Effects of Desmodium caudatum on Gastrointestinal Hormones and Intestinal Flora in Rats with Gastritis
PREPARAZIONE ISTRUTTORI
CONCETTI
Student Protocol
The procedures for the care and use of animals were approved by the Ethics Committee of Mianyang Normal University, and all relevant institutional and governmental regulations concerning the ethical use of animals were strictly followed. For the present study, SPF rats (Kunming species, both male and female, weighing 180-220 g) were utilized. The animals were obtained from a commercial source (see Table of Materials). All animals were housed in a pathogen-free environment and provided ad libitum access to food.
1. Preparation of reagents
- Prepare a 5% sodium salicylate solution by weighing 2.5 g of sodium salicylate powder (see Table of Materials) with an electronic scale. Dissolve the powder in a small amount of distilled water and dilute it to a total volume of 50 mL.
- Prepare the decoction of D. caudatum (12.8 g/kg). Weigh 128 g of D. caudatum (see Table of Materials), place it into a round-bottom flask containing 1000 mL of laboratory distilled water, and concentrate it to a final volume of 100 mL.
2. Grouping and administration
- Divide the rats into different groups and intragastrically administer the required solutions.
NOTE: For the present study, 18 rats (9 males, 9 females) were divided into three groups: the control group (Group C), the model group (Group M), and the treatment group (Group T), with 6 rats in each group. The control group received saline solution, and the model group received a 5% sodium salicylate solution for 5 weeks at a dose of 10 mL/kg/day15. The treatment group was pretreated with a 5% sodium salicylate solution for 5 weeks at a dose of 10 mL/kg/day, followed by administration of the D. caudatum decoction at a dosage of 1 mL/100 g for 10 days15,16. - Collect feces from the rats in dry and sterile microcentrifuge tubes using the tail-carrying method17 on the last day of the experiment and freeze them in a liquid nitrogen ultra-low-temperature freezer.
- After the end of sampling, sacrifice the rats by cervical dislocation (following institutionally approved protocols). Open the abdominal cavity with scissors to expose the abdominal aorta.
- Insert the needle (23-25 G) almost parallel into the abdominal aorta, collect the serum, and preserve it in an ultra-low-temperature freezer. Take out the stomachs from the abdominal cavity16 and soak in a 10% formalin solution for preservation.
NOTE: In Group M, the chronic gastritis model is considered successful if the gastric mucosa is dilated and congested with regular glandular morphology15, while observing a small amount of inflammatory cell infiltration in the lamina propria.
- Insert the needle (23-25 G) almost parallel into the abdominal aorta, collect the serum, and preserve it in an ultra-low-temperature freezer. Take out the stomachs from the abdominal cavity16 and soak in a 10% formalin solution for preservation.
3. Pathological section of gastric wall
- Take the gastric wall samples from rats in each group for pathological sections. Subsequently, perform routine hematoxylin-eosin staining to observe and analyze the pathological changes18.
4. Determination of serum gastrointestinal hormones
- In the serum sample, detect the contents of gastrin (GAS) and malondialdehyde (MDA)19 using ELISA following the manufacturer's instructions (see Table of Materials). Utilize a microplate reader for the analysis.
5. Fecal total DNA extraction and PCR amplification and purification
- Perform microbial DNA extraction utilizing the commercially available DNA isolation kit (see Table of Materials) and determine the concentration and purity using UV-Vis spectrophotometer16. Subsequently, assess DNA integrity through 1% agarose gel electrophoresis18.
- Amplify the bacteria 16S rRNA gene with primers 338F (5′-ACT CCT ACG GGA GGC AGC AG-3′) and 806R (5′-GAC TAC HVG GGG GTW TCT AT-3′) using a thermocycler PCR system17. The primers are obtained from commercial sources (see Table of Materials).
