On-Chip Crystallization and Large-Scale Serial Diffraction at Room Temperature
Summary
This contribution describes how to set up protein crystallization on crystal-on-crystal devices and how to perform automated serial data collection at room temperature using the on-chip crystallization platform.
Abstract
Biochemical reactions and biological processes can be best understood by demonstrating how proteins transition among their functional states. Since cryogenic temperatures are non-physiological and may prevent, deter, or even alter protein structural dynamics, a robust method for routine X-ray diffraction experiments at room temperature is highly desirable. The crystal-on-crystal device and its accompanying hardware and software used in this protocol are designed to enable in situ X-ray diffraction at room temperature for protein crystals of different sizes without any sample manipulation. Here we present the protocols for the key steps from device assembly, on-chip crystallization, optical scanning, crystal recognition to X-ray shot planning and automated data collection. Since this platform requires no crystal harvesting nor any other sample manipulation, hundreds to thousands of protein crystals grown on chip can be introduced into an X-ray beam in a programmable and high-throughput manner.
Introduction
Due to the ionizing effects of X-ray radiation, protein crystallography, to a great extent, has been limited to cryogenic conditions in the last three decades. Therefore, the current knowledge of protein motions during its function largely arises from comparisons between static structures observed in different states under cryogenic conditions. However, cryogenic temperatures inevitably hinder the progression of a biochemical reaction or interconversion between different conformational states while protein molecules are at work. To directly observe protein structural dynamics at atomic resolution by crystallography, robust and routine methods are needed for conducting diffraction experiments at room temperature, which calls for technical innovations in sample delivery, data collection, and posterior data analysis. To this end, recent advances in serial crystallography have offered new avenues to capture the molecular images of intermediates and short-lived structural species at room temperature1,2,3. In contrast to the "one-crystal-one-dataset" strategy widely used in conventional cryocrystallography, serial crystallography adopts a data collection strategy similar to that of single-particle cryo-electron microscopy. Specifically, the experimental data in serial crystallography are collected in small fractions from a large number of individual samples, followed by intensive data processing in which data fractions are evaluated and combined into a complete dataset for 3D structure determination4. This "one-crystal-one-shot" strategy effectively eases the X-ray radiation damage to protein crystals at room temperature via a diffraction before destruction strategy5.
Since serial crystallography requires a large number of protein crystals to complete a dataset, it poses major technical challenges for many biological systems where protein samples are limited and/or delicate crystal handling is involved. Another important consideration is how to best preserve crystal integrity in serial diffraction experiments. The in situ diffraction methods address these concerns by allowing protein crystals to diffract directly from where they grow without breaking the seal of the crystallization chamber6,7,8,9. These handling-free methods are naturally compatible with large-scale serial diffraction. We have recently reported the design and implementation of a crystallization device for in situ diffraction based on a crystal-on-crystal concept – protein crystals grown directly on monocrystalline quartz11. This "crystal-on-crystal" device offers several advantages. First, it features an X-ray and light transparent window made of a monocrystalline quartz substrate, which produces little background scattering, hence resulting in excellent signal-to-noise ratios in diffraction images from protein crystals. Second, the single-crystal quartz is an excellent vapor barrier equivalent to glass, thereby providing a stable environment for protein crystallization. In contrast, other crystallization devices using polymer-based substrates are prone to drying due to vapor permeability unless the polymer material has a substantial thickness, which consequently contributes to high background scattering10. Third, this device enables the delivery of a large number of protein crystals to the X-ray beam without any form of crystal manipulation or harvesting, which is critical for preserving crystal integrity11.
To streamline serial X-ray diffraction experiments using the crystal-on-crystal devices, we have developed a diffractometer prototype to facilitate easy switching between the optical scanning and X-ray diffraction modes12. This diffractometer has a small footprint and has been used for serial data collection at two beamlines of the Advanced Photon Source (APS) at Argonne National Laboratory. Specifically, we used BioCARS 14-ID-B for Laue diffraction and LS-CAT 21-ID-D for monochromatic oscillation. This diffractometer hardware is not required if a synchrotron or X-ray free-electron laser beamline is equipped with two key capabilities: (1) motorized sample positioning with a travel range of ±12 mm around the X-ray beam in all directions; and (2) an on-axis digital camera for crystal viewing under light illumination that is safe to protein crystals under study. The monocrystalline quartz device together with a portable diffractometer and the control software for optical scanning, crystal recognition, and automated in situ data collection collectively constitute the inSituX platform for serial crystallography. Although this development is primarily motivated by its dynamic crystallography applications using a polychromatic X-ray source, we have demonstrated the potential of this technology to support monochromatic oscillation methods10,12. With automation, this platform offers a high-throughput serial data collection method at room temperature with affordable protein consumption.
