Cone-Enriched Cultures from the Retina of Chicken Embryos to Study Rod to Cone Cellular Interactions
Summary
We describe a method to obtain primary cultures of cone photoreceptors from the retina of chicken embryos and its use for high content screening.
Abstract
Human daytime vision relies on the function of cone photoreceptors at the center of the retina, the fovea. Patients suffering from the most prevalent form of inherited retinal degeneration, retinitis pigmentosa, lose night vision because of mutation driven loss of rod photoreceptors, a phenomenon followed by a progressive loss of function and death of cones leading to blindness. Geneticists have identified many genes with mutations causing this disease, but the first mutations identified questioned the mechanisms of secondary cone degeneration and how a dominant mutation in the rhodopsin gene encoding for the visual pigment expressed exclusively in rods can trigger cone degeneration.
This result of transplantations in a genetic model of the disease led to the concept of cell interactions between rods and cones and of non-cell autonomous degeneration of cones in all genetic forms of retinitis pigmentosa.
Cones comprise 5% of all photoreceptors in humans and only 3% in the mouse, so their study is difficult in these species, but cones outnumber rods in bird species. We have adapted 96-well plates to culture retinal precursors from the retina of chicken embryos at stage 29 of their development. In these primary cultures, cones represent 80% of the cells after in vitro differentiation. The cells degenerate over a period of one week in the absence of serum. Here, we describe the methods and its standardization.
This cone-enriched culture system was used to identify the epithelium-derived cone viability factor (EdCVF) by high content screening of a rat retinal pigmented epithelium normalized cDNA library. Recombinant EdCVF prevents the degeneration of the cones.
Introduction
The retina of vertebrate species is dual, with rod photoreceptors for dim light vision and cone photoreceptors for daylight, color and acuity vision. Primate visual acuity relies on a region at the center of the retina, called the fovea, that is enriched in cones, but overall, cones represent only 5% of all photoreceptors. Consequently, the analysis of the cones in primate retina and especially the culture of cones are technically difficult. All other mammalian species have no fovea and the percentage of cones is low for rodents that are most commonly used in retinal research. This is not the case for avian species, for which cones dominate the retina of these well seeing bird species. Dinosaurs, which have dominated the ecosystem when the mammals first appeared during evolution, are at the phylogenetic origin of birds1. As a consequence of such competition between dinosaurs and early mammals, the mammals are mostly nocturnal with retinas dominated by rods. Only later during evolution did the diurnal vision of some mammalian species, among which primates belong, became an evolutionary advantage. Nevertheless, the ancestral period remains as an atavism of the nocturnal bottleneck in the evolution of mammalian vision2,3.
While studying retinal cell differentiation, Adler and Hatlee showed that photoreceptors represent approximately 70% of the retinal differentiated cells in cultures derived from chicken at the embryonic day (ED) 6 or stage 294. Because of the prevalence of cones in the chicken retina, cultures of retinal cells from ED6 chicken embryos have been developed as cone-enriched cultures5.
The importance of cone-mediated visual acuity for human is a truism. People affected by genetic or aging diseases that alter cone function are greatly handicapped. This has promoted a very large body of studies on inherited retinal degenerations (IRD) with the objective of finding treatments for these blinding diseases6,7. The first success, obtained using a recombinant adeno-associated vector (AAV) for the therapy of a severe form of IRD Leber congenital amaurosis (LCA), is a proof of concept for gene therapy8. The identification of the genes whose mutations trigger IRD opens the possibility of curing these diseases using gene therapy. Nevertheless, these diseases are resulting from mutations in more than 200 distinct genes9. Even in the case of autosomal recessive forms of IRD, when the reintroduction of the normal copy of the morbid gene could restore visual function, the economic cost of each individual development favors the most prevalent ones in detriment to the less common ones and to those for which the genetic origin remains unknown. This fact led researchers to think about more general therapies. Apoptotic cell death appeared as a common pathway, and a therapeutic target of these diseases that progress by the degeneration of photoreceptors, including for autosomal dominant forms10,11. However, the successes of such an approach are missing. For the most common form of IRD, retinitis pigmentosa (RP), the common pathway is the secondary loss of function ultimately followed by the degeneration of cones12,13. Preventing the loss of cone function will preserve central vision of the fovea independently of the causative mutations14.
In the early stage of RP, the loss of rods triggers a reduction in the expression of rod-derived cone viability factor (RdCVF), encoded by the nucleoredoxin-like 1 (NXNL1) gene, which interrupts the metabolic and redox signaling between rods and cones15. The administration of a recombinant AAV encoding the two products of the NXNL1 gene, the trophic factor RdCVF and the thioredoxin enzyme RdCVFL, could theoretically prevent cone vision loss in all genetic forms of RP16. We have shown that the NXNL1 gene product, RdCVFL, is expressed in chicken cone-enriched cultures17 and where it plays a protective role18. RdCVF and the NXNL1 gene were identified by high content screening of a retinal cDNA library using the survival of cells from a cone-enriched culture as readout19. We screened the equivalent of 210,000 individual clones of the library using 8 parallel tests for each clone. This represents a very large number of tests requiring easy access to the biological material, the retinas of chicken embryos. We found that it was relatively easy to obtain embryonated chicken eggs on a weekly basis because they are widely produced for the agro-industry for egg-laying hens and meat-producing chickens. After careful standardization of the cone-enriched cultures, the system provides an easy, robust and reproducible way of testing thousands of molecules for their ability to preserve cone viability. These cells are also amenable to genetic manipulations20 that benefit to the study of signal transduction and to biochemical analyses21,22,23.
