Laser-free Hydroxyl Radical Protein Footprinting to Perform Higher Order Structural Analysis of Proteins
Summary
This protocol presents a method to use inline radical dosimetry and a plasma light source to perform flash oxidation protein footprinting. This method replaces the hazardous UV laser to simplify and improve the reproducibility of fast photochemical oxidation of protein studies.
Abstract
Hydroxyl Radical Protein Footprinting (HRPF) is an emerging and promising higher order structural analysis technique that provides information on changes in protein structure, protein-protein interactions, or protein-ligand interactions. HRPF utilizes hydroxyl radicals (▪OH) to irreversibly label a protein’s solvent accessible surface. The inherent complexity, cost, and hazardous nature of performing HRPF have substantially limited broad-based adoption in biopharma. These factors include: 1) the use of complicated, dangerous, and expensive lasers that demand substantial safety precautions; and 2) the irreproducibility of HRPF caused by background scavenging of ▪OH that limit comparative studies. This publication provides a protocol for operation of a laser-free HRPF system. This laser-free HRPF system utilizes a high energy, high-pressure plasma light source flash oxidation technology with in-line radical dosimetry. The plasma light source is safer, easier to use, and more efficient in generating hydroxyl radicals than laser-based HRPF systems, and the in-line radical dosimeter increases the reproducibility of studies. Combined, the laser-free HRPF system addresses and surmounts the mentioned shortcomings and limitations of laser-based techniques.
Introduction
Protein conformation and associated higher order structure (HOS) are the principal determinants of proper biological function and aberrant behavior1. The same applies to biopharmaceuticals, whose structure and functional activity is dependent on various aspects of their production and environment. Biopharmaceutical change in HOS have been linked to adverse drug reactions (ADR) attributed to undesirable pharmacology and patient immunological response2,3. The appearance of ADRs has alerted the biopharmaceutical industry to the critical role that protein HOS plays in the safety and efficacy of biotherapeutics, and they have established the need for new and improved HOS analytics4.
Hydroxyl Radical Protein Footprinting (HRPF) is a promising technique to track the change in protein HOS. HRPF involves the irreversible labeling of a protein’s exterior with ▪OH followed with mass spectrometry (MS) analysis to identify the solvent accessible surface of the protein5,6,7. HRPF has successfully been used to detect defects in protein HOS and its function8,9, characterize the HOS of monoclonal antibodies (mAb)10,11,12,13, determine the binding Kd of a ligand14, and much more15,16,17,18,19. A common method to generate the ▪OH for HRPF is Fast Photochemical Oxidation of Proteins (FPOP), which employs high-energy, fast UV lasers to produce ▪OH from photolysis of H2O2. For the most part, FPOP uses expensive excimer lasers employing hazardous gas (KrF) that demands substantial safeguards to avoid respiratory and eye injury20. To avoid inhalation hazards, others have used frequency quadrupled neodymium yttrium aluminum garnet (Nd:YAG) lasers21, which eliminates the use of toxic gas but are still costly, require significant operational expertise, and demand extensive stray light controls to protect users from eye injury.
Although ample information can be obtained using HRPF, broad adoption in biopharma has not been met. Two barriers for the limited HRPF adoption include: 1) the use of dangerous and expensive lasers that demand substantial safety precautions20; and 2) the irreproducibility of HRPF caused by background scavenging of ▪OH that limit comparative studies22. To supplant laser use, a high-speed, high energy plasma flash photolysis unit was developed to safely perform FPOP in a facile manner. To improve on the irreproducibility of HRPF experiments, real-time radical dosimetry is implemented.
The practice of HRPF has been limited by irreproducibility attributed to background scavenging of ▪OH22. While ▪OH are excellent probes of protein topography, they also react with many constituents found in preparations, making it necessary to measure the effective concentration of radical available to oxidize a target protein. Variations in buffer preparation, hydrogen peroxide concentration, ligand properties, or photolysis can result in oxidation differences between control and experimental groups that create ambiguity in HOS differential studies. The addition of real-time radical dosimetry enables the adjustment of the effect ▪OH load and therefore increases the confidence and reproducibility during an HRPF experiment. The use of radical dosimetry in FPOP has been described elsewhere23,24,25, and is further discussed in detail in a recent publication26. Here, we describe the use of a novel flash photolysis system and real-time dosimetry to label equine apo-myoglobin (aMb), comparing levels of peptide oxidation in an FPOP experiment to that of obtained when using an excimer laser.
Protocol
1. Installing the capillary tube
- Using a silica cleaving stone, cleave 250 µm inner diameter (ID) silica capillary to 27 inches. Check the capillary ends for a clean, straight cut.
- Create two windows roughly 15 mm in length by burning away the polyimide coating. Starting from the “lower end” make the first photolysis window 90 mm away from the “lower end” and the second dosimeter window 225 mm away from the “lower end”.
NOTE: Once the coating is burned away the capillary is very fragile. - Unscrew the nut and ferrule at port 5 and insert the “lower end” of the capillary just beyond the conical end of the ferrule (Figure 1A).