In this contribution, we describe in detail how to set up on-chip crystallization in a wet lab and how to perform serial X-ray data collection at a synchrotron beamline using the inSituX platform.
The batch method is used to set up on-chip crystallization under a condition similar to that of the vapor diffusion method obtained for the same protein sample (Table 1). As a starting point, we recommend using precipitant at 1.2-1.5x concentration of that for the vapor diffusion method. If necessary, the batch crystallization condition can be further optimized via fine grid screening. Quartz wafers are not necessary for optimization trials; glass coverslips can be used instead (see below). Partially loaded crystallization devices are recommended to keep optimization trials on a smaller scale. A number of protein samples have been successfully crystallized on such devices using the batch method10 (Table 1).
The device itself consists of the following parts: 1) an outer ring; 2) two quartz wafers; 3) one washer-like shim of plastic or stainless steel; 4) a retaining ring; 5) microscope immersion oil as sealant (Figure 1). The total volume of the crystallization solution loaded on one chip depends on the purpose of the experiment. The capacity of the crystallization chamber can be adjusted by choosing a shim of different thicknesses and/or inner diameter. We routinely set up crystallization devices of 10-20 µL in capacity using shims of 50-100 μm in thickness. A typical device can produce tens to thousands of protein crystals adequate for serial data collection (Figure 2).
When successful, on-chip crystallization will produce tens to hundreds or even thousands of protein crystals on each quartz device ready for X-ray diffraction. At a synchrotron beamline, such a device is mounted on a three-axis translation stage of the diffractometer using a kinematic mechanism. The crystallization window of a mounted device is optically scanned and imaged in tens to hundreds of micrographs. These micrographs are then stitched into a high-resolution montage. For photosensitive crystals, optical scanning can be carried out under infrared (IR) light to avoid unintended photoactivation. A computer vision software has been developed to identify and locate protein crystals randomly distributed on the device. These crystals are then ranked according to their size, shape, and position to inform or guide the data collection strategy in serial crystallography. For example, single or multiple shots can be located on each targeted crystal. Users could plan a single pass or multiple routes through targeted crystals. We have implemented software to compute various traveling routes. For example, the shortest route is computed using algorithms that address the traveling salesman problem13. For pump-probe dynamic crystallographic applications, the timing and duration of laser (pump) and X-ray (probe) shots can be chosen. An automated serial data collection is programmed to translocate each targeted crystal into the X-ray beam one after another.
The key components of the insituX diffractometer include: 1) a device holder; 2) a three-axis translation stage; 3) a light source for optical scanning; 4) an X-ray beam stop; 5) pump lasers if light-sensitive proteins are studied; 6) Raspberry Pi microcomputer equipped with an IR-sensitive camera; 7) control software to synchronize motors, camera, light sources, pump laser, and to interface with beamline controls.
Protocol
1. Device pre-assembly
- Label the outer ring (30 mm diameter) for sample identification. If necessary, include the project name, device number, crystallization condition, and date (Figure 1A). Place the outer ring upside down on a clean surface (Figure 1B), and carefully place one quartz wafer inside the ring (Figure 1C). This first quartz wafer serves as an entrance window for the incident X-rays.
- Pour a small amount of microscope immersion oil (viscosity of 150 cSt) into a Petri dish. Dip a shim in the oil, and make sure that both sides of the shim are properly oiled (Figure 1D). Remove the excess oil by dabbing the shim on a clean surface.
- Place the oiled shim on top of the first quartz wafer (Figure 1E).
NOTE: Immersion oil is an excellent sealant that protects the crystallization chamber from potential vapor loss. Properly assembled chips usually last for weeks without visible drying. This pre-assembly step is performed under room light. For light-sensitive samples, all subsequent steps, including sample loading, device storage, and observation, must be carried out under safety light.
2. Sample loading and device assembly
- Use a pipette to thoroughly mix the protein solution and crystallization buffer on the first quartz wafer. The volume ratio between the protein sample and buffer typically ranges from 2:1 to 1:2 (Figure 1F). Make sure that the total volume of the crystallization solution does not exceed the maximum capacity of the crystallization chamber determined by the shim size and thickness. Avoid air bubbles during mixing.
NOTE: The composition of a crystallization buffer varies from one experiment to another. Refer to Table 1 for crystallization conditions. - Place the second quartz wafer over the mixed solution as the solution starts to spread out (Figure 1G). This second quartz wafer serves as the exit window of the diffracted X-rays.
- Tap the second quartz wafer lightly on the edge to help spread the oil while pushing air out. Secure the device by screwing a retaining ring into the outer ring (Figure 1H). Use a tightening tool if necessary (Figure 1I). Be mindful that over-tightening may cause delicate quartz wafers to deform or even crack.