Retinal researchers have developed alternative methods as the use of the cone cell line 661W24,25,26. Nevertheless, the identity of this cell line remains controversial27,28. The 661W cells were cloned from retinal tumors of a transgenic mouse line that expresses the SV40 large T antigen under control of the human interphotoreceptor retinol-binding protein promoter. SV40 large T antigen mediates cellular transformation and immortalization. As the consequence, signaling pathway identified using 661W cells must be reported into the context of a transformed and immortalized cell line which is distinct in many ways from cones in situ. In that respect, the cone-enriched culture system is composed of primary neurons, the cones that are more physiologically relevant.
While it is possible to obtain a pure culture of photoreceptors using vibratome sectioning of the mouse retina, the very low percentage of cones in the outer retina of rodents makes this approach unsuited for producing cone-enriched cultures29. The pig retina contains no fovea but has a region called area centralis that is very enriched in cones30. The high proportion of cones in the retina diurnal rodents, as Arvicanthis ansorgei and Psammomys obsesus31,32, offers a possible solution but requires the breeding of such exotic species. Adult pig eyes, collected from local slaughterhouses, can be used to produce a mixed culture of rods and cones that have been used to study photoreceptor survival33. An elegant solution is to pre-purify cones from the pig retina using panning with peanut agglutinin (PNA) lectin, which selectively binds to cones34. Nevertheless, this method is difficult to implement on a large scale because of its complexity.
Human induced pluripotent stem cells (iPS) offers the most promising approach to obtain a cone photoreceptor cell population that can be used for retinal transplantation but that can also be adapted to cone-enriched culture35,36. Since the transcription factor NRL is required for rod-photoreceptors37, the Nrl-/- mouse has a retina dominated by short wave cones (S-cones). The inactivation could be used to produce S-cone-enriched preparation by differentiation of human from iPS38,39. Another possible approach is to promote cone differentiation using thyroid hormone signaling40. While novel methods for producing cone-enriched cultures from human iPS are emerging, chicken embryos provide a current proven method19.
The cone-enriched culture was instrumental in the identification of RdCVF by expression cloning19. This system was also used successfully to demonstrate that RdCVF stimulates glucose uptake and its metabolism by aerobic glycolysis22. Furthermore cone-enriched culture was used to validate the protective role of RdCVFL, the second product of the NXNL1 gene23. More recently, this system was used to demonstrate the existence of protecting molecules secreted by retinal pigmented epithelial cells transduced with OTX241.
Protocol
The protocol was approved by the Committee on the Ethics of Animal Experiments of the University Pierre and Marie Curie and the French ministry of research (Permit Number: APAFIS#1028 2015070211275177). The animal experiments were performed under the following authorization: "Certificat d'autorisation d'expérimenter sur les animaux vertébrés A-75-1863. Préfecture de Police de Paris (November 9th 2011-November 8th 2016)".
1. Incubation of fertilized eggs
- Collect weekly fertilized eggs (strain I 657, red label), obtained naturally, at an industrial hatchery.
- Maintain the fertilized eggs at 17°C (their biological zero) in the laboratory after they are "laid" by the hen.
- For each culture, incubate seven fertilized eggs for 24 hours at 20 °C and then 136 hours at 37 °C with intermittent reversion of the inclination (a progressive movement for 2 hours from one side to its opposite, a 4-hours cycle) of the eggs in a humidified chamber.
2. Recovery of the chicken embryos
- Wash the surface of the seven eggs with disinfectant (e.g., Pursept A express).
- To break the eggshell, make a hole in the top of the shell with large straight pliers. Then cut the shell to remove the hat from the egg as a soft-boiled egg.
- Gently extract each embryo from the eggshell with curved forceps, and then transfer it to a Petri dish containing sterile phosphate buffer saline (PBS) previously heated to 37 °C. Gently, remove the envelope that surrounds the embryos (the chorion or chorioallantoic membrane).
- Verify the stage of development of each embryo by visual comparison to Hamburger and Hamilton42.
- Select two embryos at the 29th stage of development (Figure 1). The wings bend at the elbows. The collar stands out visibly. The bill is more prominent than at the 28th stage.
- Enucleate the eyes of these selected embryos and transfer them in CO2-independent medium (Life technologies).
CAUTION: It is very important that the embryo is at stage 29, and not at stage 28 or 30 (Figure 1); that is why it is necessary to incubate at least 7 eggs, even if only two will be finally used. The chorion is very thin, so hard to distinguish, but it is very close from the embryo, so it has to be removed without touching the embryo.
3. Dissection of the retinas
- Decapitate and enucleate the selected embryos with curved forceps.