- On the photolysis module, remove the photolysis cell cap by pulling it straight out and then remove the magnetically mounted metal mask which will hold the capillary in place.
CAUTION: Inside the photolysis cell cap is a curved mirror, do not allow anything to touch the mirror. - On the dosimeter module, open the dosimeter cell by pushing on the tab on the left and swing the dosimeter cell open to the right. Remove the magnetically mounted clips which will hold the capillary in place.
- Place the capillary into the grooved channel in the base of the photolysis cell. Center the capillary window with the photolysis cell window. While holding the capillary in position with one hand, add the magnetic mask to hold the capillary in place. Place the photolysis cap back in position.
- Place the capillary into the grooved channel in the base of the dosimeter cell. Center the second capillary window on the small aperture at the center of the dosimeter cell base. While holding the capillary in position with one hand, place the two magnetic clips in position to hold the capillary in place. Close the dosimeter cell until it clicks closed.
- Insert the capillary through the knurled nut atop the capillary lift of the product collector (Figure 1B). Extend the capillary to just above the bottom of the vial.
NOTE: It is important for the capillary to reach the bottom of the vial to ensure that the capillary is submerged in the quench solution while running an experiment.
2. Installing an injection loop
- Use Teflon tubing with an outer diameter 1/16” and an inner diameter of 0.015” (381 µm). Follow equation 1 to calculate the length of tubing needed for the desired volume using equation 1.
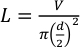
Where L is the length of the tubing in millimeters, V is the desired volume in microliters and d is the tube inner diameter in millimeters. For an injection volume of 25 µL and an inner diameter of 381 µm, the tubing length needs to be roughly 219.3 mm.
NOTE: For volumes less than 20 µL, use PTFE tubing with an inner diameter of 0.01”. - Cut the Teflon tubing to the necessary length using a tube cutter. Check the ends of the tube for a clean, straight cut.
- Insert one end of the new injection loop through one of the nuts and place a new ferrule onto the end of the tube. Hold the ferrule and nut in place and insert the tube into port 3 of the injection valve until it bottoms out in the valve. Hold the tube firmly and tighten the nut finger tight. Using a wrench, tighten the nut ¼ turn further. Remove and inspect the assembly.
NOTE: If the ferrule is not fixed in position, reinstall and tighten 1/8 turn further. Repeat with the other end of the loop. - Once both ends have a nut and fixed ferrule, loosely screw one end to port 3 and the other end to port 6 (Figure 1C). Once in position tighten both sides to finger tight then ¼ turn past finger tight with a wrench.
3. Initialize the photolysis system
- Turn on the photolysis modules in the following order: (1) Fluidics Module (2) Photolysis Module (3) Dosimetry Module (4) Product Collector, and lastly (5) the system computer and launch the control software
NOTE: Allow the Dosimeter Module at least a half-hour to warm up from a cold start. - Fully submerge tubing from the “Valve” position on the syringe pump (Figure 1D) into 10 mL of buffer. Place the tubing from the “Waste” position (Figure 1D) and tubing from port 2 on the syringe port (Figure 1C) to an empty container to collect waste.
NOTE: Use the buffer your protein will be suspended in. A recommended buffer is 10 mM NaPO4. - On the Product Collector carousel, place 1.5 mL microcentrifuge tubes at the position marked H and 6.
- If using 250 µm ID capillary, set the load flow rate to 500 µL/min, processing flow rate to 7.5 µL/min, and waste flow rate to 500 µL/min.
- Calculate the flash delay, product time, and waste time to use the Semi-Automatic Mode using equation 2.
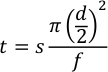
Where t is the time in minutes, s is the capillary distance in millimeters, d is the inner diameter of the capillary in millimeters, and f is the flow rate in µL/min.- For the flash delay, the distance is from the “lower end” of the capillary to the first window, which should be roughly 90 mm. Using 250 µm ID capillary and a processing flow rate of 7.5 µL/min the sample will arrive to the photolysis window in about 35 seconds. Allow two flashes at 1 Hz to occur before the samples arrive, so set the flash delay to 33 seconds.
- For the product time, enter the amount of time injected solution will take to flow from the injection valve to the end of the capillary. For a 27” long 250 µm ID capillary, set the product time to 4.5 minutes.
- For the waste time, enter the amount of time the total volume of injected solution will take to be collected. At this time, the product collector will move from the product position to the waste vial. For an injection volume of 25 µL and a flow rate of 7.5 µL/min, set the waste time to 7.8 minutes.
- Rinse out the injection loop five times by injecting 25 µL of HPLC-grade water into the injecting port with the injection valve set to the load position.
- Manually turn the injection valve up to ‘inject position’ to flush the rest of the system. Select Process (Out) on the control software to begin flowing water. While flowing, raise the product collector capillary lift by selecting Up under the product collector so that you can see the end of the capillary. Flow water through the capillary until a droplet forms.
NOTE: If a droplet does not form, check the injector valve for leaks. If there is a leak, loosen the nut, reseat the capillary, and retighten.
