Estimation of Structural Sensitivity of Intrinsically Disordered Regions in Response to Hyperosmotic Stress in Living Cells Using FRET
Summary
Intrinsically disordered regions (IDRs) are flexible protein domains that modify their conformation in response to environmental changes. Ensemble fluorescence resonance energy transfer (FRET) can estimate protein dimensions under different conditions. We present a FRET approach to assess IDR structural sensitivity in living Saccharomyces cerevisiae cells under hyperosmotic stress.
Abstract
Intrinsically disordered regions (IDRs) are protein domains that participate in crucial cellular processes. During stress conditions, the physicochemical properties of the cellular environment change, directly impacting the conformational ensemble of IDRs. IDRs are inherently sensitive to environmental perturbations. Studying how the physicochemical properties of the cell regulate the conformational ensemble of IDRs is essential for understanding the environmental control of their function. Here, we describe a step-by-step method for measuring the structural sensitivity of IDRs in living Saccharomyces cerevisiae cells in response to hyperosmotic stress conditions. We present the use of ensemble fluorescence resonance energy transfer (FRET) to estimate how the global dimensions of IDRs change during a progressive increase of hyperosmotic stress imposed on cells with any osmolyte. In addition, we provide a script for processing fluorescence measurements and comparing structural sensitivity for different IDRs. By following this method, researchers can obtain valuable insights into the conformational changes that IDRs undergo in the complex intracellular milieu upon changing environments.
Introduction
Intrinsically disordered regions (IDRs) are critical components in cellular processes1. In combination with structured domains, IDRs are essential for protein functions. The amino acid composition of IDRs is biased, represented mainly by charged, hydrophilic, and small residues. Because of this property, IDRs are considered low complexity domains2,3. Numerous IDRs have garnered attention, primarily because these regions play a crucial role in pathological conditions, particularly neurodegenerative diseases. Such diseases are characterized by self-assembly and subsequent extracellular or intracellular deposition of IDRs in neurons4. Examples of such IDRs include amyloid-β (Aβ) in Alzheimer's disease, huntingtin (HTT) in Huntington's disease, and TAR DNA-binding protein-43 (TDP-43) and fused in sarcoma (FUS) in amyotrophic lateral sclerosis and frontotemporal dementia4. The study of the structural rearrangements of IDRs in the context of disease has been significantly enhanced by spectroscopic methods, including fluorescence resonance energy transfer (FRET).
The hydrophilic and extended nature of IDRs makes them extremely sensitive to changes in the physicochemical properties of the solution environment5. The degree by which the conformational ensemble of IDRs is modified by the environment is called structural sensitivity5,6,7. Different techniques can be used to study the conformation and dynamics of IDRs, including circular dichroism (CD) and small-angle X-ray scattering (SAXS)8,9. Unfortunately, CD and SAXS require large quantities of purified proteins, so they are not appropriate for studies in cells. In contrast, FRET is a technique that measures the fluorescence intensity of two fluorescent molecules that specifically label one IDR, meaning that they can be monitored in complex mixtures such as living cells10. Dynamically measuring the structural sensitivity of IDRs in living cells is necessary for understanding how the environment regulates the conformation and function of the disordered proteome.
FRET is a powerful method for quantifying the structural sensitivity of IDRs, as well as globular and multidomain proteins in living cells. The method requires a construct consisting of an IDR of interest sandwiched between two fluorescent proteins (FP), known as a FRET pair. For this protocol, we suggest the use of mCerulean3 as the donor FP and Citrine as the acceptor FP, because of their large dynamic range, compared to other FPs reported in a previous study about IDRs sensitivity6. FRET has previously been exploited to measure the structural sensitivity of a plant IDR in different cellular contexts6. In addition, this technique has been used to characterize overall protein dimensions of IDRs by different research groups both in vitro and in vivo5,11.
Here, we describe the ensemble FRET method for studying the structural sensitivity of IDRs in living yeast (Saccharomyces cerevisiae) cells. We show representative results that are based in a plant IDR called AtLEA4-5. AtLEA4-5 is disordered in solution, but folds into α-helix when macromolecular crowding is induced in vitro12. AtLEA4-5 is a good reference model for this method because it is relatively small (158 residues), disordered and sensitive to environmental perturbation as reported in silico and in vitro6,12. The method presented here can be scaled for high throughput approaches because yeast cells are easy to grow, and the treatment is applied in small volumes. In addition, small modifications to the protocol can be applied to other cellular systems such as bacteria and plant cells6. The protocol can be performed in any molecular biology laboratory with access to a microplate reader with fluorescence mode, an equipment available in most research institutions.