NOTE: Follow the PCR program: denaturation (3 min, 95 °C), followed by 27 cycles (30 s, 95 °C; 30 s, 55 °C; 45 s, 72 °C), and a final extension (10 min, 72 °C). Conduct PCR reactions with the following components: 4 µL of 5× DNA polymerase buffer, 2 µL of 2.5 mM dNTPs, 0.8 µL of each primer (5 µM), 0.4 µL of DNA polymerase, and 10 ng of template DNA (see Table of Materials). - Extract, purify, and quantify the PCR products sequentially using 2% agarose gel, commercially available DNA gel extraction kit, and fluorometer, respectively17.
6. Sequencing
NOTE: Step 6 and step 7 are performed following previously published methods20,21.
- Utilize the Illumina platform for sequencing.
NOTE: One end of the DNA fragment complements the base of the connector embedded in the chip, securing it onto the chip. The other end randomly complements the base of another adjacent buried joint, forming a "bridge." - Perform PCR amplification to generate DNA clusters. Linearize DNA amplifiers into single strands.
- Introduce an altered DNA polymerase alongside deoxyribonucleotide triphosphates (dNTPs) featuring four distinct fluorescent markers. Synthesize only one base per cycle.
- Utilize a laser to scan the surface of the reaction plate and determine the nucleotide types polymerized during the initial reaction of each template sequence.
- Chemically cleave the "fluorescence group" and "termination group" to reestablish the 3' sticky end. Subsequently, advance to polymerizing the second nucleotide.
- Obtain the sequence of template DNA fragments by analyzing the fluorescence signal results collected in each round.
7. Processing of sequencing data
- Initially, trim the reads of raw fastq data at any site with a score < Q20 and eliminate reads with uncertain bases. Additionally, permit two mismatches in precisely matched primers. Subsequently, merge the sequences with an overlap >10 bp.
- Cluster operational taxonomic units (OTUs) with 97% similarity using UPARSE, and remove chimeric sequences with UCHIME20,21.
- Employ the RDP Classifier algorithm to analyze the taxonomy of each 16S rRNA gene sequence against the Silva (SSU123) 16S rRNA database, using a confidence threshold of 70%.
- Perform Alpha diversity analysis and Beta diversity analysis using Mothur and Qiime, respectively (see Table of Materials).
8. Statistical analysis
- Analyze the data in this study using statistical analysis software (see Table of Materials). Employ one-way analysis of variance (ANOVA) for comparisons among multiple groups and utilize the least-significant-difference (LSD) test for comparisons between two groups.
- Consider P < 0.05 as statistically significant in all comparisons. P < 0.01 is regarded as extremely significant.
Effects of Desmodium caudatum on Gastrointestinal Hormones and Intestinal Flora in Rats with Gastritis
Learning Objectives
The results of the pathological section of the stomach wall are depicted in Figure 1. In comparison to Group C, Group M exhibited mild gastric wall atrophy and mild inflammation. However, when compared to Group M, Group T showed no evident inflammation, intestinal metaplasia, or atrophy. This suggests that D. caudatum decoction can effectively improve gastritis.
The serum gastrointestinal hormone assay results are presented in Figure 2. The content of malondialdehyde (MDA) was significantly higher in Group M than in Group C. After treatment with D. caudatum decoction, it was significantly reduced. Regarding the content of gastrin (GAS), Group M showed significantly lower levels than Group C, but Group T exhibited a significant increase. These results indicate that D. caudatum decoction may regulate the level of gastrointestinal hormones to treat gastritis.
Alpha diversity analysis, as shown in Table 1 and Figure 3, reveals a significant difference between the communities of Group M and Group T. The number of bacteria in Group C and Group T is higher than that in Group M, suggesting that the number and types of intestinal bacteria in rats gradually tend to a normal level after treatment.