4. Determine actual ▪OH yield to test radical scavenging effects from the buffer.
- In the Control Software, start the flow by selecting Process (Out). In the settings tab, set the flash voltage to 0 V. Under the Manual Control tab in the dosimeter data section, select Start Data + AutoZero.
- Select the position for the product vial (H) and waste vial (6).
- Select Ready, manually turn the injection valve down to the load position, and inject 25 µL of 1 mM Adenine with 100 mM H2O2 into the injection port. Once injected, manually turn the injection valve up to the inject position.
NOTE: This automatically triggers the system to begin the flow, turn on the plasma source, and acquire the dosimetry data. - Ramp the voltage up by navigating to the settings tab, and changing the flash voltage. Repeat steps 4.2 and 4.3 using 500 V, 750 V, 1000 V, and 1250 V. Perform the reading of adenine absorbance at each voltage in triplicates.
- Select calculations tab to determine the average absorbance of each sample. First, click Select, then manually select the beginning and end of the peak absorbance. In the available space, write in a description of the sample. Repeat for all acquired data.
NOTE: Bubbles may form causing a spike in dosimeter reading. When selecting data to determine average absorbance, omit areas with bubbles. - Copy and paste data in Excel to calculate the average change in adenine absorbance for each voltage, thus determining the effective ▪OH yield.
- Repeat steps 4.1-4.6 if multiple sample conditions (different buffer/additives) are being used to normalize the effective ▪OH yield for each condition.
5. Modification of protein to detect changes in higher order structure.
- Mix 4 mM adenine with 20 µM of protein in a one-to-one ratio to make a solution containing 2 mM adenine and 10 µM protein.
- Make the quench solution using 0.3 mg/mL catalase to break down excess hydrogen peroxide and 35 mM methionine amide to quench any remaining radicals. Aliquot 25 µL of quench solution into a 200 µL microtube so that an equal volume of quench and the labeled product is mixed.
- Dilute H2O2 to 200 mM and keep on ice.
- On the Control Software in the settings tab, start the flash voltage at 0 V to determine any background oxidation.
- In the manual control tab, select Start Data + AutoZero, followed by Process (Out), then Ready, and finally turn the injection valve down to the load position.
- Place a quench solution in position 1 on the product collect carousel. On the System Control Software change the product vial to 1.
- Immediately before injection, mix 12.5 µL of the adenine and protein mixture with 12.5 µL of H2O2 to make a final concentration of 1 mM Adenine, 5 µM protein, and 100 mM H2O2. Gently pipette up and down to mix, quickly spin down, and inject 25 µL using the injection port within 10 seconds of mixing.
- Switch the injection valve to the inject position and wait while sample is being processed.
- Repeat acquisition with 500 V, 750 V, 1000 V, and 1250 V. Perform each voltage measurement in triplicate.
- Calculate the average absorbance as described in step 4.5. Copy and paste all data to Excel.
6. Shut the system down
- After all samples have been collected, flush out the syringe port and sample loop by setting the injection valve down to the load position and inject 25 µL of HPLC water five times.
- Turn the injection valve up to the inject position to flush the rest of the system with HPLC water.
- Stop the flow, close the system control software, and turn off the modules starting with the product collector, dosimeter module, photolysis module, then finally the fluidics module.
7. Sample preparation and liquid chromatography-mass spectrometry
- Denature the protein by incubating samples at 80 °C for 20 min in the presence of 50 mM Tris and 1 mM CaCl2. Cool samples to room temperature and add 1:20 trypsin to protein. Digest the protein overnight at 37 °C with sample mixing. Next morning, terminate trypsin digestion by heating samples to 95 °C for 10 minutes.
- Detect peptides using a high-resolution LC-MS/MS system connected with a UPLC system.
- Load the sample first onto trap column (300 µm ID X 5 mm 100 Å pore size, 5 µm particle size) and wash at 5.0 µL/mL for 3 min with water containing 2% solvent B (acetonitrile and 0.1% formic acid).
- Separate the peptides on a C18 nanocolumn (0.75 mm x 150 mm, 2 µm particle size, 100 Å pore size) at a flow rate of 300 nL/min with a gradient between solvent A (water containing 0.1% formic acid) and solvent B. The gradient for peptide elution consists of a linear increase from 2 to 35% B over 22 min, ramped to 95% B over 5 min and held for 3 min to wash the column, and then returned to 2% B over 3 min and held for 9 min to re-equilibrate the column.
- Acquire the data in positive ion mode. Set the spray voltage to 2400 V, and the temperature of the ion transfer tube to 300 °C.
- Acquire the full MS scans from 250-2000 m/z followed by eight subsequent data-dependent MS/MS scans on the top eight most abundant peptide ions. Use collision-induced dissociation at 35% normalized energy to fragment the peptides.
- Identify all the unmodified peptides detected from MS/MS analysis using an available protein analysis software against the necessary FASTA file containing the protein sequence and the relevant proteolytic enzyme.