3. Device storage and crystallization optimization
- Store the assembled devices (Figure 1J) in a box at room temperature or inside an incubator with temperature control.
NOTE: Protein crystals may appear in a few hours to days after a crystallization device is assembled. Typical results from on-chip crystallization are shown for several representative protein samples (Figure 2). - Monitor crystal growth by observing the crystallization device under a microscope. If necessary, optimize crystallization conditions by iterations of sections 1-3.
4. Calibration
NOTE: The programs and commands mentioned in the sections below are run in inSituX software.
- Install a thin crystal of doped yttrium aluminum garnet on the chip holder (Figure 3). Install the beam stop. Take X-ray fluorescence images of the direct beam by running the program:
burnmark.py <device>.param
where <device> is a user-selected name for the crystallization device. <device>.param is a filename that contains device-specific control parameters. The default values will be gradually replaced by specific values along the protocol. A sample <device>.param file is shown in Supplementary File 1. - Find the precise position of the direct X-ray beam by running the beam profile fitting program:
beam.py <burn image> -d <device>
where <burn image> is the filename of the X-ray fluorescence image (Figure 4).
NOTE: This program calculates the precise direct beam positions as well as the beam size. The beam position marks the translocation destination for all crystals from the same device. The beam size is also used for target planning.
5. Optical scanning
- Place a crystallization device in the chip holder and secure the device using a thumbscrew (Figure 3A).
- Mount the chip holder onto the translation stage of the diffractometer through a kinematic mechanism (Figure 3B).
- Install a proper light source for taking micrographs from the optical window of the device. White light, IR light, or other light of choice can be used depending on the light sensitivity of the protein sample as well as the purpose of the experiment.
- Run the scan program:
scan.py <device>.param
This program captures a set of micrographs that are automatically transferred to specified user computers. - Run the tiling program on a user computer:
tile.py <device> -x <x> -y <y>
where <x> and <y> are the initial values for column and row displacements of micrographs, respectively. This program stitches all micrographs into a montage of 1-3 μm/pixel resolution (Figure 5).
NOTE: Steps 5.4 and 5.5 usually take a few minutes. The total number of micrographs ranges from several tens to hundreds depending on the scan area and magnification. - Run the crystal-finding program:
findX.py <montage> -c <length> <width> -w <wedge> -x <beam size>
where <montage> is the tiled image. This program performs crystal recognition and shot planning. <length> and <width> indicate the crystal size to be found. If the user wishes to avoid smaller crystals, <width> can be used as a cutoff by setting a number larger than the sizes of unwanted small crystals. <wedge> is an angular value that sets the tolerance for crystals of irregular shape. <beam size> refers to the direct beam size obtained from the profile fitting above (step 4.2; Figure 4). Also, a nominal value can be set by users to further space out targeted shots. These key parameters enable specific crystal selection and target planning (Figure 6).
6. X-ray diffraction
- Remove the light source and install the beam stop. Set an appropriate detector distance. Follow the beamline safety protocol to search the X-ray hutch. Open the X-ray shutter and laser shutter if applicable.
- Run the data collection program for serial diffraction:
collect.py <device>.param -l <light duration>
This command triggers data collection in which all planned shots are visited one after another according to a preprogrammed sequence. Each targeted crystal is translocated to the beam position (step 4.2). At each stop, X-ray exposure is taken with or without a laser illumination at a scheduled time delay. Movie 1 shows an automated data collection sequence operated at a frequency of 1 Hz. Routinely tens to hundreds of diffraction images are collected from a single crystallization device (Movie 2).
NOTE: Section 4 calibration and section 5 optical scan are self-contained in the inSituX platform, therefore, completely transferable to another beamline. Section 6 X-ray diffraction has to involve some detail in beamline operation.
Representative Results
Several representative datasets have been published In the past few years10,12 along with the crystallographic results and scientific findings from a diverse range of protein samples, including photoreceptor proteins and enzymes, for example, a plant UV-B photoreceptor UVR8, a light-driven DNA repair photolyase PhrB10, a novel far-red-light sensing protein from a multi-domain sensory histidine kinase14, ligand/light dual-sensor domains, and the photosensory core module of a bacteriophytochrome12. As representative results, we list the on-chip crystallization conditions of these proteins in Table 1, and directly compare them with the conditions used for the vapor diffusion method. Here we show four additional case studies of on-chip crystallization (Figure 2) and a collection of in situ diffraction patterns in a movie (Movie 2). Representative in situ datasets collected using this protocol are summarized in Table 2.