Species composition analysis
According to Figure 4A, the community barplot illustrates that Lactobacillus and norank_f__Muribaculaceae account for the largest proportion in Group C. Group M contains a higher abundance of Clostridium_sensu_stricto_1 and some Helicobacter. The community composition of Group T is more similar to that of Group C, with important components being Lactobacillus and Romboutsia. Additionally, the Spearman correlation heatmap (Figure 4B) combined with the circos diagram (Figure 4C) reveals significant differences in flora composition between Group C and Group M, with distinct variations in dominant flora. After treatment, Group T tends to return to a normal flora state. Furthermore, Figure 5 indicates extreme differences in community composition between Group M and Group C, with significant changes occurring in the structure and composition of intestinal flora in rats with gastritis. After treatment, there are similarities in flora composition between Group C and Group T, as well as some similarities between Group T and Group M. These results indicate that D. caudatum decoction could restore the intestinal flora of rats with gastritis to a normal or close-to-normal state.
Species difference analysis
From the multi-species comparison column chart in Figure 6A, it can be observed that Norank_f_Muribaculaceae is abundant in Group C and significantly differs from Group M and Group T. After treatment, the number of Norank_f_Muribaculaceae in Group T shows an increasing trend. Additionally, Clostridium_sensu_stricto_1, abundant in Group M, exhibits resilience in harsh environments. After treatment, the number decreases significantly, suggesting that D. caudatum decoction improves the survival environment of intestinal flora to some extent. On the other hand, the LEfSe multi-level species hierarchy tree analysis in Figure 6B indicates 44 species with significant differences between groups. Norank_f_muribaculaceae, Clostridium_sensu_stricto_1, and Romboutsia were found to be abundant in Group C, Group M, and Group T, respectively.
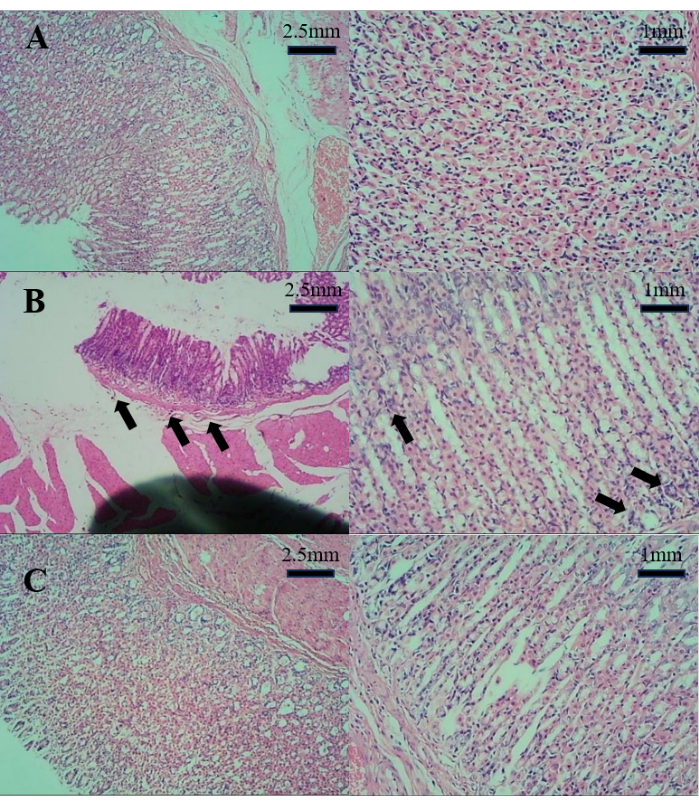
Figure 1: The results of the pathological section of the stomach wall. (A) Pathological section of the stomach in Group C. (B) Pathological section of the stomach in Group M. (C) Pathological section of the stomach in Group T. Please click here to view a larger version of this figure.