- Search and quantify modified peptides using a HRPF Data Processing Software. The extent of oxidation for each identified peptide is calculated by dividing the summed chromatographic peak area of a modified peptide by the total chromatographic peak area of that peptide modified and unmodified using equation 3.
P = [I(singly oxidized) X 1 + I (doubly oxidized) X 2 + I(triply oxidized) X 3 + …/[Iunoxidized + I(singly oxidized) + I(doubly oxidized) + I(triply oxidized) …]
where P denotes the average number of oxidation events per peptide molecule, and I represents the peak area of the unoxidized peptide (Iunoxidized) and the peptide with n oxidation events.
8. For a differential study, repeat steps 5-7 on the second condition.
NOTE: To confirm reproducibility, two biological replicates in addition to technical triplicates for each condition are recommended.
Representative Results
The high-pressure plasma source coupled with real-time dosimetry allows better control of ▪OH yield to observe changes in higher-order protein structure more accurately. The addition of adenine allows for an effective real-time radical dosimeter. Upon oxidation, adenine loses UV absorbance at 265 nm (Figure 2A). The change in adenine absorbance is directly related to the concentration of radicals available for HRPF thus providing a means to effectively monitor changes in radical concentration in the presence of radical scavengers like buffers, excipients, and ligands (Figure 2B).
Apomyoglobin (aMb) was modified in the presence of 100 mM H2O2 and 1 mM adenine at four increasing plasma voltages (Figure 3). Six peptides were detected with a linear increase in oxidation with the change of adenine UV 265 nm absorbance. Adenine UV absorbance is a surrogate for effective ▪OH concentration. The linear change in oxidation versus the change in radical concentration confirms the absence of artifactual change in the protein’s higher-order structure during labeling. Furthermore, the extent of oxidation detected with the high-pressure plasma source was much higher than the laser-based method. This increase arises from the high-pressure plasma source’s broad-spectrum UV emission spectrum (Figure 4). By producing a broad UV spectrum, the plasma source better overlaps the absorbance domain of H2O2 providing more efficient production of ▪OH through the photolysis of H2O2 (Figure 5). This is in direct comparison to the KrF Excimer laser and Nd:YAG laser, which are common sources that photolyze H2O2 in HRPF studies21,27,28. These lasers have a very narrow bandwidth at the lower end of H2O2’s photometric absorbance range. In comparison to the KrF excimer laser, the high pressure plasma source significantly increases the production of ▪OHs while concomitantly significantly decreasing the demand of required optical fluence (Figure 6).
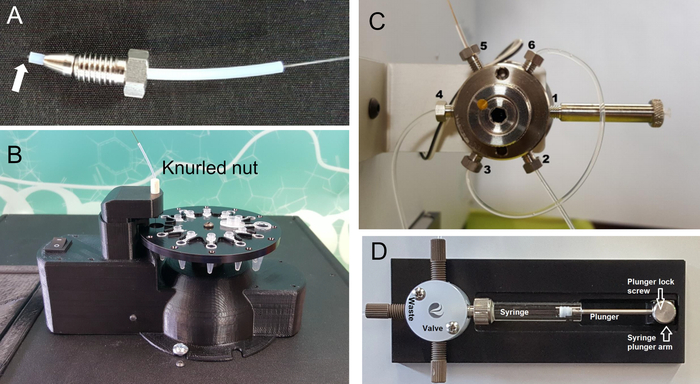
Figure 1: Components of the flash photolysis platform. (A) Assembled capillary tube, union tube, nut and ferrule. The tip of the “lower end” of the capillary extends barely beyond the union tube. (B) Knurled nut on the product collector to insert the end of the capillary. (C) Labeled six-port injection valve. (D) Syringe pump with the syringe. Please click here to view a larger version of this figure.
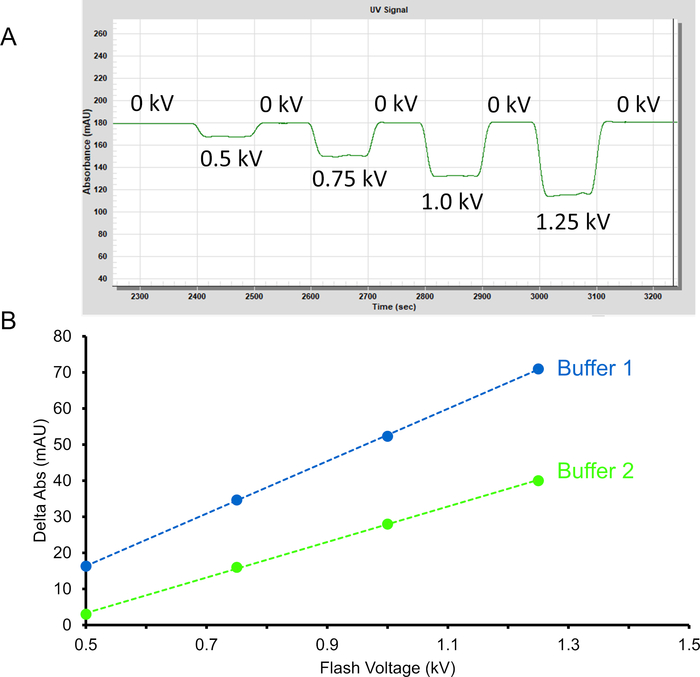
Figure 2: Change in adenine absorbance used for dosimetry. (A) Upon oxidation, adenine decreases in UV absorbance at 265 nm. (B) Change in adenine absorbance at 265 nm at different flash voltages for two buffers. Buffer 2 contains a radical scavenger which will decrease the change in adenine absorbance compared to buffer 1. To overcome the radical scavenging effects from buffer 2, increased flash voltage is required. Please click here to view a larger version of this figure.