- Transfer the four eyes into CO2-independent medium. This medium contains 0.9 mM CaCl2 and 0.65 mM MgCl2.
- Position the eye with the cornea face down, the optic nerve facing the experimenter. Drill a hole in the optic nerve using two straight forceps.
- Insert a branch of each forceps between the retina and the pigment epithelium (Figure 2). Pull on each forceps and rotate the eye to detach the epithelium from the retina. Remove the cornea followed by the lens and the vitreous.
- Transfer the four retinas in a Petri dish containing Ringer's medium at pH 7.2.
CAUTION: Make sure that only the retina remains and remove any traces of vitreous and remaining retinal pigmented epithelium.
4. Preparing the retinal cell suspension
- Cut the four retinas in very small pieces using two straight pliers.
- Wash the retinal pieces twice with Ringer's medium.
- After the second wash with Ringer's medium, let the pieces of retina fall on the bottom of the tube and remove the Ringer's medium. Treat the retinal pieces for 20 minutes at 37 °C with a solution of trypsin (0.25% w/v).
- Disperse the solution after 10 minutes by successive suction. Discharge using a Pasteur pipette and check for dissociation of the retinal pieces. Stop the reaction by adding culture media supplemented with 10% inactivated foetal calf serum.
- Incubate the cell suspension with 0.05 mg of DNase I. Dissociate the cell clusters and the DNA by successive suction and discharge using a Pasteur pipette immediately after adding the DNase.
- Wash the retinal cell suspension twice with chemical defined culture medium (CDCM): an equal volume of Dulbecco's Modified Eagle Medium and M199 media supplemented with 100 µg/mL linoleic acid/BSA, 0.86 µM insulin, 0.07 µM transferrin, 2.0 µM progesterone, 0.28 µM prostaglandin, 0.29 µM Na2SeO3, 182 µM putrescine, 3 mM taurine, 4.7 µM cytidine 5′-diphosphocholin, 2.7 µM cytidine 5′-diphosphoethanolamine, 0.55 µM hydrocortisone, 0.03 µM triiodothyronine, 1 mM sodium pyruvate and 20 µM gentamycin.
5. Retinal cell seeding
- Treat two black 96-well culture plates with transparent bottom for 2 hours at 37 °C with poly-L-lysine at 32.25 µg/cm2.
- Rinse these plates twice with M199 culture medium. Resuspend the cell pellet in 1 mL of CDCM.
- Add to an aliquot of 10 µL of the cell suspension trypan blue to stain the living cells. Add the cells suspension specimen to a haemocytometer (cell counting-chamber of Malassez).
- Under a microscope, count the stained cells for four rows of the haemocytometer (i.e., 40 squares), and then calculate the average cell number for one row. Calculate the concentration of cells of the suspension by applying the following method: Number of cells/mL of suspension = average number of cells in a row (10 squares) x 10 the total number of squares of the haemocytometer x dilution with trypan blue x 1,000 (to express the result as cells/mL).
- Bring the cell suspension to two concentrations (5.6 x 104 cells/mL and 1.12 x 105 cells/mL) corresponding to the two plating densities (1 x 105 cells/cm2 and 2 x 105 cells/cm2) using CDCM.
- Seed 50 µL of the two cell suspensions into the two pretreated black 96-well culture plates. Distribute the cells in the plates with a multichannel pipette from the right of the plate to the left, homogenizing between each column, so that the distribution of the cells is homogeneous.
- Add 50 µL of the library of molecules (e.g., the conditioned media from a cDNA library, see below) to be screened using a predefined pattern (Table 1).
- Incubate the plates for seven days at 37°C under 5% CO2 with no change of media.
6. Counting viable cells
- To each well of the plate, add 2.7 µM of calcein AM and 0.3 mM of ethidium homodimer.
NOTE: Calcein penetrates the cells that are impermeable to large molecules as ethidium homodimer. In the cytoplasm of living cells, calcein is hydrolyzed by endogenous esterases, becomes fluorescent by emitting at 520 nm when excited at 485 nm. Ethidium homodimer bind to DNA on dead cells. Ethidium DNA-homodimer binding causes red fluorescence to be emitted at 635 nm after excitation at 520 nm. - Incubate the plates for 1 hour at room temperature in the absence of light.
- Read the fluorescence on an automated plate reader composed of an inverted microscope equipped with a mercury lamp with two excitation filters at 485 and 520 nm, two filters of emission at 520 and 635 nm, an objective (x10), a motorized stage controlled by a processor and a charge-coupled device camera (CCD). This counting platform is controlled by the Metamorph software19. It makes it possible to acquire fluorescence images of living cells and dead cells simultaneously in each well of the 96-well plate, starting from wells A1 towards well A12, then B12 towards B1, C1 towards C12 and so on (Table I).
- Calculate the average area A of a single cell using the 18 wells of the negative controls (Table I).
- Count the cells in each well of the plate and apply the following empirical formula A x 29 / 20.7 to prevent that cell doublets (grouping of two cells) are counted. Score the protective effect of the cones by molecules as the ratio between the average cell number in the 4 wells where we tested the molecule versus the average cell number in the 18 wells of the negative control (see Table 1 and Supplementary Figure 1).
