纯培养和条纹电镀:从混合样品中分离单一细菌菌落
English
Condividere
Panoramica
资料来源:蒂尔德·安徒森1号,罗尔夫·卢德1
1临床科学隆德系,感染医学系,生物医学中心,隆德大学,221 00 隆德,瑞典
似乎无法确定,微生物生物多样性确实令人震惊,估计有一万亿种物种并存(1,2)。虽然气候特别恶劣,如人类胃的酸性环境(3)或南极洲的冰下湖泊(4),可能由特定物种主导,但细菌通常存在于混合培养物中。由于每种菌株都可能影响另一种(5)的生长,分离和培养”纯”(仅包含一种类型)菌落的能力在临床和学术环境中都变得至关重要。纯培养能进一步进行遗传(6)和蛋白细胞学检查(7),分析样品纯度,也许更值得注意的是,从临床样本中鉴定和鉴定传染性病原体。
细菌具有广泛的生长要求,有许多类型的营养介质,旨在维持不苛求和挑剔的物种 (8)。生长介质可以以液体形式(作为肉汤)或以典型的琼脂基(一种从红藻中提取的凝胶剂)固体形式制备。直接接种到肉汤有产生基因多样性甚至混合细菌群的风险,电镀和再循环创造了一种更纯净的培养,每个细胞都有高度相似的遗传组成。条纹板技术基于样品的逐行稀释(图1),目的是将单个细胞彼此分离。任何由介质和指定环境维持的存活细胞(以下简称细胞形成单元,CFU)随后可以通过二元裂变找到孤立的子细胞群落。尽管细菌群落内突变率很快,但通常认为该细胞群为克隆细胞。因此,采集和重新采集这一种群可确保后续工作只涉及一种细菌类型。
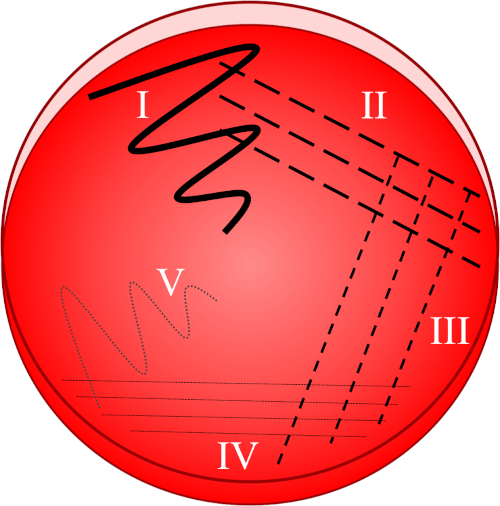
图1:条纹板基于原始样品的逐行稀释。I)接种最初使用锯齿形运动进行分散,从而形成细菌相对密集的区域。II-IV)条纹从前面的区域绘制,每次使用无菌接种循环,直到达到第四个象限。V)最后一个指向板块中间的锯齿状运动形成了一个区域,其中接种已明显稀释,使菌落彼此分开出现。
条纹板技术也可以与选择性和/或差分介质的使用相结合。选择性培养基会抑制某些生物体的生长(例如通过添加抗生素),而差别培养基将仅有助于区分另一种(例如,通过血液琼脂板上的溶血)。
微生物学的所有工作的基础是使用无菌(无菌)技术。每种细菌培养物都应被视为潜在的致病性,因为存在危险菌株意外生长、气溶胶形成和设备/人员污染的风险。为了尽量减少这些风险,所有介质、塑料、金属和玻璃器皿通常在使用前后通过高压灭菌进行灭菌,使其在 121°C 左右受到高压饱和蒸汽的影响,从而有效清除任何残留的细胞。工作空间通常在使用之前和之后使用乙醇进行消毒。在与传染性病原体一起工作时,始终佩戴实验室外套和手套。
Procedura
Risultati
The initial streak-plate may contain colonies originating from cells with different genetic makeup or (depending on sample purity) from different bacterial species (Figure 2A).
Through subsequent isolation of a single colony, where all units are derived from a common mother-cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth (Figure 2B).
