Isolation and Biophysical Study of Fruit Cuticles
Summary
Aerial plant organs are protected by the cuticle, a supramolecular biopolyester-wax assembly. We present protocols to monitor selective removal of epi- and intracuticular waxes from tomato fruit cuticles on molecular and micro scales by solid-state NMR and atomic force microscopy, respectively, and to assess the cross-linking capacity of engineered cuticular biopolyesters.
Abstract
The cuticle, a hydrophobic protective layer on the aerial parts of terrestrial plants, functions as a versatile defensive barrier to various biotic and abiotic stresses and also regulates water flow from the external environment.1 A biopolyester (cutin) and long-chain fatty acids (waxes) form the principal structural framework of the cuticle; the functional integrity of the cuticular layer depends on the outer ‘epicuticular’ layer as well as the blend consisting of the cutin biopolymer and ‘intracuticular’ waxes.2 Herein, we describe a comprehensive protocol to extract waxes exhaustively from commercial tomato (Solanum lycopersicum) fruit cuticles or to remove epicuticular and intracuticular waxes sequentially and selectively from the cuticle composite. The method of Jetter and Schäffer (2001) was adapted for the stepwise extraction of epicuticular and intracuticular waxes from the fruit cuticle.3,4 To monitor the process of sequential wax removal, solid-state cross-polarization magic-angle-spinning (CPMAS) 13C NMR spectroscopy was used in parallel with atomic force microscopy (AFM), providing molecular-level structural profiles of the bulk materials complemented by information on the microscale topography and roughness of the cuticular surfaces. To evaluate the cross-linking capabilities of dewaxed cuticles from cultivated wild-type and single-gene mutant tomato fruits, MAS 13C NMR was used to compare the relative proportions of oxygenated aliphatic (CHO and CH2O) chemical moieties.
Exhaustive dewaxing by stepwise Soxhlet extraction with a panel of solvents of varying polarity provides an effective means to isolate wax moieties based on the hydrophobic characteristics of their aliphatic and aromatic constituents, while preserving the chemical structure of the cutin biopolyester. The mechanical extraction of epicuticular waxes and selective removal of intracuticular waxes, when monitored by complementary physical methodologies, provides an unprecedented means to investigate the cuticle assembly: this approach reveals the supramolecular organization and structural integration of various types of waxes, the architecture of the cutin-wax matrix, and the chemical composition of each constituent. In addition, solid-state 13C NMR reveals differences in the relative numbers of CHO and CH2O chemical moieties for wild-type and mutant red ripe tomato fruits. The NMR techniques offer exceptional tools to fingerprint the molecular structure of cuticular materials that are insoluble, amorphous, and chemically heterogeneous. As a noninvasive surface-selective imaging technique, AFM furnishes an effective and direct means to probe the structural organization of the cuticular assembly on the nm-μm length scale.
Protocol
1. Enzymatic Isolation of Tomato Cuticles5
- Place several commercial or cultivated tomatoes in a bowl. Peel the skin from the fruits in large sections; discard the inner pericarp tissue. Wash the tomato skins with deionized water and preserve them under water in a beaker.
- Prepare a 50 mM pH 4.0 sodium acetate buffer (at 31 °C) by putting 1.22 g of sodium acetate trihydrate (Mr. 136.08 g/mol) and 2.34 ml glacial acetic acid (17.485 M) in a beaker, adding 200 ml of deionized water, and then adjusting the pH to 4.0 at 31 °C. Prepare a mixture containing 4 ml of pectinase (EC 3.2.1.15; 10 U ml-1, TCI America), 0.2 g of cellulase (EC 232.734.4; 1.3 units/mg solid, Sigma Aldrich), and 13 mg NaN3, and then add 196 ml of sodium acetate buffer to the enzyme mixture to obtain 200 ml of the final enzyme cocktail.5 Completely immerse the peeled tomato skin in the enzyme cocktail and incubate at 31 °C for 24 hours with constant shaking (G24 Environmental Incubator Shaker; New Brunswick Scientific Co.).
- Collect the tomato skins using a kitchen strainer or Büchner funnel and wash them with deionized water. Thereafter, place them in a vacuum oven at room temperature for an hour. Preserve the dried tomato skins in a labeled and capped bottle for subsequent dewaxing procedures.
2. Exhaustive Dewaxing by Soxhlet Extraction6
- The equipment used for exhaustive dewaxing consists of a heat source (heating mantle and Variac controller), round bottom flask for a solvent reservoir, Soxhlet extractor, sintered-glass thimble or disposable extraction thimble, anti-bumping chips and a condenser (see Fig. 1). Note that the narrow siphon arm (parts 6 and 7 in Fig. 1) is very delicate and prone to breakage, requiring careful handling.