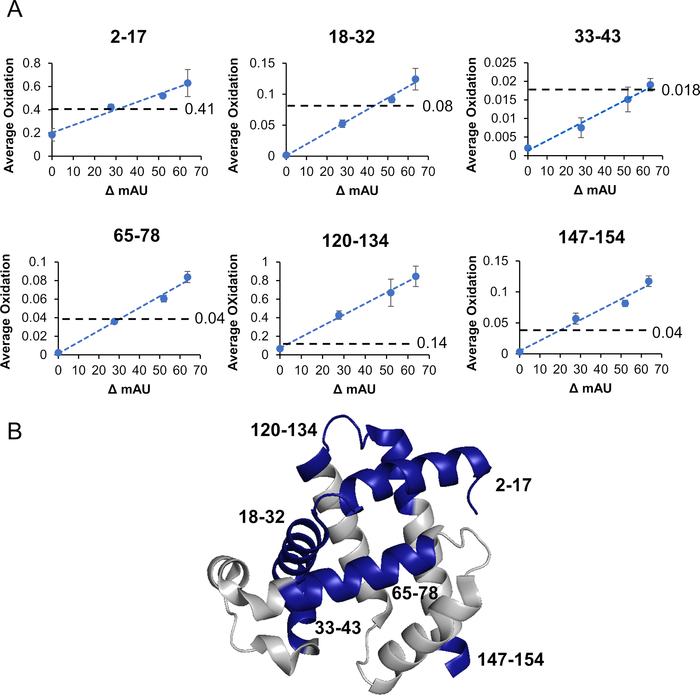
Figure 3: Apomyoglobin dose response curve. (A) Average oxidation of six peptides vs adenine change in absorbance is shown. FPOP oxidation levels are shown in the dashed lines. (B) The six oxidized peptides are labeled on a crystal structure of myoglobin (PDB: 3RGK). Please click here to view a larger version of this figure.
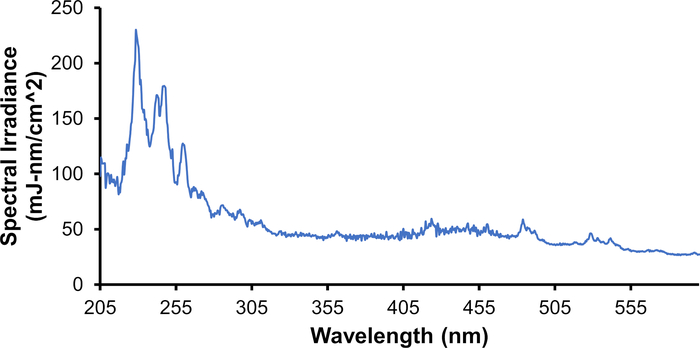
Figure 4: Spectral Irradiance of the plasma lamp. The high-pressure Xe gas lamp emits broad spectrum UV radiation from 200-300 nm along with visible light. Please click here to view a larger version of this figure.
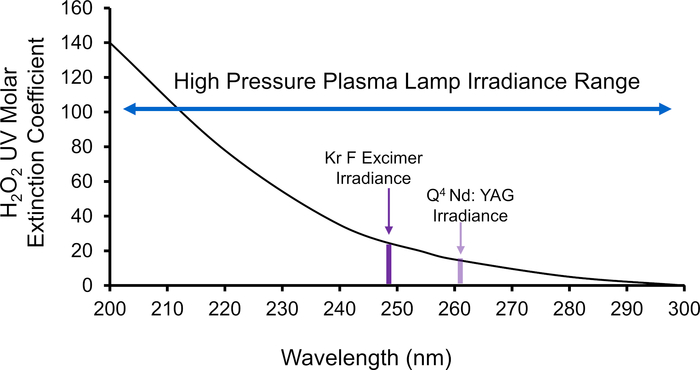
Figure 5: H2O2 photometric absorbance spectra. The black trace is the UV absorbance spectrum of H2O229. Highlighted in purple is the narrow wavelengths of the KrF excimer laser (248 nm) and Nd:YAG laser (265 nm) produces while the blue arrow represents the broad-spectrum plasma source which extends the range of useful H2O2 absorbance. Please click here to view a larger version of this figure.