In a representative case, cryocrystallography gave rise to poor diffraction for a far-red-light sensing photoreceptor protein likely due to light sensitivity and high solvent content (~80%) of these crystals14. The electron densities obtained from the cryocrystallography data were too smeared to resolve the chromophore conformation, which is at the center of our scientific question. Using the in situ protocol, we were able to avoid unintended light activation before diffraction and obtained a dark dataset at room temperature from more than 800 crystals. This dark dataset from in situ serial Laue diffraction resulted in better resolved electron densities, allowing confident model building of a bilin chromophore that exhibits a hitherto unknown all-Z,syn conformation (Figure 7A)12,14. Our dynamic crystallography experiments have further revealed light-induced changes in this far-red photoreceptor protein by comparing data from 4,352 crystals in dark and 8,287 crystals after light illumination (Figure 7). A preliminary analysis of the light-induced difference maps has revealed concerted motions in the central β sheet, suggesting the importance of the π-π stacking between the pyrrole rings of the chromophore and several aromatic residues (Figure 7B,C). An in-depth analysis and scientific findings will be presented elsewhere.
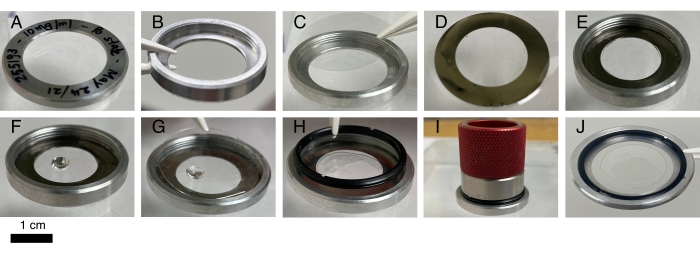
Figure 1: Crystallization device assembly. Each assembly is estimated to cost US$30 with two monocrystalline quartz wafers or US$10 with two glass coverslips. Hardware components except the shim are reusable. (A) The flat side of the outer ring is labeled for identification purposes. (B) The outer ring is placed upside down on a clean surface. (C) A quartz wafer of 1 inch in diameter is carefully placed inside. A glass chip could also be used instead during crystallization trials but is not compatible with X-ray diffraction. (D) Both sides of the shim are oiled. (E) The oiled shim is placed on the first quartz chip. (F) Protein and crystallization solutions are pipetted to the center of the chip and mixed. (G) A second quartz or glass chip covers the drop so that it evenly spreads over the chip. (H) A retaining ring is screwed over the second quartz wafer. (I) A tightening tool is used to tighten the retainer ring gently. (J) A fully assembled device. Please click here to view a larger version of this figure.
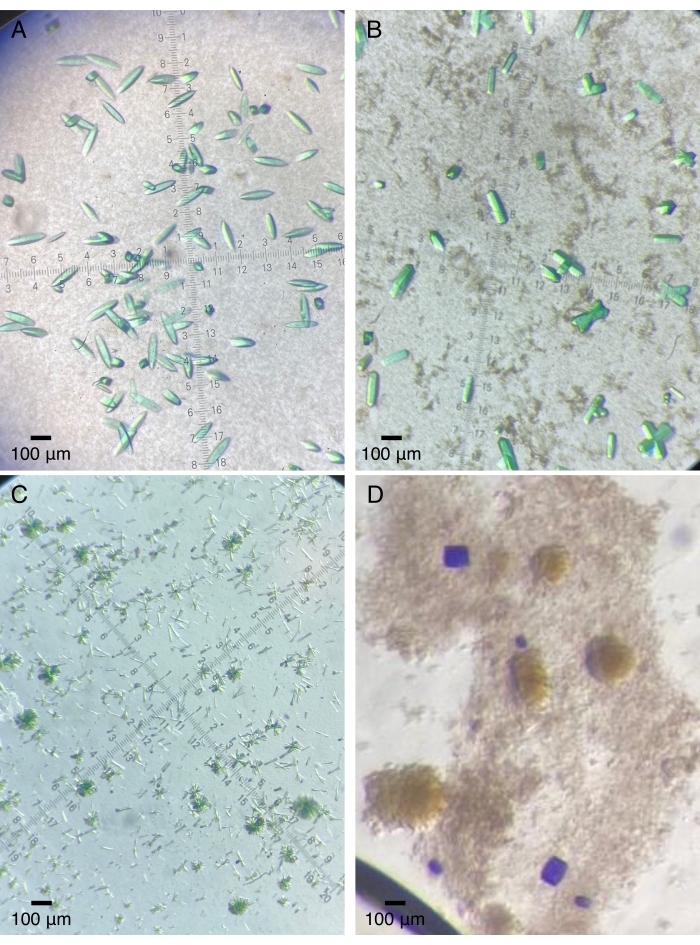
Figure 2: Representative protein crystals grown on quartz devices. (A) The photosensory core module of a bacteriophytochrome (Pa497 in Table 1). (B,C) Different constructs of the third GAF domain from a multi-domain sensory histidine kinase (2551g3 and 2551g3Δα1 in Table 1). (D) The tandem sensory domains from a dual-sensor histidine kinase (RECGAF in Table 1). Please click here to view a larger version of this figure.