- Combine the results of the plate seeded at 1 x 105 cells/cm2 with the one seeded at 2 x 105 cells/cm2 to evaluate the potential protection (viability ratio) by each molecule screened.
Representative Results
We describe here how the cone-enriched culture system can be used to identify novel cone protecting proteins. We used this protocol to screen a normalized cDNA library made of choroid and retinal pigmented epithelium from 400 eyes of 8 weeks old Long-Evans rats43.
This library contains 6.0 x 106 independent colonies forming units (CFUs) and has an average cloned insert size of 2.1 kilobase (kb), with greater than 99% of recombinant clones. Pools of 100 clones from that library were transiently transfected (0.1 µg of plasmid DNA) into COS-1 cells and the conditioned medium (CM) of COS-1 were harvested after incubation for 48 hours in DMEM without serum. Membranes or exosomes were not removed by ultracentrifugation. Fifty microliters of each CM were added to 4 wells of two 96-well plates: one seeded at 2 x 105 cells/cm2, the other one at 4 x 105 cells/cm2. CM from COS-1 cells transfected with the empty vector pcDNA3.1 used to construct the library was used as negative control (Table 1). A total of 2,112 sets of 100 clones corresponding to 211,200 individual clones were evaluated in four culture wells and for two seeding conditions of cone-enriched cultures. The two conditions correspond to two slightly different inoculation densities which makes it possible to assess the protective activity more precisely, for a total of 1,689,600 culture wells.
Among the 42 pools of clones with a ratio greater than 2, pools 0080 and 0073 have viability ratio 16 and 14 times higher after 7 days of culture than the negative control, pcDNA3.1 (Supplementary Figure 1). This analysis is essential to identify the pools of interest. Each selected pool of 100 clones was subdivided into 16 sets of 10 clones from their glycerol stock. These sub-pools were prepared and tested according to the same method in a second round of screening (i.e., a total of 3,200 culture wells). The sub-pool 0073-09 gave the strongest viability ratio (Supplementary Figure 2A) and was subdivided to produce 16 individual clones that were tested in a third round of screening on cone-enriched cultures. The clone 0073-09-37 clone clearly stands out from the others with a viability ratio equal to 2.5 (Supplementary Figure 2B). The y axis has a different scale even if the seeding density was the same than in Supplementary figure 2A. We have seen this commonly when the assays are repeated weekly for months. After analysis, these results confirm that clone 0073-09-37 has a robust and reproducible effect on cone survival. The test was repeated independently (Figure 3A), and the insert of 1.8 kb was sequenced (Figure 3B).
A bioinformatic analysis revealed that the clone 0073-09-37, that we named epithelium-derived cone viability factor (EdCVF), contains three open reading frames (ORFs), the one the most upstream (ORF1) encodes for 84 residues of the C-terminal part of the rat protein zinc finger protein-180 (ZFP180, NP_653358) of 727 amino acids44. The other two ORFs (ORF2 and ORF3) are much less well conserved in mice and absent in other mammals. When tested independently, only ORF1 exerts a protective effect on the cones (Supplementary Figure 3). ORF1 was produced as a glutathione S-transferase (GST) fusion protein (Figure 4A). The EdCVF protein was purified and the GST tag removed (Figure 4B). EdCVF is able to prevent cone degeneration in the cone-enriched culture system (Figure 4C).
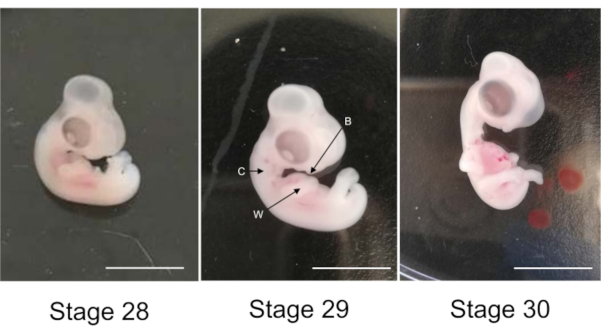
Figure 1: Chicken embryos at stages 28th, 29th and 30th of development. Arrows W: wing, C: collar and B: bill. Scale bar 1 cm. Please click here to view a larger version of this figure.
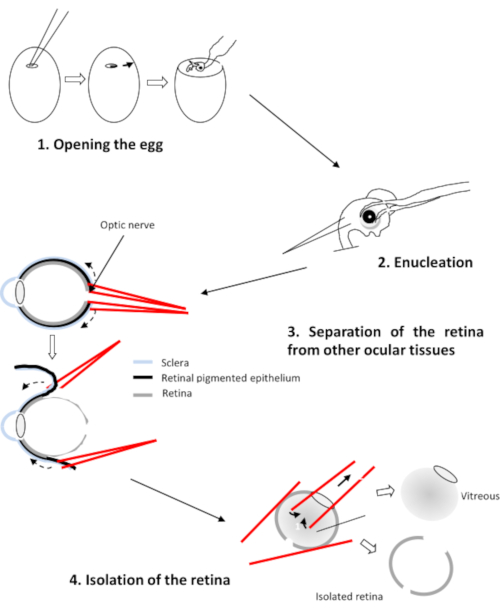
Figure 2: Dissection of the retina of the chicken embryo Please click here to view a larger version of this figure.