Protocol
1. Plasmid construct
- Amplify the open reading frame (ORF) that codes for the desired IDR by polymerase chain reaction (PCR). Do not include the stop codon since the ORF will be flanked by the genes that encode the fluorescent proteins. For the amplification, design primers with the SacI (5') and BglII (3') restriction sites.
NOTE: For the representative results section, we used AtLEA4-5 as the selected IDR. We amplified the ORF of AtLEA4-5 from the pTrc99A-AtLEA4-5 plasmid12. - Digest the ORF PCR product by consecutive restriction with SacI and BglII enzymes13.
- Obtain the commercial plasmid pDRFLIP38-AtLEA4-5 (#178189) from Addgene (https://www.addgene.org/178189/).
- Conduct a consecutive restriction using SacI and BglII enzymes to remove the AtLEA4-5 ORF from the pDRFLIP38-AtLEA4-5 plasmid, leaving an open plasmid containing the FRET pair13.
- Purify the digested DNA fragments using gel recovery from an agarose gel electrophoresis14. Ligate the restricted fragments using DNA ligase15.
- Transform the cloning reaction into competent Escherichia coli cells (DH5α or related strains) and select in Luria-Bertani (LB) plates containing 50 µg/mL ampicillin16. Grow overnight (ON) at 37 °C.
- Isolate and grow at least five transformed colonies in a new plate and genotype by PCR17. Purify the plasmid DNA from a positive transformed colony using standard miniprep methods18.
- Verify the extraction and integrity of the plasmid DNA using agarose gel electrophoresis19. Verify the correct cloning of the construct using Sanger sequencing20.
2. Plasmid expression in yeast cells
NOTE: Use standard aseptic techniques to perform the following steps. Use a culture laminar flow hood or a lighter.
- Inoculate a streak of S. cerevisiae BJ5465 (ATCC: 208289) strain into 3 mL of Yeast Peptone Dextrose (YPD) medium and grow ON at 30 °C and 200 rpm.
NOTE: BJ5465 is a protease-deficient (CH1) strain recommended for working with IDRs. Other S. cerevisiae strains can be tested and used. - Centrifuge 1 mL of the overnight saturated (OD600 ~ 3-4) yeast culture at 14,000 x g for 1 min. Carefully remove the supernatant by pipetting.
- Resuspend the pellet in 1 mL of Tris-EDTA (TE) buffer (pH 7.5) by gently flicking the tube. Centrifuge at 14,000 x g for 1 min and carefully remove the supernatant.
- Resuspend the pellet in 500 µL of Lazy Bones buffer (40% w/v PEG 3,350; 100 mM lithium acetate; 10 mM Tris-HCl; 1 mM EDTA; pH 7.5) by pipetting. Add 25 µL of boiled (100 °C for 5 min, followed by cooling on ice for 5 min) salmon sperm DNA (2 mg/mL) to the resuspended yeast cells.
- Add 100 ng of the plasmid DNA to the mixture and vortex the mixture for 1 min to ensure thorough mixing. Allow the mixture to stand at room temperature for 1-2 h.
- Heat shock the yeast mixture at 42 °C for 12 min, then cool on ice for another 12 min. Centrifuge the tube at 14 000 x g for 1 min and carefully remove the supernatant.
- Resuspend the pellet in 1 mL of TE buffer (pH 7.5). Plate 100 µL of the transformed cells onto Yeast Synthetic-dropout Medium without uracil (SD-ura) plates supplemented with 15 g/L agar.
- Incubate plates at 30 °C for 2-3 days (Figure 1). Streak at least five yeast transformants in a new plate and grow ON.
3. Validation of yeast transformants
- Pick a small portion of each transformant candidate by gently scraping and resuspending in 5 µL of 20 mM NaOH into individual PCR tubes.
- Heat the sample at 99 °C for 10 min in a thermal cycler to lyse the cells and release the DNA into the solution. This step is crucial for preparing the DNA template for PCR.
- Transfer 1 µL of the boiled sample to a separate PCR reaction mix containing the appropriate primers and PCR reagents for genotyping. We used the following primers: Forward primer: 5´-AAATATACCCCAGCCTCGATCTAGA-3´. Reverse primer: 5´-GTAATACGACTCACTATAGGGCG-3´.
- Set up a PCR reaction and validate the presence of the correct plasmid using agarose gel electrophoresis. Select one transformant for further experiments.