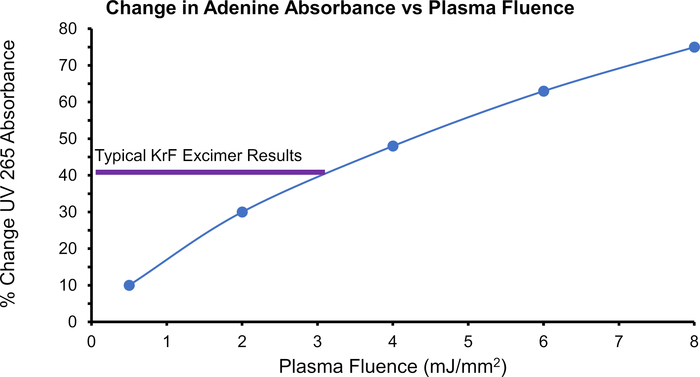
Figure 6: Efficacy of the plasma source to photolyze H2O2. The change in adenine absorbance over increased plasma fluence demonstrates the concentration of ▪OHs generated. Highlighted in purple is a typical max change in absorbance produced from a KrF excimer laser25. Please click here to view a larger version of this figure.
Discussion
There are several critical steps to ensure proper labeling of proteins during any HRPF experiment. First, an appropriate flow rate and source flash rate are selected to make certain each bolus of the sample is irradiated once. This ensures that the protein is exposed to a single bolus of newly formed ▪OH. Once a protein is oxidized, the higher order protein structure can be altered. To be confident the native protein structure is probed, each protein molecule must be modified in a single instant. Dosimetry can be used to test if the native protein structure is probed. As the concentration of hydroxyl radical increases the extent of oxidation linearly increases if the native protein structure is probed. However, if a large concentration of hydroxyl radicals causes the protein to partially unfold during labeling, then the extent of oxidation will significantly increase thus providing an effective means to ensure the native protein structure is probed.
It is also critical to ensure the reaction is quenched appropriately. This includes incorporating a proper radical scavenger mixed with the protein during the labeling process. For this method, adenine is not only used for dosimetry but also scavenges ▪OH limiting the lifetime of the radical as it labels the protein. By limiting the lifetime of the radical, confidence that the native protein structure is being modified is gained. Following the labeling process, it is important to collect the sample in a quench solution containing catalase and methionine amide which will break down excess H2O2 and scavenge ▪OH. Limiting the protein’s exposure to H2O2 ensures H2O2 induced oxidation is minimized.
It is also critical to appropriately select a protease for peptide digestion. Many enzymes cleave the protein at specific locations. For example, trypsin cleaves on the carboxyl side of arginine and lysine residues. This allows for simple data analysis, but if the protein of interest has a limited number of arginines and lysines, an alternative protease or a mixture of proteases might be required for acceptable peptide coverage during bottom-up proteomics. It is also advised to digest the modified protein before long term storage to avoid loss of oxidized protein due to aggregation and/or solubility issues.
Using HRPF to study changes in protein structure and protein interactions has been very successful11,30,31,32,33, but there are added complications while labeling heme-binding proteins or glycoproteins. While both heme binding34 and glycoproteins35 have been successfully probed using HRPF, thorough troubleshooting in the concentration of H2O2, radical scavenger, and the timeframe of H2O2 exposure must be balanced. Additionally, some buffers extensively quench ▪OHs. Therefore, it is vital to select an appropriate buffer and limit excipients like DTT and DMSO. Typical buffers used during an HRPF experiment include acetate, sodium phosphate, phosphate buffered saline, or low concentrations of Tris buffer. Interestingly, Tris scavenges some ▪OH, but following oxidation, it increases in photometric absorbance at 265 nm and can be used in place of adenine for dosimetry36. Although some buffers might decrease the effective concentration of ▪OH, by incorporating an in-line radical dosimeter these challenges can be surmounted.
HRPF utilizes non-specific irreversible labeling by ▪OH to probe solvent accessibility. The non-specific nature of the label provides increased overall coverage compared to other more targeted footprinting methods including N-ethylmaleimide37 and glycine ethyl ester38. Since the modification is irreversible, ample time is available during sample handling. This allows for complete protein digestion and a long LC gradient for improved MS/MS analysis. For HRPF there are several methods to generate ▪OH. A popular method utilizes a laser to photolyze H2O2 into ▪OH. Laser based platforms have been very successful in probing native protein structure, but they require extensive safety precautions, rigorous maintenance, and a substantial amount of space.
With the use of a high-pressure plasma source described here, there is no need for harmful gases and no fear of stray UV radiation. The lifetime of each high-pressure plasma source is enough to label between 180-200 samples, and besides monitoring the dosimeter response, no additional maintenance is required. The plasma source is under high-pressure, so while handling the plasma source, wear safety glasses. The plasma source also photolyzes H2O2 more efficiently, allowing for increased protein modification. If insufficient labeling is observed, H2O2 concentration can be increased along with increasing the lamp energy for addition radical production. Additionally, with the incorporation of real-time dosimetry, one can increase the reproducibility of experiments. Altogether, with the use of a plasma source coupled with real-time dosimetry and software for automated FPOP data calculation provides an advantageous package to perform HRPF experiments in a safe, easier to operate, and with improved reproducibility, as compared to standard laser-based methods.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was funded by the National Institute of General Medical Sciences (R43GM125420 and R44GM125420).