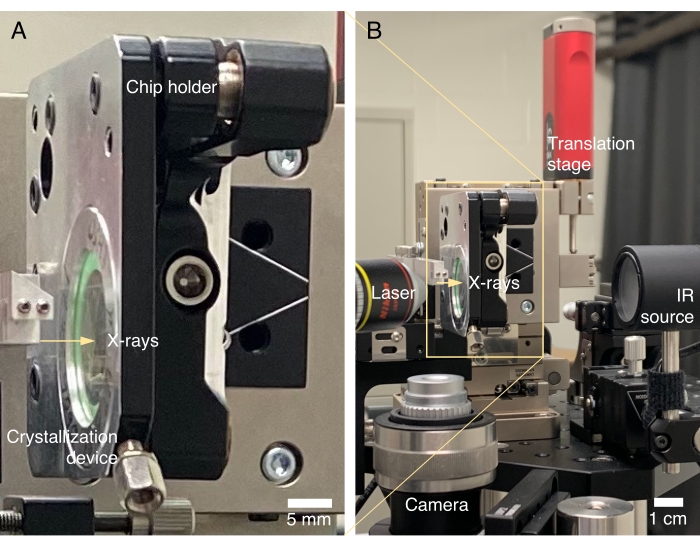
Figure 3: inSituX diffractometer. (A) A crystallization device is mounted in the chip holder. Although the device is mounted vertically, crystals grown on chip are not going to fall, primarily because the liquid layer in an assembled device is very thin, and crystals are anchored to their nuclei when growing. (B) An IR light source is installed for the optical scan. The camera captures the inline view of the protein crystals along the X-ray beam through a prism mirror (not visible in the picture). Please click here to view a larger version of this figure.
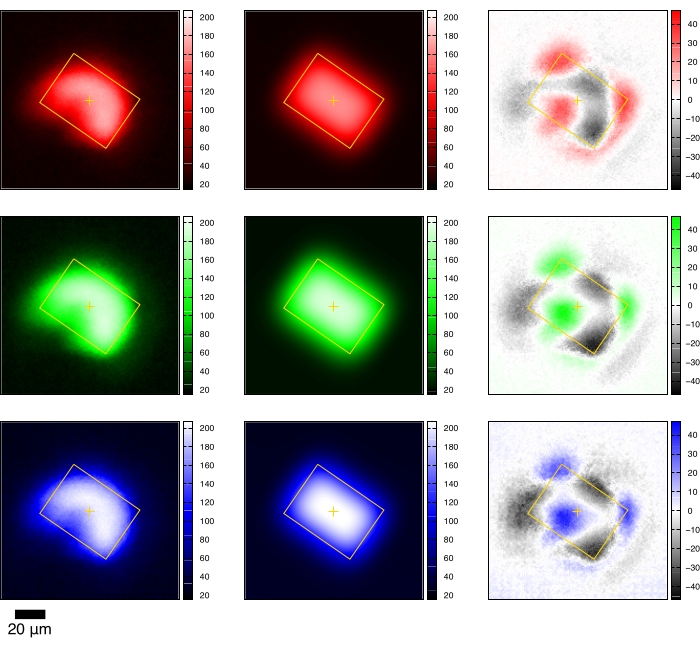
Figure 4: Direct beam profile fitting. The red, green, and blue channels of the X-ray fluorescence image are used to fit a two-dimensional Gaussian function. The left column shows the raw image of the red, green, and blue channels. The middle column is the fitting result with the precise beam position and size. The right column displays the fitting residuals. If the amplitude of the fitting residuals spans a small fraction of the raw image, the profile fitting of the direct beam is successful. Please click here to view a larger version of this figure.