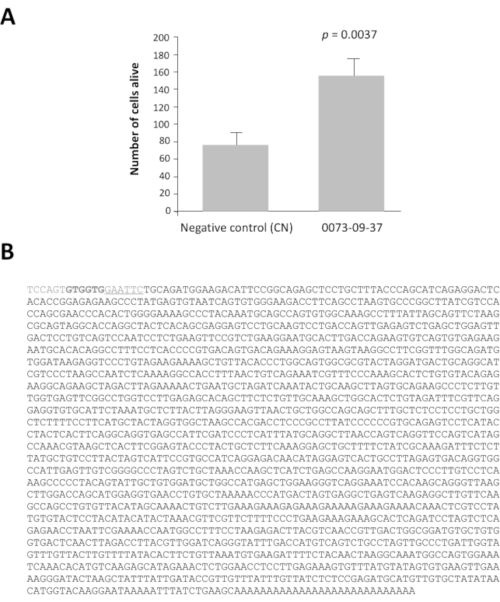
Figure 3: Epithelium-derived cone viability factor (EdCVF), clone 0073-09-37. A. Increased viability on cone-enriched culture. B. The sequence of the cDNA clone 0073-09-37. The underlined sequence GAATTC is the EcoRI restriction site used to construct the library. The two codons GTG in bold are uncommon translation initiation sites for EdCVF originating from the vector. Statistical analysis by Student's test. Please click here to view a larger version of this figure.
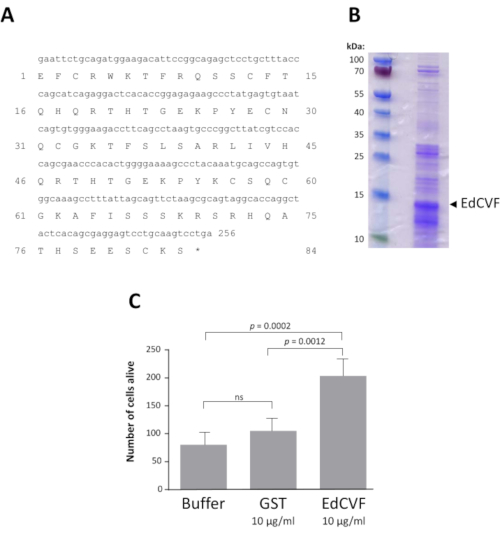
Figure 4: Recombinant EdCVF activity. A. Sequence of EdCVF in the fusion with glutathione S-transferase (GST). B. The purified recombinant EdCVF protein. C. Trophic activity of GST-EdCVF on cone in culture. Statistical analysis using Tukey's test. Please click here to view a larger version of this figure.
Supplementary Figure 1: Ratio of the average cell number for a cDNA pool and the negative control, the conditioned medium of COS-1 cells transfected with the empty vector pcDNA3.1 during the first round of screening. Please click here to download this file.
Supplementary Figure 2: Number of cells alive. A. The second round of screening with sub-pools of 0073 pool. B. The third round of screening with isolated clones. CN: the conditioned medium of COS-1 cells transfected with the empty vector, pcDNA3.1. Please click here to download this file.
Supplementary Figure 3: Trophic activity of the three open reading frames of the isolated clone 0073-09-37. Statistical analysis using Dunnett's test. Please click here to download this file.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 1 | 1 | 1 | 1 | 2 | NC | 2 | 2 | 2 | 3 | 3 | NC |
| B | NC | 3 | 3 | 4 | 4 | 4 | NC | 4 | 5 | 5 | 5 | 5 |
| C | 6 | NC | 6 | 6 | 6 | 7 | 7 | NC | 7 | 7 | 8 | 8 |
| D | 8 | 8 | NC | 9 | 9 | 9 | 9 | 10 | NC | 10 | 10 | 10 |
| E | 11 | 11 | 11 | NC | 11 | 12 | 12 | 12 | 12 | NC | 13 | 13 |
| F | 13 | 13 | 14 | 14 | NC | 14 | 14 | 15 | 15 | 15 | NC | 15 |
| G | 16 | 16 | 16 | 16 | 17 | NC | 17 | 17 | 17 | 18 | 18 | NC |
| H | 18 | 18 | 19 | 19 | 19 | 19 | NC | 20 | 20 | 20 | NC | 20 |
| NC negative controls | ||||||||||||
| 1-20 positions of the pools tested in quadruplicate | ||||||||||||
Table 1: Plan of the 96-well plate for high content screening
Discussion
Among the many parameters that might limit the production of a cone-enriched culture from chicken embryos, the first critical step is to accurately identify the stage of development of the embryos in the hatched eggs. It has been observed that culture of cells from the retinas of embryos at ED8 (34th stage) produces only 35% photoreceptors, with the remaining 65% are made of other neurons4. Whatever the logistic applied to get the hatched eggs, it is necessary to fine-tune the temperature and the incubation time, and to examine carefully the embryos as compared to the reference pictures of all stages of development42,45.