- Put 0.5-1 g of tomato skin (obtained in step 1.) in a mortar, and grind the sample to a coarse powder with a pestle unless the samples are to be used for AFM measurements (Section 5). Fill a sintered-glass or disposable thimble approximately halfway with the sample and use tweezers to place it carefully at the base of the extraction column.
- Attach the condenser and wrap it with aluminum foil. Heat the methanol solvent (ACS grade) in the presence of a few anti-bumping chips until it boils gently and refluxes on the walls of the flask. Cover the solvent reservoir with both glass wool and aluminum foil. Check the process for an hour, adjusting the Variac voltage so that the reservoir accumulates about one drop per second and siphoning occurs within the Soxhlet apparatus when the thimble is full.
- Continue the extraction process for 12 h, then lower the heating mantle and allow the apparatus to cool down. Remove the extractor and reservoir as a single unit to dispose of the solvent. Use tweezers to raise the thimble to just below the neck of the extraction column, drain excess solvent from it, and place the thimble on a clean surface. Tilt the extraction column to allow siphoning into the flask below. Disconnect the flask and pour the waste into a labeled waste solvent receptacle.
- Repeat steps 2.3 and 2.4 for successive solvents of progressively diminishing polarity, e.g., chloroform and hexane for 12 h in each case.
- Allow the tomato cutin sample to dry inside the thimble, either by blowing a stream of nitrogen gas over it or by placing it in a vacuum oven at room temperature. Finally, measure the mass of the dry sample and store it at room temperature in a screw-top jar sealed with parafilm.
3. Selective Isolation of Epicuticular and Intracuticular Waxes3,4
- First, wash whole tomatoes (a separate batch of tomatoes from those described in 1.) with distilled water. Dry them with paper towels and Kimwipes, and place them stem downward on a piece of aluminum foil.
- Paint the whole tomatoes with 120% (w/w, mass ratio) gum arabic aqueous solution in a top down fashion and allow about an hour for the gum arabic to dry on the fruit’s skin, leaving a thin film. Remove this film using tweezers, being careful not to puncture the tomato skin. Repeat the procedure once more.
- Add the films to vials containing 1:1 (v/v) chloroform-water and mix for three minutes. After vigorous agitation and phase separation, pipette out the chloroform fractions and evaporate them in separate uncovered vials, leaving epicuticular wax. The tomatoes will remain physically intact. Dip them into chloroform for two minutes at room temperature and collect the intracuticular wax after evaporating the solvent. Now, peel the tomatoes and treat them enzymatically (with cellulase and pectinase in sodium acetate buffer) to remove cellulose and pectin, respectively (as described in 1).
- If desired, perform exhaustive dewaxing on these enzymatically isolated cuticles by Soxhlet extraction (see step 2), using three solvents of varying polarity (methanol, chloroform, and hexane, respectively).
4. Molecular Characterization of Tomato Fruit Cutin by Cross-polarization Magic-angle Spinning Solid-state Nuclear Magnetic Resonance (CPMAS ssNMR)6
- Place 4-6 mg of fully dewaxed tomato cuticles (cutins) in a 1.6 mm fastMAS zirconia rotor using the vendor-supplied packing tool. (Either ground dewaxed tomato cuticles or very small pieces of partially dewaxed cuticles are suitable.) Ensure that the sample is packed uniformly, but not too tightly, in the rotor. After putting on the top cap, paint half of the cap with a black-ink marker pen to facilitate measurements of the spin rate.
- Adjust the shimming of the NMR spectrometer for minimum spectral line width at half-height and calibrate the proton (1H) and carbon (13C) 90 ° pulse widths using a standard compound such as adamantane.
- Using glycine or glutamine as model compounds, obtain maximum signal intensity by optimizing all parameters (Hartmann-Hahn matching power levels, 1H – 13C contact time, heteronuclear 1H decoupling strength) of the cross-polarization magic-angle spinning (CPMAS) experiment. For spectra acquired at a 1H frequency of 600 MHz, recommended conditions include 10 kHz or 15 kHz spinning frequency, a 3-sec delay between acquisitions, and SPINAL heteronuclear proton decoupling7 at a magnetic field strength equivalent to a frequency of 185 kHz.
- Insert the cutin-packed rotor into the probe. Then, place the probe into the magnet. Increase the spinning speed up to 10 kHz gradually to check for good sample packing and rotor stability. Verify the final spinning stability of the rotor to within ±20 Hz.
- Adjust the tuning and matching capacitors of the probe iteratively to achieve minimum power reflection at both 1H and 13C NMR frequencies. Set the experimental temperature to 25 °C (or room temperature).