4. Preparation of yeast cell culture for FRET assay
- Inoculate a streak of the selected yeast transformant into 3 mL of liquid 1x SD-Ura medium.
NOTE: Do this procedure in a laminar flow hood and sterile material. - Grow at 30 °C, 200 rpm for at least 12 h to reach saturation. Measure the OD600. Growth should fall between OD600 = 1.0-2.0.
NOTE: If OD600 is lower than 1.0, give cells more incubation time to grow. If OD600 exceeds 2.0, do not proceed and restart from step 4.1.
5. Yeast cell preparation for FRET assay
- Collect 2 mL of the overnight yeast culture and centrifuge at 14,000 x g at room temperature. Carefully remove the supernatant by pipetting.
- Resuspend the pellet in 1 mL of 50 mM 2-(N-Morpholino) ethanesulfonic acid (MES) buffer (pH 6).
- Centrifuge at 14,000 x g at room temperature. Remove the supernatant.
- Repeat steps 5.2 and 5.3. Resuspend yeast cells in 2 mL of MES buffer, pH 6 and pour the volume into a reagent reservoir.
6. Setting up the fluorescence measurements
NOTE: The present scanning is considered to be performed in a microplate reader with fluorescence mode.
- For basic settings, set the instrument as follows: Measurement type: Fluorescence Intensity (FI) spectrum; Microplate name: Greiner 96 F-bottom.
- For optic settings, set the instrument as follows: No. of wavelength scan points: 91; Excitation wavelength: 433 nm; Excitation bandwidth: 10 nm; Emission wavelength: 460 – 550; Step width: 1 nm; Emission bandwidth: 10 nm; Focal height obtained by autofocus; Focal height: 5 mm; Wavelength used for gain: 490 nm.
- For general settings, set the instrument as follows: Settling time: 0.1 s; Reading direction: unidirectional, horizontal left to right, top to bottom.
NOTE: The manually entered gain must be adjusted for every measurement using the well expected to display the highest fluorescence intensity levels throughout the wavelength scan.
7. Preparation of hypertonic solutions and fluorescence measurements
- Prepare solutions of 0 M, 0.2 M, 0.4 M, 0.6 M, 0.8 M, 1 M, 1.5 M, and 2 M NaCl in a 96-well clear-bottom black plate. The final volume in each well will be 200 µL (150 µL of osmolyte solution + 50 µL of yeast cell suspension).
NOTE: Other osmolytes can be used to induce hyperosmotic stress. All the solutions must be prepared in 50 mM MES buffer pH 6. - Load 150 µL of 50 mM MES buffer, pH 6 in first three wells of row A (A1, A2, A3) and in the subsequent wells, add 150 µL of the corresponding solution in increasing order from left to right (Figure 2). To test all the concentrations suggested, continue the loading in row B.
NOTE: This setting considers the measurements of 3 technical replicates for each concentration. - Use a 12-channel micropipette to transfer 50 µL of washed yeast cells to each well. Yeast cells tend to sediment to the bottom of the reservoir. Make sure to resuspend the yeast suspension thoroughly by pipetting up and down before transferring the cells.
- Load cells into the 96-well plate containing the different solutions and mix well by pipetting up and down at least 4x.
- Measure fluorescence intensity emission spectra immediately for the desired wells in a microplate reader with the specification settings of step 6. Repeat the procedure for every IDR to be tested in the FRET assay.
- Optional step. If available, measure a donor-only construct (pDRFLIP38-mCerulean3) following the steps described here.
NOTE: We recommended performing this step to calculate each condition's FRET efficiency. If the reader cannot perform this step, FRET can be calculated using the FRET ratio method. Both methods for FRET calculations are explained in steps 8 and 9. - Perform at least three replicates on three independent days for every construct.
8. Data processing using the FRET efficiency method
NOTE: For readers with knowledge of the R programming language, we provide a set of R scripts to perform data processing described in this section. Scripts can be found at https://github.com/Kaz-bits/cuevaslab-procotols/tree/main/FRET. Follow the instructions in the README file.
- Export data as .xlsx files from the microplate reader.
- Normalize fluorescence intensity values at every scan wavelength to the isosbestic point of the FRET pair used.
NOTE: This step is necessary for comparing the spectra of all conditions tested. The isosbestic point of mCerulean3 and Citrine FRET pair is 515 nm. For example, obtain the ratio of fluorescence intensity values of 460 nm/515 nm, 461 nm/515 nm, 462 nm/515 nm, and so on. - Calculate the mean for every technical replicate once the fluorescence values are normalized. In total, there should be a column for every condition of hyperosmotic stress.