Originally, the cone-enriched culture system was developed using the White Leghorn strain4. The white color of the eggs of that strain is not particularly appreciated in France, so we used a strain of chicken that produce brown eggs. We utilized the I 657 strain, which is made by crossing I 66 roosters with JA57 hens5. We were able to reproduce the characteristics of the original cultures. This shows that the genetic background of the chicken is not critical to obtain cone-enriched cultures.
We have not tested the effect of individual removal of the supplements in the culture medium, but have observed that insulin plays a critical role in accordance to the effect of insulin on the survival of cones in the rd1 mouse, a model of autosomal recessive RP46. Triiodothyronine (T3) may also participate in the differentiation of retinal precursor cells of the chicken embryo into cones according to the role of thyroid hormone receptor in retinal cell fate during development40. Consequently, the cone-enriched culture system cannot be used to identify insulin by expression cloning46.
The cone-enriched culture system relies on the culture of primary neurons and is much more appropriate tha methods relying on the use of immortalized cells as the cell line 661W24,25,26.
The method described here can be modified by performing a prior electroporation with plasmid DNA20. Before preparing the retinal cell suspension, the entire retina is placed in the chamber of a custom-made electroporator with 120 µL of 0.5 µg/µL of plasmid DNA in 10 mM Tris-HCl pH 8.0, 1 mM EDTA. Five pulses of 15 V for 50 ms each are applied separated by 950 ms interval22. Attempts to deliver interfering RNA (RNAi) using the replication competent avian splice (RCAS) retroviruses into cone-enriched cultures were unsuccessful47. This is certainly due to the fact that in the absence of serum and in cultures of low density, retinal precursor cells are not replicative, a requisite for the propagation of retroviruses.
We developed the cone-enriched culture system to identify trophic factors that promote cone survival using expression cloning19. In order to make it feasible, we performed a first step of high content screening using conditioned medium from pools of 100 clones. Even if the cDNAs from the library are expressed under the control of a strong CMV promoter after transfection of COS-1 cells, it does not offer a guaranty that all the proteins encoded by individual cDNAs reach a concentration sufficient to be scored positive by the viability assay. This is a major limitation. In that sense, any screening is not really exhaustive. In addition, even if membranous proteins were not removed from the conditioned medium, the configuration of the assay is unfavorable to the identification of non-diffusive factors. An alternative would be to screen individual clones after having obtained the sequence of the cDNAs in order to avoid duplications in assaying many times the same candidate protein. This was initiated by sequencing the retinal cDNA libraries we used43. While rational, this approach has also its limitations. The bioinformatic analysis of the cDNA sequences will irresistibly impose, beside the reduction of the redundancy, the prioritization of screening certain clones based on knowledge. This will not be detrimental if finally, the whole library would be screened even if the time required to do it will be significantly lengthened. But invariably, the identity of the sequence will influence our way of looking at the results. This will not be neutral since the interpretation of the sequence will naturally be in competition with the experimental data.
The identification of EdCVF also show that high content screening entails technical limitations. From the first round of screening, we identified two pools with high activity (Supplementary Figure 1). The pool 0073 led to the successful identification of EdCVF, while pool 0080 did not conduct to such finding. We have not solved the problem that could result from the loss of the active clone during the preparation of sub-pools. Alternatively, it is not excluded, even if not statistically favorable, that among the cDNAs of pool 0080, two proteins were acting synergistically and their activity could not be observed as individual clones.
The identification of molecules protecting cones by screening small molecules is a future application of the cone-enriched culture system. Such molecules will be invaluable for the treatment of retinal pathologies for which gene therapy is not the most appropriate approach as age-related macular degeneration.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors thank Jacques Bellalou, Lorelei Fournier, Emmanuelle Clérin, Frédéric Blond and the entreprise agricole à responsabilité limitée (EARL Morizeau Dangers, France) for their invaluable help. This work was supported by Inserm, Sorbonne University, the Agence Nationale pour la Recherche (ANR, Labex Lifesenses), Foundation Fighting Blindness (USA) and IHU FOReSIGHT [ANR-18-IAHU-0001] supported by French state funds managed by the ANR within the Investissements d'Avenir program.