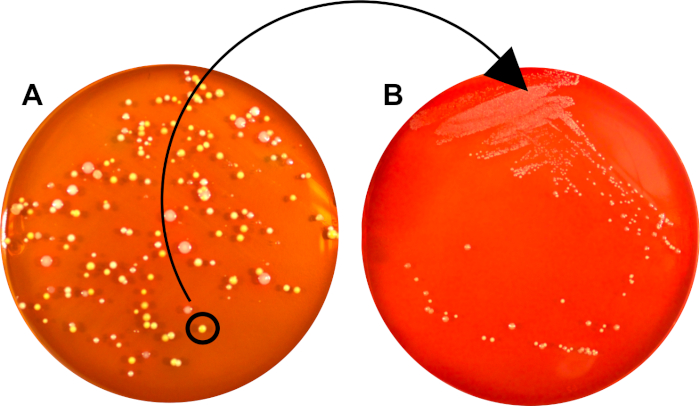
Figure 2: A pure culture can be generated from a mixed sample through isolation of a single, secluded colony. A) Growth of a single bacterial cell (CFU) generated a clonal colony, separated from those of other species and strains. This CFU was used for subsequent streaking onto a new plate B) A second plate, where the bacterial population consists solely of cells derived from the initial CFU.
Applications and Summary
The ability to obtain and cultivate a pure bacterial colony is essential, both in clinical and academic settings. Streak plating enables the isolation of a relatively clonal cell population, originating from a shared CFU, that may be of particular interest during diagnosis or for additional characterization of the isolate. A sample is streaked onto a suitable agar-based nutrient medium and incubated until colonies become visible. An isolated colony is subsequently harvested and re-streaked onto a second plate.
Riferimenti
- The Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 486:207-214. (2012)
- Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 113 (21) 5970-5975 (2016)
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infection and Immunity. 66:4517-21. (1998)
- Mikucki JA, Auken E, Tulaczyk S, Virginia RA, Schamper C, Sørensen KI, Doran PT, Dugan H, Foley N. Deep groundwater and potential subsurface habitats beneath an Antarctic dry valley. Nature Communications. 6:6831. (2015)
- Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nature Microbiology. 3:132-140. (2018)
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infectious Diseases. 7:711-23 (2007)
- Yao Z, Li W, Lin Y, Wu Q, Yu F, Lin W, Lin X. Proteomic Analysis Reveals That Metabolic Flows Affect the Susceptibility of Aeromonas hydrophila to Antibiotics. Scientific Reports. 6:39413 (2016)
- Medina D, Walke JB, Gajewski Z, Becker MH, Swartwout MC, Belden LK. Culture Media and Individual Hosts Affect the Recovery of Culturable Bacterial Diversity from Amphibian Skin. Frontiers in Microbiology. 8:1574 (2017)
Trascrizione
On a Petri dish, if a single bacterium undergoes multiple rounds of asexual reproduction, it will lead to the formation of a clonal colony. However, obtaining a single bacterium from a mixed sample, such as a soil suspension, can be difficult. If one loopful of this heterogeneous culture is taken, it can contain as many as one trillion individual bacteria. To spread this many bacteria out onto the surface of an agar plate and obtain a single colony, even using a zig-zag pattern, the loop would need to be dragged continuously over the surface of enough plates set side-by-side to encircle the entirety of Liberty Island. Obviously, scientists do not really use that many plates. Instead, they use a technique called streak plating.
The streak plate technique is based on progressive dilution of a bacterial sample, and it is performed over the solid media surface of a single Petri dish. To begin, the media surface is visually divided into five sections by assigning four fragments of the circumference as the first four sections, and the plate’s center as the fifth. This will effectively create five media plates out of a single Petri dish. Next, using a loopful of desired inoculum, the first section is streaked using a zig-zag pattern. Then, either a new disposable loop is used, or in the case of a wire loop, it is sterilized with a Bunsen burner, flaming it until it is red hot along the length of the wire. This use of a new loop, or flame sterilize loop, removes any remaining bacterial cells, assisting in the dilution of the bacteria. The hot loop is then cooled in the air for a few seconds before being dragged through the first section to create three to four separate lines, each carrying only a fraction of bacteria into the second section. The remaining sections are streaked in the same manner, using a sterile loop each time, and a single pass through the previous streak.