- Visualize all the fluorescence intensity spectra in the same plot (Figure 3). Each spectrum should display a combination of the fluorescence emission spectra of individual mCerulean3 and Citrine. In rare cases, where the FRET efficiency is 0, only the donor fluorescence emission spectrum will be observed, or when FRET efficiency is 1, only the acceptor fluorescence will be observed. The fluorescence emission peak for mCerulean3 (donor) is 475 nm, and the fluorescence emission peak for Citrine (acceptor) is 525 nm.
- Determine if the fluorescence measurement displays a typical FRET change in response to hyperosmotic stress. If the conformation of the IDR is sensitive to the treatment, this will reflect as a change in FRET efficiency. A typical change in FRET efficiency will couple a decrease in the fluorescence intensity of the donor with an increase in the fluorescence intensity of the acceptor or vice versa.
- Quantify the FRET efficiency (EFRET). Obtain the donor (mCerulean3) normalized peak values (475 nm/515 nm) for the IDR construct and the donor-only construct for every condition of hyperosmotic stress. Use the mean normalized peak values of the technical replicates. Perform this operation for each of the three independent replicates. Calculate EFRET with the following formula:
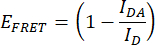
Where IDA is the ratio 475 nm/515 nm from the IDR construct (FRET pair construct), and ID is the ratio 475 nm/515 nm from the donor-only construct. - Compare FRET efficiencies. Normalize the EFRET values of every hyperosmotic stress condition to the EFRET of the non-stress condition (for example, 0 M NaCl). Make comparisons using a box plot or a smooth curve plot. Perform a One-way ANOVA followed by a post hoc Tukey statistical test (p-values: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
9. Data processing using the FRET ratio method
NOTE: If donor-only measurements cannot be acquired, perform the FRET ratio method. This method compares the FRET ratio across the different hyperosmotic stress conditions. Steps 7.1 to 7.5 should be performed before the steps of this section to validate a typical FRET behavior. For readers with knowledge of the R programming language, we provide a set of R scripts to perform data processing described in this section. Scripts can be found at https://github.com/Kaz-bits/cuevaslab-procotols/tree/main/FRET. Follow the instructions in the README file.
- Extract data at 475 nm and 525 nm from the xlsx file previously exported from the microplate reader. These values correspond to the fluorescence emission peak for mCerulean3 (donor, DxDm) and the fluorescence emission peak for Citrine (acceptor, DxAm) when the donor fluorophore is excited (433 nm).
- Normalize fluorescence intensity values at 475 nm and 525 nm to the isosbestic point of the FRET pair used. The isosbestic point of mCerulean3 and Citrine FRET pair is 515 nm. Obtain the FRET ratio (DxAm/DxDm).
- Compare FRET ratios. Normalize the DxAm/DxDm values of every hyperosmotic stress condition to the mean DxAm/DxDm of the non-stress condition (for example, 0 M NaCl). Make comparisons using a box plot or a smooth curve plot (Figure 4 and Figure 5). Perform a One-way ANOVA followed by a post hoc Tukey statistical test (p-values: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Representative Results
After transforming yeast cells with pDRFLIP38-AtLEA4-5 plasmid, the fluorescence of the positive transformants was observed with a blue light transilluminator and a filter (Figure 1). Preparing the different solutions to induce hyperosmotic stress is time-consuming, so we suggest following the 96-well template of Figure 2. Immediately after the hyperosmotic stress treatment with varying concentrations of sodium chloride, the fluorescence emission spectra were acquired (Figure 3). The fluorescence emission spectra of every condition were obtained to validate a typical FRET change behavior. Normalization of the fluorescence intensity values to the isosbestic point is required to correct experimental errors. For these representative results, we tested the previously reported AtLEA4-5, which is a highly sensitive plant IDR6. As a reference, we also tested the slightly insensitive ABP globular protein (Figure 3). It can be observed that both constructs display a combination of mCerulean3 and Citrine emission spectra (peaks for the donor and acceptor FPs), indicating some degree of basal FRET efficiencies under standard conditions (0 M NaCl). For AtLEA4-5, hyperosmotic stress caused an increase in the fluorescence intensity of the acceptor coupled with a decrease in the fluorescence intensity of the donor, indicating hyperosmotic stress-induced compaction of the IDR (Figure 3A). This trend was not observed in ABP (Figure 3B). To quantify differences in structural sensitivity across the different hyperosmotic stress treatments, we plotted the FRET ratio (DxAm/DxDm) for each condition and performed a One-way ANOVA statistical test (Figure 4). Finally, we compared the structural sensitivity of AtLE4-5 and ABP. We observed that AtLEA4-5 displayed a significantly higher sensitivity than ABP (Figure 5).