Materials
| 96 black plates (Clear Button with lid Tissue culture treated) | Corning | 3603 | |
| Calcein AM | Thermo-Fisher scientific | C1430 | |
| CCD Camera | Photometrics | CoolSnap FX HQ | |
| CDPC (Cytidine 5′-diphosphocholine sodium salt dihydrate) | Sigma-Aldrich | C0256 | |
| CO2 Independant | Thermo-Fisher scientific | 18045-054 | |
| Curved forceps | Dutscher | 005093 | |
| DMEM Media | Thermo-Fisher scientific | 41966-029 | |
| DNAse | Sigma-Aldrich | D4263 | |
| Eggs incubator | FarmLine | M08 01 3100 | |
| Ethidium Homodimer | Thermo-Fisher scientific | E1169 | |
| Fœtal bovine serum | Thermo-Fisher scientific | 10270-098 | |
| Gentamycin | Thermo-Fisher scientific | 15710-049 | |
| Hydrocortisone | Sigma-Aldrich | H0880 | |
| ITS (insulin Transferine selenium) | Sigma-Aldrich | I1884 | |
| large straight pliers | Dutscher | 005074 | |
| Linoleic acid | Sigma-Aldrich | L8384 | |
| M199 medium | Thermo-Fisher scientific | 31150-022 | |
| Metamorph software | Metamorph | ||
| Microscope | NIKON | Eclipse TE2000 | |
| Motorized stage | Martzauzer | Mutlicontrol 2000 | |
| Optical filter switch | Shutter Instrument company | Lambda 10-2 | |
| PBS 1X | Thermo-Fisher scientific | 14190-086 | |
| Poly-L-lysine | Sigma-Aldrich | P6282 | |
| Progesterone | Sigma-Aldrich | P7556 | |
| Pursept A express | Fisher scientific | 11814110 | |
| Putriscine | Sigma-Aldrich | P5780 | |
| Sodium pyruvate | Sigma-Aldrich | S8636 | |
| straight forceps | Dutscher | 005092 | |
| Taurine | Sigma-Aldrich | T8691 | |
| Triiodothyronine | Sigma-Aldrich | T6397 | |
| Trypan blue | Thermo-Fisher scientific | 15250-061 | |
| Trypsine 0.25 % | Thermo-Fisher scientific | 25200-056 |
Riferimenti
- Brownstein, C. D. . The Implicit Mind. , (2019).
- Heesy, C. P., Hall, M. I. The nocturnal bottleneck and the evolution of mammalian vision. Brain, Behavior and Evolution. 75 (3), 195-203 (2010).
- Borges, R., et al. Adaptive genomic evolution of opsins reveals that early mammals flourished in nocturnal environments. BMC Genomics. 19 (1), 121 (2018).
- Adler, R., Hatlee, M. Plasticity and differentiation of embryonic retinal cells after terminal mitosis. Science. 243 (4889), 391-393 (1989).
- Fintz, A. C., et al. Partial characterization of retina-derived cone neuroprotection in two culture models of photoreceptor degeneration. Investigative Ophthalmology & Visual Science. 44 (2), 818-825 (2003).
- Roska, B., Sahel, J. A. Restoring vision. Nature. 557 (7705), 359-367 (2018).
- Duncan, J. L., et al. Inherited Retinal Degenerations: Current Landscape and Knowledge Gaps. Translational Vision Science & Technology. 7 (4), 6 (2018).
- Russell, S., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 390 (10097), 849-860 (2017).
- RetNet. . Retinal Information Network. , (2020).
- Doonan, F., Donovan, M., Cotter, T. G. Activation of multiple pathways during photoreceptor apoptosis in the rd mouse. Investigative Ophthalmology & Visual Science. 46 (10), 3530-3538 (2005).
- Comitato, A., et al. Dominant and recessive mutations in rhodopsin activate different cell death pathways. Human Molecular Genetics. 25 (13), 2801-2812 (2016).
- Cronin, T., Leveillard, T., Sahel, J. A. Retinal degenerations: from cell signaling to cell therapy; pre-clinical and clinical issues. Current Gene Therapy. 7 (2), 121-129 (2007).
- Baumgartner, W. A., Baumgartner, A. M. Accounting for disagreements on average cone loss rates in retinitis pigmentosa with a new kinetic model: Its relevance for clinical trials. Medical Hypotheses. 89, 107-114 (2016).
- Leveillard, T., Sahel, J. A. Rod-derived cone viability factor for treating blinding diseases: from clinic to redox signaling. Science Translational Medicine. 2 (26), (2010).
- Leveillard, T., Sahel, J. A. Metabolic and redox signaling in the retina. Cellular and Molecular Life Sciences. 74 (20), 3649-3665 (2017).
- Clerin, E., Marussig, M., Sahel, J. A., Leveillard, T. Metabolic and Redox Signaling of the Nucleoredoxin-Like-1 Gene for the Treatment of Genetic Retinal Diseases. International Journal of Molecular Sciences. 21 (5), (2020).
- Fridlich, R., et al. The Thioredoxin-like Protein Rod-derived Cone Viability Factor (RdCVFL) Interacts with TAU and Inhibits Its Phosphorylation in the Retina. Molecular & Cellular Proteomics. 8 (6), 1206-1218 (2009).
- Mei, X., et al. The Thioredoxin Encoded by the Rod-Derived Cone Viability Factor Gene Protects Cone Photoreceptors Against Oxidative Stress. Antioxidants, Redox Signaling. 24 (16), 909-923 (2016).
- Leveillard, T., et al. Identification and characterization of rod-derived cone viability factor. Nature Genetics. 36 (7), 755-759 (2004).
- Vergara, M. N., Gutierrez, C., Canto-Soler, M. V. Efficient Gene Transfer in Chick Retinas for Primary Cell Culture Studies: An Ex-ovo Electroporation Approach. Journal of Visualized Experiments. (105), e52002 (2015).