Materials
| 15 mL Conical Centrifuge Tubes | Fisher Scientific | 14-959-53A | any brand is sufficient |
| 50 µL SGE Gastight Syringes | Fisher Scientific | SG-00723 | |
| Acclaim PepMap 100 C18 nanocolumn (0.75 mm X 150 mm, 2 µm) | Thermo Scientific | ||
| Acetonitrile with 0.1% Formic Acid (v/v), LC/MS Grade | Fisher Scientific | LS120-500 | |
| Apomyoglobin | Sigma-Aldrich | ||
| Catalase | Sigma-Aldrich | C9322 | |
| Centrifuge | Eppendorf | 022625501 | |
| Delicate Task Wipers | Fisher Scientific | 06-666A | |
| Hydrogen Peroxide | Fisher Scientific | H325-100 | any 30% hydrogen peroxide is sufficient |
| Methionine amide | Chem-Impex | 03109 | |
| Microcentrifuge | Thermo Scientific | 75002436 | |
| Orbitrap Fusion Lumos Tribrid Mass Spectrometer | Thermo Scientific | Orbitrap Fusion Lumos Tribrid Mass Spectrometer | other high resolution instruments (e.g. Q exactive Orbitrap or Orbitrap Fusion) can be used |
| Pierce Trypsin Protease, MS Grade | Thermo Scientific | 90058 | |
| Polymicro Cleaving Stone, 1" x 1" x 1/32” | Molex | 1068680064 | any capillary tubing cutter is sufficient |
| UPLC | Thermo Scientific | ||
| Water with 0.1% Formic Acid (v/v), LC/MS Grade | Fisher Scientific | LS118-500 | |
| Water, LC/MS Grade | Fisher Scientific | W6-4 |
Riferimenti
- Nagarkar, R. P., Murphy, B. M., Yu, X., Manning, M. C., Al-Azzam, W. A. Characterization of protein higher order structure using vibrational circular dichroism spectroscopy. Current Pharmaceutical Biotechnology. 14 (2), 199-208 (2013).
- Giezen, T. J., Schneider, C. K. Safety assessment of biosimilars in Europe: a regulatory perspective. Generics and Biosimilars Initiative Journal. , 1-8 (2014).
- Giezen, T. J., Mantel-Teeuwisse, A. K., Strauss, S. Safety-related regulatory actions for biologicals approved in the United States and the Europena Union. Journal of the American Medical Society. 300 (16), 1887-1896 (2008).
- Gabrielson, J. P., Weiss, W. F. Technical decision-making with higher order structure data: starting a new dialogue. Journal of Pharmaceutical Sciences. 104 (1), 1240-1245 (2015).
- Brenowitz, M., Erie, D. A., Chance, M. R. Catching RNA polymerase in the act of binding: intermediates in transcription illuminated by synchrotron footprinting. Proceedings of the National Academy of Sciences U S A. 102 (13), 4659-4660 (2005).
- Guan, J. Q., Takamoto, K., Almo, S. C., Reisler, E., Chance, M. R. Structure and dynamics of the actin filament. Biochimica. 44 (9), 3166-3175 (2005).
- Hambly, D. M., Gross, M. L. Laser flash photochemical oxidataion to locate heme binding and conformataional changes in myoglobin. International Journal of Mass Spectrometry. 259 (2007), 124-129 (2007).
- Li, K. S., Shi, L., Gross, M. L. Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Accounts of Chemical Research. 51 (3), 736-744 (2018).
- Watson, C., Sharp, J. S. Conformational Analysis of Therapeutic Proteins by Hydroxyl Radical Protein Footprinting. American Association of Pharmaceutical Scientists Journal. 14 (2), 206-217 (2012).
- Deperalta, G., et al. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. mAbs. 5 (1), 86-101 (2013).
- Jones, L. M., et al. Complementary MS methods assist conformational characterization of antibodies with altered S-S bonding networks. Journal of American Society of Mass Spectrometry. 24 (6), 835-845 (2013).
- Storek, K. M., et al. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proceedings of the National Academy of Sciences. , (2018).
- Vij, R., et al. A targeted boost-and-sort immunization strategy using Escherichia coli BamA identifies rare growth inhibitory antibodies. Scientific Reports. 8 (1), 7136 (2018).
- Liu, X. R., Zhang, M. M., Rempel, D. L., Gross, M. L. A Single Approach Reveals the Composite Conformational Changes, Order of Binding, and Affinities for Calcium Binding to Calmodulin. Analytical Chemistry. 91 (9), 5508-5512 (2019).
- Lu, Y., et al. Fast Photochemical Oxidation of Proteins Maps the Topology of Intrinsic Membrane Proteins: Light-Harvesting Complex 2 in a Nanodisc. Analytical Chemistry. 88 (17), 8827-8834 (2016).