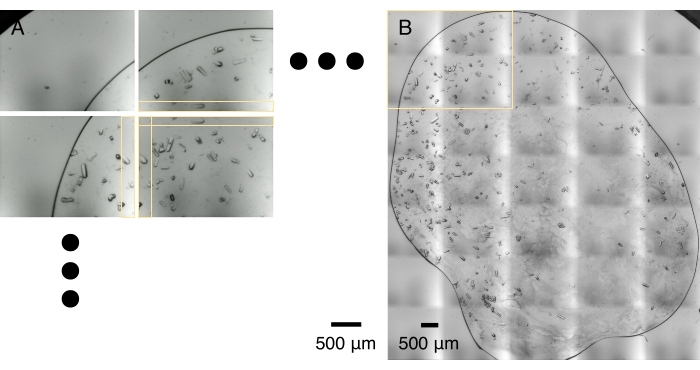
Figure 5: Image tiling. (A) An array of crystal micrographs is captured during an optical scan. The optical scan and data transfer usually take 1-2 min. Adjacent micrographs share a strip of overlapping area, horizontally and vertically, as marked by yellow boxes. (B) Micrographs are stitched together to make a high-resolution montage based on the optimal correlation in the overlapping areas. This process usually takes a minute on a laptop computer. The yellow box outlines the area captured by 2 x 2 micrographs shown in (A). Please click here to view a larger version of this figure.
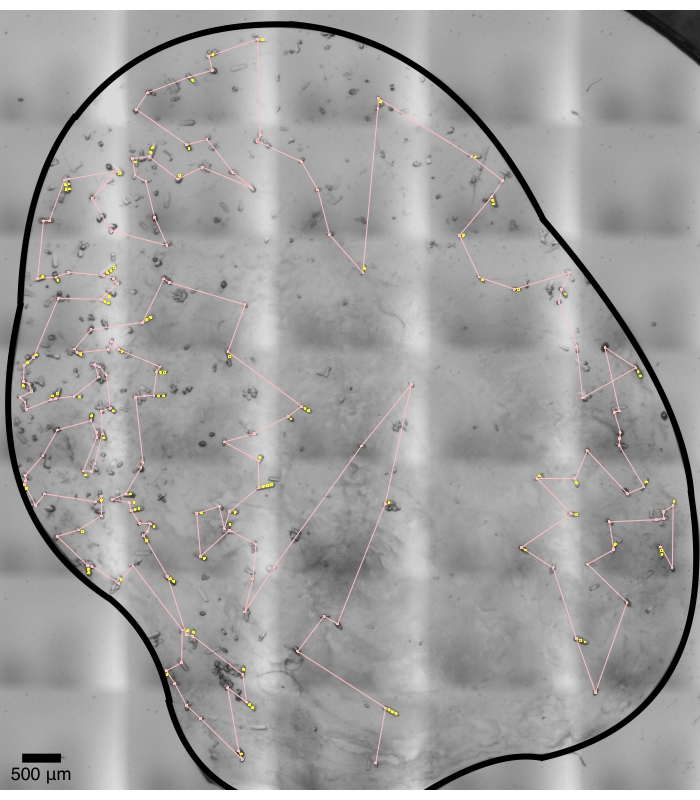
Figure 6: Crystal recognition and shot planning. Each pink circle marks the primary shot of a crystal. The yellow circles mark additional shots if a crystal is long enough to position these shots. The pink lines mark a route as a solution to the traveling salesman problem. Clustered crystals and smaller crystals are largely avoided. The aggressiveness of crystal finding can be adjusted as an option to findX.py (step 5.6). A brute force "overkill" strategy would leave no crystal un-shot but could produce lots of diffraction images, but not processable12. Please click here to view a larger version of this figure.
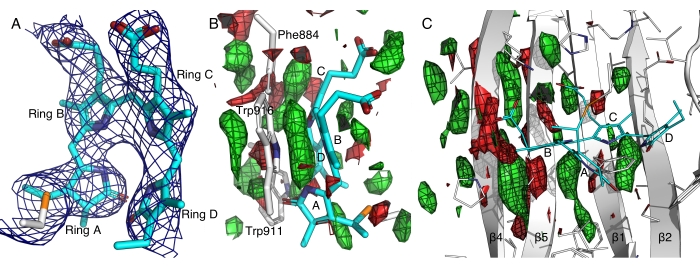
Figure 7: Electron density maps of the far-red-light sensing domain of a histidine kinase. (A) The 2Fo-Fc map contoured at 2.5σ shows the electron densities associated with the bilin chromophore in an all-Z,syn conformation14. Pyrrole rings A through D are marked. (B and C) Light-dark difference maps contoured at ±2.5σ in green and red, respectively, highlight the gain and loss of electron densities. Please click here to view a larger version of this figure.
Table 1: Comparison of crystallization conditions between vapor diffusion and on-chip batch method. Vapor diffusion and batch methods of crystallization are highly correlated10,14,15,16,17,18,19. Starting from a vapor diffusion condition, a similar condition can be optimized for on-chip crystallization. Please click here to download this Table.
Table 2: Summary of in situ datasets collected directly from quartz devices. Thousands of Laue diffraction patterns can be collected from several crystallization devices. Please click here to download this Table.