Figure 1: Fluorescence emission of S. cerevisiae transformants. SD-ura Petri dish plated with S. cerevisiae transformed with the pDRFLIP38-AtLEA4-5 plasmid. Fluorescence was observed using a blue light transilluminator and an orange filter. Please click here to view a larger version of this figure.
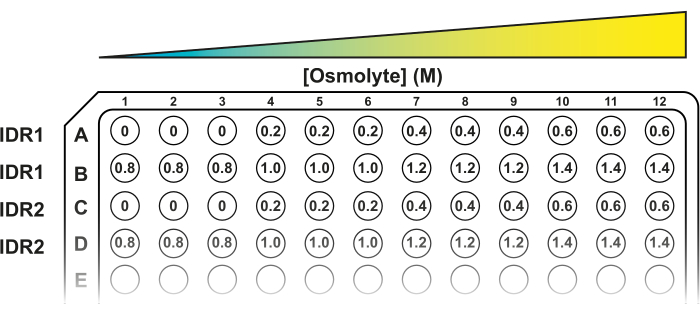
Figure 2: A 96-well plate template for the FRET system. Three technical replicates for each hyperosmotic stress condition are placed in consecutive wells. The first two rows (A and B) fit eight conditions for one construct (IDR1). The following rows can accommodate new constructs (IDR2, IDR3, etc.). Please click here to view a larger version of this figure.
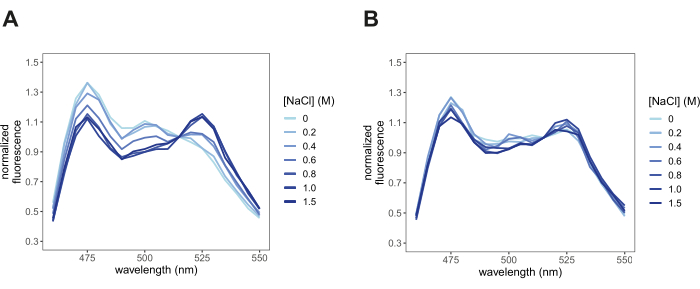
Figure 3: Normalized fluorescence emission spectra under different hyperosmotic stress conditions. Spectra validated a typical FRET behavior in response to hyperosmotic stress (NaCl) treatment. (A) AtLEA4-5, a highly sensitive IDR. The increase in the fluorescence intensity of the acceptor is coupled with the decrease in the fluorescence intensity of the donor. (B) ABP, a slightly insensitive globular protein. Peaks are observed at 475 nm and 525 nm according to the FRET pair mCerulean3 and Citrine. Please click here to view a larger version of this figure.
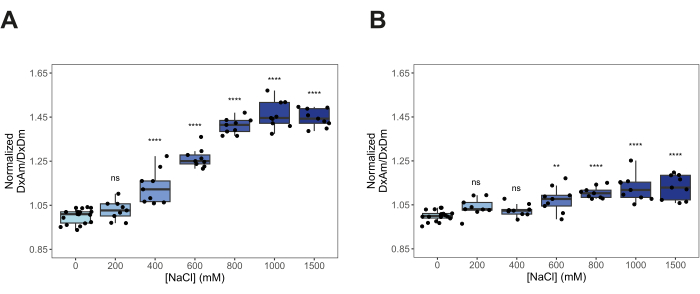
Figure 4: Quantification of structural sensitivity across different hyperosmotic stress conditions. Normalized FRET ratio (DxAm/DxDm) of live yeast cells treated with different concentrations of NaCl. (A) AtLEA4-5. (B) Arabinose-binding protein (ABP). n = 9 independent measurements. One-way ANOVA. ns: non-significant. p-values: *p < 0.05, **p < 0.01, ***p < 0.001, ****p< 0.0001. Boxes represent 25th-75th percentile (line at median). Please click here to view a larger version of this figure.