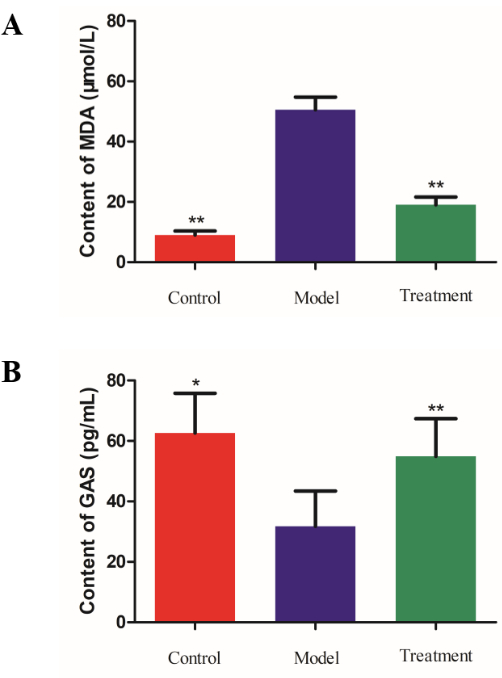
Figure 2: The contents of serum gastrointestinal hormones. (A) Bar analysis chart of MDA (malondialdehyde) determination results. (B) Bar analysis chart of GAS determination results. *P < 0.05, **P < 0.01. Please click here to view a larger version of this figure.
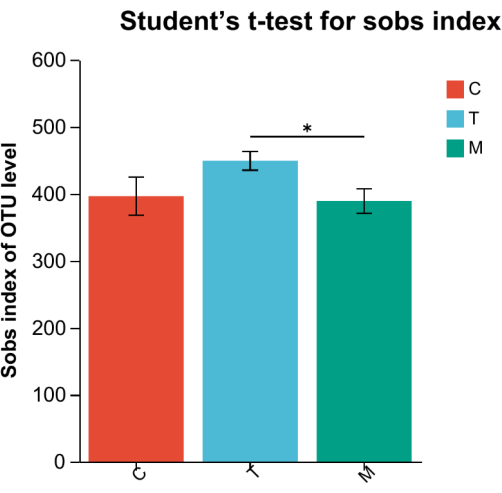
Figure 3: Column gram of diversity index T. Red bar represents Group C, blue represents Group T, while green represents Group M.*P < 0.05, **P < 0.01. Please click here to view a larger version of this figure.
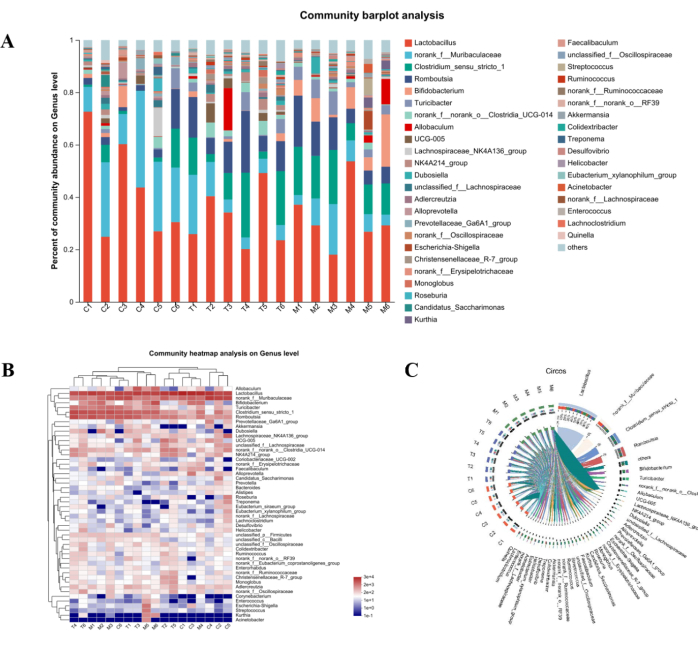
Figure 4: Community composition analysis. (A) Community histogram analysis. (B) Community heatmap analysis on the level of Genus. (C) Circos diagram analysis of relationship between samples and species. Please click here to view a larger version of this figure.