Movie 1: A mock data collection. Targeted crystals are translocated into the X-ray beam as marked by a red circle. The sequence of the targeted crystals in this movie does not follow a solution to the traveling salesman problem. Laser and X-ray exposures are fired at each stop with a programmed delay. Diffraction images are collected. Please click here to download this Movie.
Movie 2: Diffraction images. Hundreds of diffraction images can be collected from a single crystallization device. Several devices are sufficient to produce a complete and highly redundant dataset (Table 2). Please click here to download this Movie.
Supplementary File 1: Sample <device>.param file. A small text file collects some control parameters specific to each crystallization device. These parameters start with their default values and will be modified accordingly in sections 4, 5, and 6 as the protocol proceeds. Please click here to download this File.
Discussion
Protein crystallography in the early years conducted at room temperature experienced tremendous difficulty in battling X-ray radiation damage. Thus, it has been superseded by the more robust cryocrystallography method as synchrotron X-ray sources became readily available20. With the advent of X-ray free-electron lasers, room-temperature protein crystallography has been revived in recent years, with many new developments driven by a desire to observe protein structural dynamics at a physiologically relevant temperature2,21. The development of the inSituX platform based on the crystal-on-crystal devices has been motivated by the same ambition, that is, to establish routine and robust data collection methods for dynamic crystallography studies at room temperature. This automated serial X-ray diffraction method is also applicable to static structure determination for protein crystals unamenable to freezing14. In this protocol, we present key technical considerations along with critical steps needed to deliver room-temperature data collection using this platform. This method is particularly suited for fragile protein crystals that are sensitive to mechanical handling, X-ray radiation damage, or air exposure.
The platform prototype has been extensively tested at two protein crystallography beamlines at the Advanced Photon Source (APS) of Argonne National Laboratory. While it is rather straightforward to set up on-chip crystallization according to this protocol, the data collection step involves several custom-made hardware and software components. As a result, its application and implementation of project-specific data collection strategies may require close collaboration between users and beamline scientists. In other words, this technology in its current form is limited to those users who have adequate access to synchrotrons such as APS. Nevertheless, the overall workflow and key steps described in this protocol would serve as a reference or guide for any research group interested in room-temperature protein crystallography.
The most significant advantage of this platform is that no crystal manipulation such as mounting or freezing is required such that delicate protein crystals are diffracted in pristine conditions. Another major advantage is that the use of the monocrystalline quartz substrate gives rise to very little background scattering to protein diffraction images while offering stable environments for protein crystallization over a long time (weeks to months). However, this platform is not suited for sparse-matrix crystal screening as it is intended for large-scale crystal production. As such, prior knowledge of crystallization conditions is required to set up initial on-chip crystallization trials for a given protein sample.
In practice, we find that some device assembly steps, such as how to oil a shim (step 1.2) and how to seal a device (step 2.3), as trivial as they seem, often directly affect the outcomes of crystallization. A device may dry out quickly if oiling is not done properly. In addition, over-tightening of the device in the final step of assembly may deform the quartz wafers, while under-tightening leads to potential leaks and/or uncontrolled evaporation from the device. Another critical step is planning X-ray shots. One must carefully deal with clustered or crowded crystals to avoid overlapping diffraction patterns that are often difficult to process. This problem can be alleviated by the use of a micro-focusing X-ray beam. Potentially, a complete dataset could be difficult to obtain if the crystal morphology is a large thin plate so that most of the plates are parallel to the quartz windows. In addition, the single-crystal quartz chips can be recycled and reused after a cleaning procedure involving soap and organic solvents that remove oil and protein debris. Usually, about 80-90% of these delicate chips can be cleaned without damage for the next experiments. In the case of small crystals on a micro-focused beamline, when better precision must be achieved in crystal positioning, several hardware components could be upgraded, such as finer motors, better camera and optics, greater magnification, etc. However, none of these are close to the state-of-the-art limitations. Therefore, plenty of room is available for improvement without much difficulty.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Use of Advanced Photon Source, an Office of Science User Facility operated for US Department of Energy by Argonne National Laboratory, was supported by contract DE-AC02-06CH11357. Use of BioCARS was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number R24GM111072. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Use of LS-CAT Sector 21 was supported by Michigan Economic Development Corporation and Michigan Technology Tri-Corridor grant 085P1000817. This work is supported by grants from the University of Illinois at Chicago, National Institutes of Health (R01EY024363), and National Science Foundation (MCB 2017274) to XY.