Using this cycle of streaking and sterilizing, the bacterial concentration in every subsequent section should be diluted so that the final section contains only a few discretely located bacteria. Upon incubation, these discrete bacteria multiply to produce isolated clonal colonies of daughter cells, which are referred to as Colony Forming Units, or CFUs. These can be harvested and re-streaked to ensure that subsequent work involves only a single bacterial type, referred to as a pure culture. As well as isolating single colonies from a mixed-bacterial culture, the streak plating technique is also used to select media-specific strains, determine bacterial colony morphology, or identify different bacterial species. In this video, we will demonstrate how to isolate single-bacterial colonies from a mixed-bacterial sample suspension via streak plating technique.
To begin, put on laboratory gloves and a lab coat. Next, sterilize the workspace using 70% ethanol. Next, select a suitable medium that will sustain the utilized bacterial species or strain and begin preparing the media. Here, common LB agar is prepared by weighing out ten grams of pre-formulated, powdered media and 7.5 grams of agar. Add the weighed, dried components to a glass bottle which is able to hold twice the final volume to avoid overflow. Then, add 500 milliliters of water to the bottle, and cap it semi-tightly. Sterilize the media by placing the bottle in an autoclave set to 121 degrees Celsius for twenty minutes. After completion, use heat-proof gloves or a hot pad to remove the media from the machine and then immediately twist the bottle cap to close it tightly.
For the same-day use, let the media cool down by placing the bottle into a water bath heated to approximately 45 degrees Celsius, to preserve the media in a liquid state. Alternatively, the media can be left at room temperature to store at solid state. When needed, microwave the bottle with the lid slightly open to melt the media, and allow the media to cool using a 45 degree Celsius water bath.
Next, take a sleeve of sterile Petri dishes, and with a permanent marker, label them with the investigator and media names as well as the date. Then, transfer the required volume of media into a sterile vessel, and add antibiotics or other sensitive components if necessary. Here, 50 milliliters of media is mixed with 100 microliters of Kanamycin for a final concentration of 25 micrograms per milliliter. Swirl the tube to ensure even distribution of the added components throughout the media. Slowly, so as to avoid bubble formation, pour 20 to 25 milliliters of approximately 45 degree Celsius culture medium into each of the plates. If bubbles or foam appear, swiftly remove using a regular pipette and a sterile tip. Then, immediately replace all lids to prevent contamination. Allow the agar to solidify at room temperature for at least two hours or overnight. Once solidified, store the culture plates upside down at four degrees Celsius to minimize condensation on the medium’s surface.
To streak the culture of choice, first take a clean culture plate and remove the lid. Working quickly, submerge a disposable, sterile loop into the desired inoculum and then immediately swab the loop over the first quadrant of the plate using a zig-zag motion. Replace the lid of the dish, discard the used inoculation loop, and then select a new sterile loop. Using the new loop, make three to four strokes crossing the original swab line radiating from the first quadrant, which should contain a relatively dense bacteria population into the second quadrant. Close the lid once more, and discard the loop. With a new loop, repeat this action again, but this time streaking from the second into the third quadrant. Then, with a new loop again, make another streak from the third into the fourth section of the plate. Finally, with a fresh loop, make one last stroke in a zig-zag pattern from the fourth quadrant towards the center of the plate. The bacterial prevalence will be lower in this area, ideally allowing individual colonies to be established from a single viable mother cell.
Replace the plate lid, and if appropriate for the bacterial species, seal the plate with para film to prevent airflow. Turn the culture plate upside down to prevent condensation drips, and then place at a suitable temperature for growth. Here, an incubator is set to 37 degrees Celsius. Allow the plate to incubate until bacterial colonies are visible. To generate a clonal bacterial population, select one discrete colony from this plate. Now, with the sterile loop, touch the target colony, and as before, make a streak in the first quadrant of a new plate. Continue to alternately sterilize the loop and streak the remaining quadrants of the plate as previously demonstrated, ending with the zig-zag to the center. Close the plate, and place it to incubate until discrete colonies form. Once these colonies are grown, they will typically represent pure clonal strains.
The initial streak plate may contain colonies originating from cells from different bacterial species or cells with different genetic makeup, depending upon the sample purity. Through subsequent isolation of a single colony, where all units are derived from a common mother cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth.