- Fridlich, R., et al. The thioredoxin-like protein rod-derived cone viability factor (RdCVFL) interacts with TAU and inhibits its phosphorylation in the retina. Molecular & Cellular Proteomics. 8 (6), 1206-1218 (2009).
- Ait-Ali, N., et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 161 (4), 817-832 (2015).
- Mei, X., et al. The Thioredoxin Encoded by the Rod-Derived Cone Viability Factor Gene Protects Cone Photoreceptors Against Oxidative Stress. Antioxidants & Redox Signaling. 24 (16), 909-923 (2016).
- Tan, E., et al. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Investigative Ophthalmology & Visual Science. 45 (3), 764-768 (2004).
- Wang, X. W., Tan, B. Z., Sun, M., Ho, B., Ding, J. L. Thioredoxin-like 6 protects retinal cell line from photooxidative damage by upregulating NF-kappaB activity. Free Radical Biology and Medicine. 45 (3), 336-344 (2008).
- Perron, N. R., Beeson, C., Rohrer, B. Early alterations in mitochondrial reserve capacity; a means to predict subsequent photoreceptor cell death. Journal of Bioenergetics and Biomembranes. 45 (1-2), 101-109 (2013).
- Krishnamoorthy, R. R., Clark, A. F., Daudt, D., Vishwanatha, J. K., Yorio, T. A forensic path to RGC-5 cell line identification: lessons learned. Investigative Ophthalmology & Visual Science. 54 (8), 5712-5719 (2013).
- Al-Ubaidi, M. R. RGC-5: are they really 661W? The saga continues. Experimental Eye Research. 119, 115 (2014).
- Clerin, E., et al. Vibratome sectioning mouse retina to prepare photoreceptor cultures. Journal of Visualized Experiments. (94), (2014).
- Hendrickson, A., Hicks, D. Distribution and density of medium- and short-wavelength selective cones in the domestic pig retina. Experimental Eye Research. 74 (4), 435-444 (2002).
- Bobu, C., Craft, C. M., Masson-Pevet, M., Hicks, D. Photoreceptor organization and rhythmic phagocytosis in the nile rat Arvicanthis ansorgei: a novel diurnal rodent model for the study of cone pathophysiology. Investigative Ophthalmology & Visual Science. 47 (7), 3109-3118 (2006).
- Saidi, T., Mbarek, S., Chaouacha-Chekir, R. B., Hicks, D. Diurnal rodents as animal models of human central vision: characterisation of the retina of the sand rat Psammomys obsesus. Graefes Arch Clin Exp Ophthalmol. 249 (7), 1029-1037 (2011).
- Traverso, V., Kinkl, N., Grimm, L., Sahel, J., Hicks, D. Basic fibroblast and epidermal growth factors stimulate survival in adult porcine photoreceptor cell cultures. Investigative Ophthalmology & Visual Science. 44 (10), 4550-4558 (2003).
- Balse, E., et al. Purification of mammalian cone photoreceptors by lectin panning and the enhancement of their survival in glia-conditioned medium. Investigative Ophthalmology & Visual Science. 46 (1), 367-374 (2005).
- Gagliardi, G., et al. Characterization and Transplantation of CD73-Positive Photoreceptors Isolated from Human iPSC-Derived Retinal Organoids. Stem Cell Reports. 11 (3), 665-680 (2018).
- Gagliardi, G., Ben M’Barek, K., Goureau, O. Photoreceptor cell replacement in macular degeneration and retinitis pigmentosa: A pluripotent stem cell-based approach. Progress in Retinal and Eye Research. 71, 1-25 (2019).
- Mears, A. J., et al. Nrl is required for rod photoreceptor development. Nature Genetics. 29 (4), 447-452 (2001).
- Swaroop, A., Kim, D., Forrest, D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nature Reviews Neuroscience. 11 (8), 563-576 (2010).
- Kallman, A., et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Nature Communications. 3 (1), 82 (2020).
- Eldred, K. C., et al. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 362 (6411), (2018).
- Kole, C., et al. Otx2-Genetically Modified Retinal Pigment Epithelial Cells Rescue Photoreceptors after Transplantation. Molecular Therapy. 26 (1), 219-237 (2018).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chick embryo. Journal of Morphology. 88 (1), 49-92 (1951).
- Kole, C., et al. Identification of an Alternative Splicing Product of the Otx2 Gene Expressed in the Neural Retina and Retinal Pigmented Epithelial Cells. PLoS One. 11 (3), 0150758 (2016).
- Shannon, M., Hamilton, A. T., Gordon, L., Branscomb, E., Stubbs, L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Research. 13 (6), 1097-1110 (2003).
- Stern, C. D., Holland, P. W. . Essential developmental biology: a practical approach. , (1993).
- Punzo, C., Kornacker, K., Cepko, C. L. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nature Neuroscience. 12 (1), 44-52 (2009).
- Harpavat, S., Cepko, C. L. RCAS-RNAi: a loss-of-function method for the developing chick retina. BMC Developmental Biology. 6, 2 (2006).