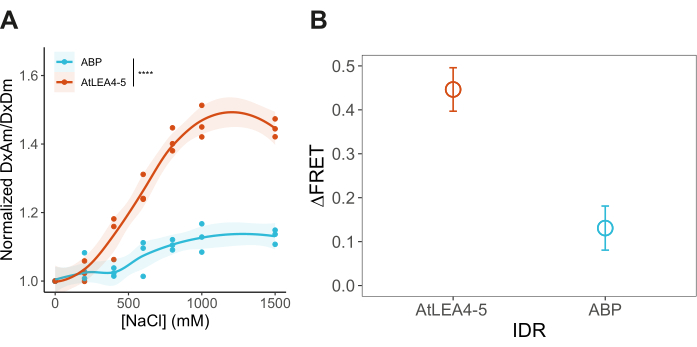
Figure 5: Comparison of the structural sensitivity of AtLEA4-5 and ABP. (A) Normalized FRET ratio (DxAm/DxDm) under different hyperosmotic stress conditions for AtLEA4-5 and ABP. n = 3 independent measurements. (B) Delta FRET value (1.5 M – 0 M NaCl) for AtLEA4-5 and ABP. Change is reported as the structural sensitivity of the constructs measured in the FRET system at 1.5 M NaCl. n = 3 independent measurements. Mean ± SD. Two-way ANOVA using the following p-values: *p < 0.05, **p < 0.01, ***p < 0.001, ****p< 0.0001. Please click here to view a larger version of this figure.
Discussion
The method presented here offers a way to obtain insights into how the global dimensions of the ensemble of IDRs sense and respond to environmental perturbations. This method relies on a genetically encoded construct and requires no additional components beyond a plasmid stable expression in yeast cells, making it adaptable for potential applications in other cell types. Moreover, it is versatile for exploring other physicochemical perturbations that eukaryotic cells experience during their life cycle21. The resolution of FRET is notable, with energy transfer occurring only within proximity of less than 10 nm between the donor and acceptor FPs. However, the main limitation is the absence of precise structural information that cannot be obtained following the ensemble FRET method.
The inherent heterogeneity and flexibility of IDRs pose challenges when studying their ensemble in a cellular context, making techniques such as X-ray crystallography impractical. NMR has been considered for studying disordered proteins in cells, but changing conditions can be an issue because of signal interference22. Implementing the FRET technique for studying IDR conformational ensembles offers unique capabilities, allowing the mapping of the population distribution within living cells. Additionally, it can be complemented with in vitro methods such as single-molecule FRET (smFRET), showcasing its adaptability to various approaches and experimental conditions23. This protocol uses mCerulean3 as the donor FP and Citrine as the acceptor FP, providing a straightforward and easy-to-track system. Selecting FRET pairs must carefully consider spectral overlap to mitigate the cross-talk between signals24,25. When reporting changes in structural sensitivity using this method, choosing an appropriate FRET metric, such as FRET efficiency (EFRET), is crucial for reliable signal interpretation. Alternatively, structural sensitivity can be analyzed and compared using the FRET ratio (DxAm/DxDm), but this requires a careful correction for donor-acceptor fluorescence cross-talk25. Interpreting fluorescence data obtained from microplate readers can be influenced by various factors, including the properties of the fluorophores, dipole orientation, intermolecular-FRET versus intramolecular-FRET systems, sample preparation and data analysis25.
Intermolecular-FRET is a major limitation of the ensemble FRET method because it is not possible to discard it from the initial analysis of the data. This poses and important consideration for the reader, because some IDRs are known to recruit into biomolecular condensates26,27. The local concentration of macromolecules in biomolecular condensates is high, which could induce intermolecular interactions and thus impact the FRET readout. For AtLEA4-5, it was shown that it self-assembles into discrete punctate structures upon hyperosmotic stress in yeast cells6. To rule out the effect of intermolecular-FRET, authors incubated the cells with 1,6-hexanediol (1,6-HD), a molecule that disrupts biomolecular condensates. 1,6-HD treatment dissolved the AtLEA4-5 condensates, but the FRET levels were not reduced, indicating that protein condensation is not causing undesired intermolecular FRET. Because this effect will depend on each IDR, readers should perform additional approaches such as acceptor photobleaching or co-expression of donor-only and acceptor-only constructs6. Yet, the information acquired from ensemble FRET in living cells is of significance relevance to characterize the structural sensitivity of IDRs.
An intricate aspect of the protocol presented here is that it utilizes an organism that exhibits responsive behavior to hyperosmotic stress. In the presence of high external concentrations of sodium chloride, water leaves the cell, increasing total protein concentration and establishing a crowded environment28. IDRs are sensitive to changes of macromolecular crowding5,6,11,12. Specifically, AtLEA4-5, the protein used as a model in this protocol, was sensitive to different molecular weight crowders, but not to high concentrations of salts or glycerol in vitro6. Since the accumulation of glycerol is important for the response and acclimation of yeast cells to hyperosmotic stress29, it is relevant to highlight that the main contributor to AtLEA4-5 compaction is macromolecular crowding during early events of osmo-sensing. How the structure of AtLEA4-5 is altered during subsequent stages of acclimation, when glycerol accumulates, requires further investigation.