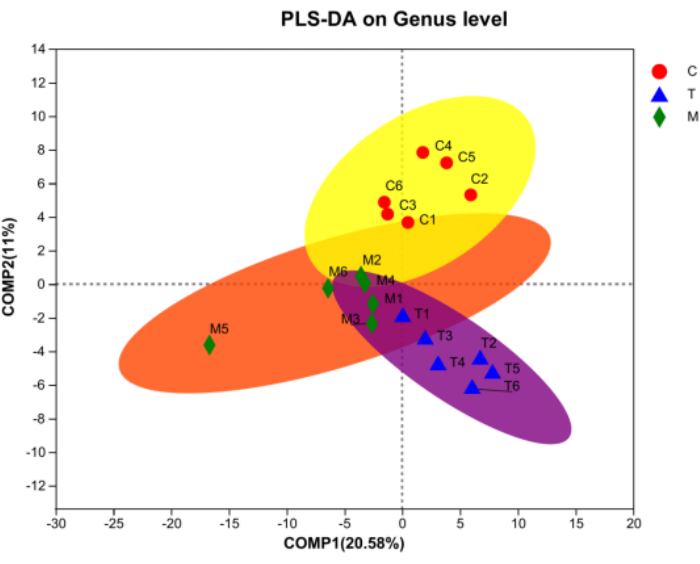
Figure 5: PLS-DA analysis on the level of Genus. Red represents Group C, blue represents Group T, and green represents Group M. Please click here to view a larger version of this figure.
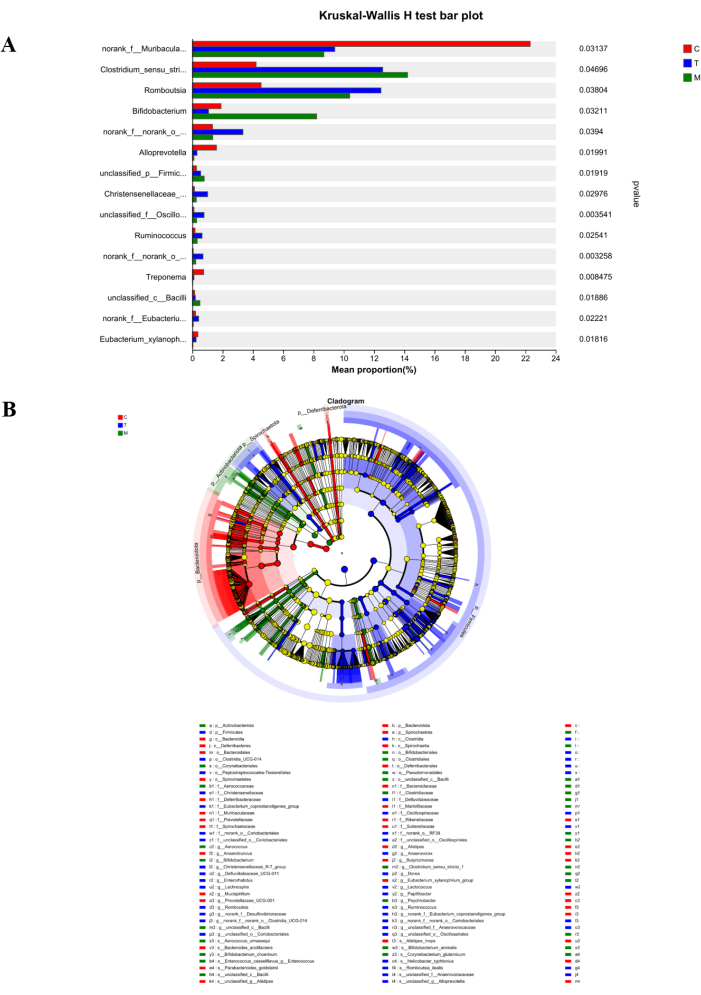
Figure 6: Species difference analysis. (A) Kruskal-Wallis H test bar plot. (B) Tree map of LefSe multi-level species. Red represents Group C, blue represents Group T, and green represents Group M. Please click here to view a larger version of this figure.
| Group | Mean value | Standard deviation |
| C | 3.5201 | 0.63641 |
| M | 3.2755 | 0.28494 |
| T | 3.5388 | 0.29945 |
Table 1: Alpha diversity index results.