- Marty, M., Zhang, H., Cui, W., Gross, M., Sligar, S. Interpretation and Deconvolution of Nanodisc Native Mass Spectra. Journal of American Society of Mass Spectrometry. 25, (2013).
- Johnson, D. T., Di Stefano, L. H., Jones, L. M. Fast photochemical oxidation of proteins(FPOP): A powerful mass spectrometry based structural proteomics tool. Journal of Biological Chemistry. , (2019).
- Chea, E. E., Jones, L. M. Analyzing the structure of macromolecules in their native cellular environment using hydroxyl radical footprinting. Analyst. 143 (4), 798-807 (2018).
- Aprahamian, M. L., Chea, E. E., Jones, L. M., Lindert, S. Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data. Analytical chemistry. 90 (12), 7721-7729 (2018).
- Linde. . Linde Specialty Gases of North America. , (2009).
- Aye, T. T., Low, T. Y., Sze, S. K. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Analytical Chemistry. 77 (18), 5814-5822 (2005).
- Niu, B., Zhang, H., Giblin, D., Rempel, D. L., Gross, M. L. Dosimetry determines the initial OH radical concentration in fast photochemical oxidation of proteins (FPOP). Journal of American Society of Mass Spectrometry. 26 (5), 843-846 (2015).
- Misra, S. K., Orlando, R., Weinberger, S. R., Sharp, J. S. Compensated Hydroxyl Radical Protein Footprinting Measures Buffer and Excipient Effects on Conformation and Aggregation in an Adalimumab Biosimilar. American Association of Pharmaceutical Scientists Journal. 21 (5), 87 (2019).
- Olson, L. J., Misra, S. K., Ishihara, M., Battaile, K. P., Grant, O. C., Sood, A., Woods, R. J., Kim, J. P., Tiemeyer, M., Ren, G., Sharp, J. S., Dahms, N. M. Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor. Communications Biology. 3 (1), 498 (2020).
- Sharp, J. S., Misra, S. K., Persoff, J. J., Egan, R. W., Weinberger, S. R. Real Time Normalization of Fast Photochemical Oxidation of Proteins Experiments by Inline Adenine Radical Dosimetry. Analytical Chemistry. 90 (21), 12625-12630 (2018).
- Misra, S. K., Sharp, J. S. Enabling Real-Time Compensation in Fast Photochemical Oxidations of Proteins for the Determination of Protein Topography Changes. Journal of Visualized Experiments. 163, (2020).
- Hambly, D. M., Gross, M. L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. Journal of American Society of Mass Spectrometry. 16 (12), 2057-2063 (2005).
- Liu, X. R., Zhang, M. M., Gross, M. L. Mass Spectrometry-Based Protein Footprinting for Higher-Order Structure Analysis: Fundamentals and Applications. Chemical Reviews. 120 (10), 4355 (2020).
- Jones, L. M., Sperry, B. J., Carroll, A. J., Gross, M. L. Fast photochemical oxidation of proteins for epitope mapping. Analytical chemistry. 83 (20), 7657-7661 (2011).
- Li, J., et al. Mapping the Energetic Epitope of an Antibody/Interleukin-23 Interaction with Hydrogen/Deuterium Exchange, Fast Photochemical Oxidation of Proteins Mass Spectrometry, and Alanine Shave Mutagenesis. Analytical chemistry. 89 (4), 2250-2258 (2017).
- Liu, X. R., Zhang, M. M., Rempel, D. L., Gross, M. L. A Single Approach Reveals the Composite Conformational Changes, Order of Binding, and Affinities for Calcium Binding to Calmodulin. Analytical Chemistry. 91 (9), 5508-5512 (2019).
- Kiselar, J. G., Janmey, P. A., Almo, S. C., Chance, M. R. Structural analysis of gelsolin using synchrotron protein footprinting. Molecular and Cellular Proteomics. 2 (10), 1120-1132 (2003).
- Chea, E. E., Deredge, D. J., Jones, L. M. Insights on the Conformational Ensemble of Cyt C Reveal a Compact State during Peroxidase Activity. Biophysical Journal. 118 (1), 128-137 (2020).
- Poor, T. A., et al. Probing the paramyxovirus fusion (F) protein-refolding event from pre- to postfusion by oxidative footprinting. Proceedings of the National Academy of Sciences U S A. 111 (25), 2596-2605 (2014).
- Roush, A. E., Riaz, M., Misra, S. K., Weinberger, S. R., Sharp, J. S. Intrinsic Buffer Hydroxyl Radical Dosimetry Using Tris(hydroxymethyl)aminomethane. Journal of American Society of Mass Spectrometry. 31 (2), 169-172 (2020).
- Everett, E. A., Falick, A. M., Reich, N. O. Identification of a critical cysteine in EcoRI DNA methyltransferase by mass spectrometry. Journal of Biological Chemistry. 265 (29), 17713-17719 (1990).
- Sanderson, R. J., Mosbaugh, D. W. Identification of specific carboxyl groups on uracil-DNA glycosylase inhibitor protein that are required for activity. Journal of Biological Chemistry. 271 (46), 29170-29181 (1996).

