devices to study microbial transport in porous media (A) Illustration of a fluidic device milled from PMMA. The porous matrix is milled into the base layer of the device, the lid is closed using screws. A cross section shows the arrangement of the pillars within the fluidic device. The insert shows a porous matrix with a regular spaced grid of pillars and the respective velocity flow field. (B) The PDMS device is mounted onto a microscopy glass slide. Shown are the in- and outflow, connected to the medium reservoir and the syringe pump, respectively. The observation chamber for microscopic counting is placed as a separate chamber without a porous matrix onto the same microscope slide. The insert shows a porous matrix with a random array of pillars (in diameter and spacing). Please click here to view a larger version of this figure.
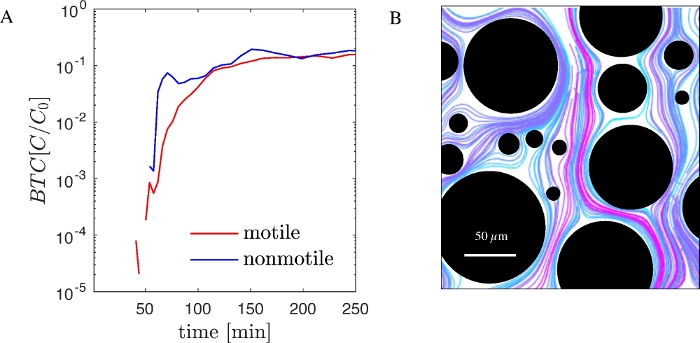
Figure 2: Bacterial transport at channel and pore scale in the PDMS fluidic device (A) BTCs of motile and non-motile P. putida KT2440 (GFP tagged) obtained with a PDMS microfluidic device and microscopic counting. (B) Trajectories of non-motile cells at the pore scale. Colors are chosen to enhance differentiation of trajectories. Please click here to view a larger version of this figure.
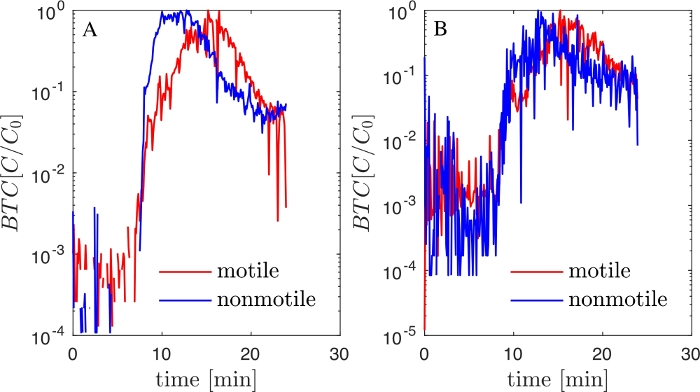
Figure 3: Bacterial transport at channel and pore scale in the PMMA fluidic device (A) BTCs of motile and non-motile P. putida KT2440 (non tagged) obtained using a PMMA fluidic device and flow-cytometry counting. (B) The fluidic device was colonized by a natural stream community for 2 days. Please click here to view a larger version of this figure.
Supplementary Figure 1: Technical drawings of the PMMA fluidic device. The device is composed of a base unit containing the porous matrix and a lid unit featuring the holes for the inlet and outlet. The device is sealed using 12 screws and an O-ring. Please click here to download this figure.
Discussion
Here we suggest two means to study the transport of microbes through porous systems at the single-cell and population level. While the study of transport phenomena using BTC modeling has provided valuable insights into the spread of pathogens or contaminants at the ecosystem scales, difficulties to scale from laboratory experiments to field conditions still exist. The tools described here allow researchers to experimentally resolve the spatial and temporal scales in order to better understand the ecological strategies of microbes relevant for transport in porous environments. Experimenters may use or modify these systems to study other microbial traits than motility, such as chemotaxis or quorum sensing or modify the geometry or other habitat characteristics of the porous matrix. Moreover, using these systems the bacterial transport behavior can be readily coupled to deposition profiles, which provide important insights into colonization patterns and are critical to understand how biofilms modify local flow fields. We anticipate that a better understanding of microbial strategies to disperse and colonize porous media will improve model predictions and thus contribute to the management of pathogen spread or contaminant containment. Further modifications of the system may also contribute to the development of novel filtration devices or biotechnology tools in which cells need to be physically separated.
We recommend PMMA-based devices for large and long-term experiments and PDMS based devices for smaller, shorter term experiments or when high temporal resolution is critical. It has to be kept in mind that the two materials have different properties. For instance PDMS is permeable to gas like oxygen, while PMMA is gas tight. This difference might be used to study gas consumption in the PMMA scenario, while PDMS might be more suitable for experiment where oxygen limitations related to bacterial respiration are undesired.
In general, the protocols described here are easily reproducible and data obtained using these tools consistently reveals differences in the transport of motile and non-motile bacteria. The self-made liquid dispenser may be replaced by a commercially available alternative. However, for reasons of versatility and cost-effectiveness we recommend the one described here. Critical steps in the protocol mainly concern the handling of the fluidic devices and experience with image processing. The quality of data obtained through image analysis critically depends on image quality (mainly determined by focus and exposure time) and an appropriate thresholding strategy. Data quality obtained by flow-cytometric counting critically depends on effective fixing and staining of the cells and expertise in the interpretation of flow-cytometry results.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We acknowledge the help of Antoine Wiedmer with the setup of the robotic dispenser and the dispenser.py script.