Exploring conformational dynamics in disordered proteins encompasses a variety of techniques. In this context, FRET is a pivotal method. Depending on the specific research inquiries and features under consideration, investigators can employ in vivo NMR spectroscopy to characterize IDRs30. Additionally, smFRET is a valuable tool for in vivo studies23. Electron paramagnetic resonance (EPR) spectroscopy is another technique utilized to quantitatively study IDRs in a cellular context31. Mass spectrometry-based methods are also powerful approaches, since they can provide structural information of the cellular conformation of proteins32. These methods include native mass spectrometry (native-MS) and ion-mobility mass spectrometry (IM-MS)32,33. All these methods enable researchers to delve into the multiple conformations of an IDR and its dynamics, shedding light on their contributions to cell biology. The protocol described in this work can be employed to perform an initial screening of structural sensitivity in a high throughput manner. Follow-up studies can focus on testing the structural sensitivity in a desired cell type or with the potential to obtain mechanistic insight through in vitro methods such as smFRET, CD, or SAXS. The combination of methods is necessary for getting a clear picture of the structural sensitivity of IDR in the changing environments of cells.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank members of the Cuevas-Velazquez lab for the critical review of the manuscript. This work was supported by Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (UNAM-PAPIIT) project number IA203422; Consejo Nacional de Humanidades, Ciencias y Tecnología (CONAHCYT), project number 252952; and Programa de Apoyo a la Investigación y el Posgrado, Facultad de Química, Universidad Nacional Autónoma de México, Grant 5000-9182. CET (CVU 1083636) and CAPD (CVU 1269643) acknowledge CONAHCYT for their M.Sc. Scholarship.
Materials
| 96-well plate | Greiner Bio-One | 655096 | |
| Agar | Sigma-Aldrich | 5040 | |
| BglII | New England BioLabs | R0144S | |
| BJ5465 cells | American Type Culture Collection | 208289 | |
| Buffer MES 50 mM | Sigma-Aldrich | M8250 | |
| Buffer Tris-HCl 10 mM | Invitrogen | 15506017 | |
| EDTA 1 mM | Merck | 108452 | |
| Falcon tubes | Corning | 352057 | |
| LB media | Sigma-Aldrich | L2897 | |
| Lithium acetate 0.1 M | Sigma-Aldrich | L6883 | |
| Low Melt Agarose | GOLDBIO | A-204-25 | |
| Microcentrifuge | eppendorf | 5452000010 | |
| Miniprep kit | ZymoPure | D4210 | |
| NaOH 0.02 M | Merck | 106462 | |
| PEG 3,350 40% | Sigma-Aldrich | 1546547 | |
| plasmid pDRFLIP38-AtLEA4-5 | addgene | 178189 | |
| Plate reader | BMG LABTECH | CLARIOstar Plus | |
| SacI | New England BioLabs | R3156S | |
| Salmon sperm DNA 2 mg/mL | Thermo Fisher Scientific | 15632011 | |
| SD-Ura | Sigma-Aldrich | Y1501 | |
| Sodium cloride | Sigma-Aldrich | S9888 | |
| Taq polymesare | Promega | M5123 | |
| Transiluminator | Accuris instruments | E4000 | |
| UV-Visible spectrophotometer | Thermo Fisher Scientific | Biomate3 | |
| YPD media | Sigma-Aldrich | Y1500 |
Riferimenti
- Wright, P. E., Dyson, H. J. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 16 (1), 18-29 (2015).
- Covarrubias, A. A., Romero-Pérez, P. S., Cuevas-Velazquez, C. L., Rendón-Luna, D. F. The functional diversity of structural disorder in plant proteins. Arch Biochem Biophys. 680, 108229 (2019).
- Ahmed, S. S., et al. Characterization of intrinsically disordered regions in proteins informed by human genetic diversity. PLoS Comput Biol. 18 (3), e1009911 (2022).
- Birol, M., Melo, A. M. Untangling the conformational polymorphism of disordered proteins associated with neurodegeneration at the single-molecule level. Front Mol Neurosci. 12, 309 (2019).