Materials
| Analysis software | In-house developed | ||
| Cerium doped yttrium aluminum garnet | MSE Supplies | Ce:Y3Al5O12, YAG single crystal substrates | |
| Chip holder | In-house developed | ||
| Control software | In-house developed | ||
| Immersion oil | Cargille Laboratories | 16482 | Type A low viscosity 150 cSt |
| inSituX platform | In-house developed | ||
| IR light source | Thorlabs Incorporated | LED1085L | LED with a Glass Lens, 1085 nm, 5 mW, TO-18 |
| Microscope | Zeiss | SteREO Discovery V8 | |
| Outer ring | In-house developed | ||
| Petri dish | Fisher Scietific | FB0875713 | |
| Pipette | Pipetman | F167380 | P10 |
| Pump lasers | Thorlabs Incorporated | LD785-SE400 | 785 nm, 400 mW, Ø9 mm, E Pin Code, Laser Diode |
| Raspberry Pi | Raspberry Pi Fundation | ||
| Retaining ring | Thorlabs Incorporated | SM1RR | SM1 retaining ring for Ø1" lens tubes and mounts |
| Seedless quartz crystal | University Wafers, Inc. | U01-W2-L-190514 | 25.4 mm diameter Z-cut 0.05 mm thickness double side polish 8 mm on -X |
| Shim | In-house developed | ||
| X-ray beam stop | In-house developed |
Riferimenti
- Brändén, G., Neutze, R. Advances and challenges in time-resolved macromolecular crystallography. Science. 373, (2021).
- Fischer, M. Macromolecular room temperature crystallography. Quarterly Reviews of Biophysics. 54, (2021).
- Schaffer, J. E., Kukshal, V., Miller, J. J., Kitainda, V., Jez, J. M. Beyond X-rays: an overview of emerging structural biology methods. Emerging Topics in Life Sciences. 5 (2), 221-230 (2021).
- Nogales, E., Scheres, S. H. W. Cryo-EM: A unique tool for the visualization of macromolecular complexity. Molecular Cell. 58, 677-689 (2015).
- Chapman, H. N., Caleman, C., Timneanu, N. Diffraction before destruction. Philosophical Transactions of the Royal Society B Biological Sciences. 369, 20130313 (2014).
- Kisselman, G., et al. X-CHIP: an integrated platform for high-throughput protein crystallization and on-the-chip X-ray diffraction data collection. Acta Crystallographica Section D Biological Crystallography. 67 (6), 533-539 (2011).
- Liang, M., et al. Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline. Acta Crystallographica Section F Structural Biology Communications. 77, 319-327 (2021).
- le Maire, A., et al. In-plate protein crystallization, in situ ligand soaking and X-ray diffraction. Acta Crystallographica Section D Biological Crystallography. 67, 747-755 (2011).
- Perry, S. L., et al. In situ serial Laue diffraction on a microfluidic crystallization device. Journal of Applied Crystallography. 47, 1975-1982 (2014).
- Ren, Z., et al. Crystal-on-crystal chips for in situ serial diffraction at room temperature. Lab on a Chip. 18, 2246-2256 (2018).
- Ren, Z. Single crystal quartz chips for protein crystallization and X-ray diffraction data collection and related methods. US patent. , (2017).
- Ren, Z., et al. An automated platform for in situ serial crystallography at room temperature. IUCrJ. 7, 1009-1018 (2020).
- Croes, G. A. A method for solving traveling salesman problems. Operations Research. 6, 791-812 (1958).
- Bandara, S., et al. Crystal structure of a far-red-sensing cyanobacteriochrome reveals an atypical bilin conformation and spectral tuning mechanism. Proceedings of the National Academy of Sciences of the United States of America. 118, 2025094118 (2021).
- Shin, H., Ren, Z., Zeng, X., Bandara, S., Yang, X. Structural basis of molecular logic OR in a dual-sensor histidine kinase. Proceedings of the National Academy of Sciences of the United States of America. 116, 19973-19982 (2019).
- Yang, X., Ren, Z., Kuk, J., Moffat, K. Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature. 479, 428-432 (2011).
- Zhang, F., Scheerer, P., Oberpichler, I., Lamparter, T., Krauss, N. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proceedings of the National Academy of Sciences of the United States of America. 110, 7217-7222 (2013).
- Zeng, X., et al. Dynamic crystallography reveals early signaling events in ultraviolet photoreceptor UVR8. Nature Plants. 1, 14006 (2015).
- Wang, M., et al. Insights into base selectivity from the 1.8 Å resolution structure of an RB69 DNA polymerase ternary complex. Biochimica. 50, 581-590 (2011).
- Rodgrs, D. W. Cryocrystallography. Structure. 2, 1135-1140 (1994).
- Zhao, F. -. Z., et al. A guide to sample delivery systems for serial crystallography. TheFEBS Journal. 286, 4402-4417 (2019).