List of Materials
| Alpha diversity analysis | Mothur 1.30.2 | ||
| AxyPrep deoxyribonucleic acid (DNA) gel extraction kit | Axygen Biosciences | AP-GX-50 | |
| Beta diversity analysis | Qiime 1.9.1 | ||
| Cryogenic refrigerator | Forma-86C ULT freezer | ||
| Desmodium caudatum | The Traditional Chinese Medicine Hospital of Beichuan Qiang Autonomous County | ||
| E.Z.N.A. soil kit | Omega Bio-tek | D5625-01 | |
| Illumina MiSeq Platform | Illumina Miseq PE300/NovaSeq PE250 | ||
| Multiskan Spectrum | spectraMax i3 | ||
| OTU clustering | Uparse 7.0.1090 | ||
| OTU statistics | Usearch 7.0 | ||
| PCR instrument | TransGen AP221-02 | ||
| PCR instrument | ABI GeneAmp 9700 | ||
| QuantiFluor-ST double-stranded DNA (dsDNA) system | Promega Corp. | ||
| Sequence classification annotation | RDP Classifier 2.11 | ||
| Sodium salicylate | Sichuan Xilong Chemical Co., Ltd | 54-21-7 | |
| SPF rats | Chengdu Dashuo Experimental Animal Co., Ltd | ||
| SPSS 18.0 | IBM |
Lab Prep
In order to preliminarily explore the effects of Desmodium caudatum on gastritis and intestinal flora in rats, a chronic gastritis rat model was established using the classic sodium salicylate method. Eighteen SPF rats were divided into three groups: the control group (Group C), the model group (Group M), and the treatment group (Group T). Pathological sections of the gastric wall were taken from rats in each group. Furthermore, the concentrations of gastrin and malondialdehyde in the serum of rats in each group were determined by ELISA. Additionally, the effects of D. caudatum on the intestinal flora of rats with gastritis were explored through a detailed comparison of gut bacterial communities in the three groups, employing Illumina-based 16S rRNA gene sequencing. The results indicated that D. caudatum decoction could reduce the malondialdehyde content and increase the gastrin content. Moreover, D. caudatum decoction was found to enhance the diversity and abundance of intestinal flora, exerting a positive impact on the treatment of gastritis by regulating and restoring the intestinal flora.
In order to preliminarily explore the effects of Desmodium caudatum on gastritis and intestinal flora in rats, a chronic gastritis rat model was established using the classic sodium salicylate method. Eighteen SPF rats were divided into three groups: the control group (Group C), the model group (Group M), and the treatment group (Group T). Pathological sections of the gastric wall were taken from rats in each group. Furthermore, the concentrations of gastrin and malondialdehyde in the serum of rats in each group were determined by ELISA. Additionally, the effects of D. caudatum on the intestinal flora of rats with gastritis were explored through a detailed comparison of gut bacterial communities in the three groups, employing Illumina-based 16S rRNA gene sequencing. The results indicated that D. caudatum decoction could reduce the malondialdehyde content and increase the gastrin content. Moreover, D. caudatum decoction was found to enhance the diversity and abundance of intestinal flora, exerting a positive impact on the treatment of gastritis by regulating and restoring the intestinal flora.
Procedura
In order to preliminarily explore the effects of Desmodium caudatum on gastritis and intestinal flora in rats, a chronic gastritis rat model was established using the classic sodium salicylate method. Eighteen SPF rats were divided into three groups: the control group (Group C), the model group (Group M), and the treatment group (Group T). Pathological sections of the gastric wall were taken from rats in each group. Furthermore, the concentrations of gastrin and malondialdehyde in the serum of rats in each group were determined by ELISA. Additionally, the effects of D. caudatum on the intestinal flora of rats with gastritis were explored through a detailed comparison of gut bacterial communities in the three groups, employing Illumina-based 16S rRNA gene sequencing. The results indicated that D. caudatum decoction could reduce the malondialdehyde content and increase the gastrin content. Moreover, D. caudatum decoction was found to enhance the diversity and abundance of intestinal flora, exerting a positive impact on the treatment of gastritis by regulating and restoring the intestinal flora.