- Moses, D., et al. Revealing the hidden sensitivity of intrinsically disordered proteins to their chemical environment. J Phys Chem Lett. 11 (23), 10131-10136 (2020).
- Cuevas-Velazquez, C. L., et al. Intrinsically disordered protein biosensor tracks the physical-chemical effects of osmotic stress on cells. Nat Commun. 12 (1), 5438 (2021).
- Holehouse, A. S., Sukenik, S. Controlling structural bias in intrinsically disordered proteins using solution space scanning. J Chem Theory Comput. 16 (3), 1794-1805 (2020).
- Martin, E. W., Hopkins, J. B., Mittag, T. Small-angle X-ray scattering experiments of monodisperse intrinsically disordered protein samples close to the solubility limit. Methods Enzymol. 646, 185-222 (2021).
- Miles, A. J., Drew, E. D., Wallace, B. A. DichroIDP: a method for analyses of intrinsically disordered proteins using circular dichroism spectroscopy. Commun Biol. 6 (1), 823 (2023).
- Kaminski, C. F., Rees, E. J., Schierle, G. S. K. A quantitative protocol for intensity-based live cell FRET imaging. Method Mol Biol. 1076, 445-454 (2014).
- Moses, D., et al. Structural biases in disordered proteins are prevalent in the cell. bioRxiv. , (2022).
- Cuevas-Velazquez, C. L., Saab-Rincón, G., Reyes, J. L., Covarrubias, A. A. The Unstructured N-terminal Region of Arabidopsis Group 4 Late Embryogenesis Abundant (LEA) Proteins Is Required for Folding and for Chaperone-like Activity under Water Deficit. J Biol Chem. 291 (20), 10893-10903 (2016).
- JoVE Science Education Database. . Restriction enzyme digests. , (2023).
- JoVE Science Education Database. . Gel purification. , (2023).
- JoVE Science Education Database. . DNA ligation reactions. , (2023).
- JoVE Science Education Database. . Bacterial transformation using heat Ssock and competent cells. , (2023).
- JoVE Science Education Database. . PCR: Principle, instrumentation, and applications. , (2023).
- JoVE Science Education Database. . Plasmid purification. , (2023).
- JoVE Science Education Database. . DNA gel electrophoresis. , (2023).
- JoVE Core Molecular Biology. Sanger/chain termination sequencing using dideoxynucleotides – Concept Available from: https://app.jove.com/science-education/v/12020/sanger-sequencing (2023)
- Theillet, F. X., et al. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem Rev. 114 (13), 6661-6714 (2014).
- Brutscher, B., et al. NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: General overview and practical guidelines. Adv Exp Med Biol. 870, 49-122 (2015).
- Metskas, L. A., Rhoades, E. Single-molecule FRET of intrinsically disordered proteins. Annu Rev Phys Chem. 71, 391-414 (2020).
- Roebroek, T., et al. Simultaneous readout of multiple FRET pairs using photochromism. Nat Commun. 12 (1), 2005 (2021).
- Algar, W. R., Hildebrandt, N., Vogel, S. S., Medintz, I. L. FRET as a biomolecular research tool – understanding its potential while avoiding pitfalls. Nat Methods. 16 (9), 815-829 (2019).
- Lyon, A. S., Peeples, W. B., Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol. 22 (3), 215-235 (2021).
- Belott, C., Janis, B., Menze, M. A. Liquid-liquid phase separation promotes animal desiccation tolerance. Proc Natl Acad Sci U S A. 117 (44), 27676-27684 (2020).
- Miermont, A., et al. Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proc Natl Acad Sci U S A. 110 (14), 5725-5730 (2013).
- Saito, H., Posas, F. Response to hyperosmotic stress. Genetica. 192 (2), 289-318 (2012).
- Selenko, P., Wagner, G. Looking into live cells with in-cell NMR spectroscopy. J Struct Biol. 158 (2), 244-253 (2007).
- Cattani, J., Subramaniam, V., Drescher, M. Room-temperature in-cell EPR spectroscopy: alpha-Synuclein disease variants remain intrinsically disordered in the cell. Phys Chem Chem Phys. 19 (28), 18147-18151 (2017).
- Beveridge, R., Chappuis, Q., Macphee, C., Barran, P. Mass spectrometry methods for intrinsically disordered proteins. Analyst. 138 (1), 32-42 (2013).
- Beveridge, R., et al. Ion mobility mass spectrometry uncovers the impact of the patterning of oppositely charged residues on the conformational distributions of intrinsically disordered proteins. J Am Chem Soc. 141 (12), 4908-4918 (2019).