Materials
| EDTA | Sigma | ||
| Elastomer Sylgard 184 | Dowsil | 101697 | |
| Flow cytometer NovoCyte | Acea | ||
| Glucose | Sigma | https://www.makeblock.com/project/xy-plotter-robot-kit | |
| LB broth | BD | ||
| Liquid dispenser, XY Plotter Robot Kit | makeblock | ||
| Microscope Axio Imager | Zeiss | ||
| Microscope AxioZoom v16 | Zeiss | ||
| Microscope slides, 75 mm × 25 mm | Corning | ||
| Minipuls 3 peristaltic pump | Gilson | ||
| Plasma bonder Corona SB | BlackHole Lab | ||
| Potassium phosphate | Sigma | ||
| Syringe pump New Era NE 4000 | New Era | ||
| Syto 13 Green Fluorescent Nucleic Acid Stain | Molecular Probes, Invitrogen | ||
| Tygon tubing | Ismatec | ||
| WF31SA universal milling machine | Mikron |
Riferimenti
- Stevik, K., Aa, K., Ausland, G., Fredrik Hanssen, J. Retention and removal of pathogenic bacteria in wastewater percolating through porous media: a review. Water Research. 38 (6), 1355-1367 (2004).
- Ribet, D., Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes and Infection. 17 (3), 173-183 (2015).
- Ginn, T. R., Wood, B. D., Nelson, K. E., Scheibe, T. D., Murphy, E. M., Clement, T. P. Processes in microbial transport in the natural subsurface. Advances in Water Resources. 25 (8), 1017-1042 (2002).
- Tufenkji, N. Modeling microbial transport in porous media: Traditional approaches and recent developments. Advances in Water Resources. 30 (6-7), 1455-1469 (2007).
- Foppen, J. W., Van, M. H., Schijven, J. Measuring and modelling straining of Escherichia coli in saturated porous media. Journal of contaminant hydrology. 93 (1-4), 236-254 (2007).
- Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M., Packmann, A. I. The ecology and biogeochemistry of stream biofilms. Nature Reviews Microbiology. 14 (4), 251-263 (2016).
- Scheidweiler, D., Peter, H., Pramateftaki, P., Anna, P., de Battin, T. J. Unraveling the biophysical underpinnings to the success of multispecies biofilms in porous environments. The ISME Journal. 1, (2019).
- Carrel, M., et al. Biofilms in 3D porous media: Delineating the influence of the pore network geometry, flow and mass transfer on biofilm development. Water Research. 134, 280-291 (2018).
- Bhattacharjee, T., Datta, S. S. Bacterial hopping and trapping in porous media. Nature Communications. 10 (1), 2075 (2019).
- Scheidweiler, D., Miele, F., Peter, H., Battin, T. J., de Anna, P. Trait-specific dispersal of bacteria in heterogeneous porous environments: from pore to porous medium scale. Journal of The Royal Society Interface. 17 (164), 20200046 (2020).
- Morales, V. L., Parlange, J. Y., Steenhuis, T. S. Are preferential flow paths perpetuated by microbial activity in the soil matrix? A review. Journal of Hydrology. 393 (1), 29-36 (2010).
- Creppy, A., Clément, E., Douarche, C., D’Angelo, M. V., Auradou, H. Effect of motility on the transport of bacteria populations through a porous medium. Physical Review Fluids. 4 (1), 013102 (2019).
- Camesano, T. A., Logan, B. E. Influence of Fluid Velocity and Cell Concentration on the Transport of Motile and Nonmotile Bacteria in Porous Media. Environmental Science & Technology. 32 (11), 1699-1708 (1998).
- Lutterodt, G., Basnet, M., Foppen, J. W. A., Uhlenbrook, S. The effect of surface characteristics on the transport of multiple Escherichia coli isolates in large scale columns of quartz sand. Water Research. 43 (3), 595-604 (2009).
- Bozorg, A., Gates, I. D., Sen, A. Impact of biofilm on bacterial transport and deposition in porous media. Journal of Contaminant Hydrology. 183 (Supplement C), 109-120 (2015).
- Long, T., Ford, R. M. Enhanced Transverse Migration of Bacteria by Chemotaxis in a Porous T-Sensor. Environmental Science & Technology. 43 (5), 1546-1552 (2009).
- Rusconi, R., Garren, M., Stocker, R. Microfluidics Expanding the Frontiers of Microbial Ecology. Annual Review of Biophysics. 43 (1), 65-91 (2014).
- Xia, Y., Whitesides, G. M. Soft Lithography. Annual Review of Materials Science. 28 (1), 153-184 (1998).
- Crocker, J. C., Grier, D. G. Methods of Digital Video Microscopy for Colloidal Studies. Journal of Colloid and Interface Science. 179 (1), 298-310 (1996).
- del Giorgio, P. A., Bird, D. F., Prairie, Y. T., Planas, D. Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnology and Oceanography. 41 (4), 783-789 (1996).