- Start the pre-optimized CPMAS experiment corresponding to the Hartmann-Hahn matching condition determined at the 10 kHz spinning frequency.
- Acquire 4096 transients, condition the spectrum with exponential (Lorentzian) line broadening of 50-100 Hz, and do a Fourier transform to generate an NMR spectrum of signal intensity vs. chemical shielding (ppm).
- Reference the 13C chemical shifts externally using adamantane set at 38.4 ppm (-CH2– group)8 as a standard.
- Increase the rotor spinning frequency to 15 kHz and repeat the CPMAS measurement (steps 4.6-4.8) corresponding to the Hartmann-Hahn matching condition determined at this latter spinning frequency.
- Repeat the CPMAS experiments (steps 4.1-4.9) with natural (waxy) and partially dewaxed fruit cuticle samples.
5. Probing the Tomato Cuticle Surface with Atomic Force Microscopy (Digital Instruments Nanoscope IIIa; procedures will vary slightly among microscopes)6
- Turn on the scanning probe microscope (SPM) (Fig. 2) and make sure that the microscope mode toggle switch is set to the contact Atomic Force Microscopy (AFM) mode.
- Manually raise the SPM head by turning its two user-adjustable front knobs. Detach the AFM tipholder from the SPM head by turning the clamping screw at the back of the head.
- Use tweezers to remove the existing AFM cantilever from the tipholder, then carefully grab a new silicon nitride cantilever (AFM probe) from its package and install it in place of the old cantilever. Use a light microscope to verify that the newly installed AFM cantilever is not broken.
- Attach the tomato cuticle sample (a section of partially dewaxed tomato cuticle ~10 mm x 10 mm) to a stainless steel disk (sample puck) with double-sided tape. Use a light microscope to verify that the cuticle surface remains flat and smooth after placement of the sample on the puck.
- Place the puck with the tomato cuticle sample onto the magnetic region at the top of the SPM scanner.
- Set the front two manual adjustment screws of the scanner high by turning the knobs; set the motorized back adjustment screw to approximately the same level as the other two front screws. Make sure that all three screws are set high enough to avoid breaking the AFM tip when placing the tipholder into the SPM head.
- Reinsert the tipholder into the SPM head and secure it by tightening the clamping screw at the back of the head.
- After turning on the laser, align the laser spot on the AFM cantilever using the central (y) and right (x) laser adjustment knobs on the top of the head. Monitor the reflected laser beam on a piece of paper to position the laser spot exactly at the end of the AFM cantilever.
- Adjust the position of the movable mirror iteratively using the photodetector adjustment knobs to achieve maximum signal (SUM signal), thus ensuring that the reflected laser beam is being received evenly by the four quadrants of the photodetector.
- Lower the AFM tip by retracting the manual adjustment front screws and the motorized back adjustment screw of the SPM scanner, visually monitoring the approach of the AFM tip toward the sample surface with an inverted microscope. Make sure that all three screws are at the same level to avoid artifacts from imaging a tilted sample. Bring the tip toward the sample, but not so close that the tip touches or breaks through the sample surface.
- At this point, the laser light reflected off the AFM cantilever will bounce off the movable mirror to the photodetector. For the contact AFM mode with a silicon nitride AFM tip, set the OUTPUT signal (Vertical Deflection) voltage to -2 V for a 0 V Setpoint and the DIFF Signal (vertical/horizontal difference) to 0 V by adjusting the position of the mirror.
- Using the Nanoscope software, click on the microscope icon and select the appropriate profile (Contact mode AFM).
- Using the “Scan Controls Settings” panel, set the scan rate and scan size e.g., 10 micron scan size and 2 Hz scan rate.
- Allow the AFM tip to engage the sample surface by clicking the engage-tip icon. By controlling the motorized back screw of the SPM base, the program will now lower the tip until it engages the sample surface. The scanning process will automatically start once the tip has engaged successfully.
- Monitor the scanning process using both image and scope modes of the software, adjusting parameters such as setpoint, integral gain, proportional gain, scan size, scan rate, lines and samples/line iteratively to achieve the highest resolution images. Start scanning with a large z-axis range (data scale); then carefully reduce the data scale value to observe the best contrast of surface features on the image. Minimize the occurrence of white areas in the scanned image, which indicate that the height of the surface features exceeds the available z-axis range.
- Capture the image to save the data file; then process the data using flattening functions to remove image artifacts due to vertical (Z) scanner drift, image bows, and vertical offset between scan lines;9 finally calculate the average roughness. After saving the data, retract the AFM tip by reversing the action of the computer-controlled screw motor that was used to engage it. Image processing may be performed in offline image analysis mode and/or using a different computer.
- Use the front bottom gray metal knobs of the tip holder to change the x and y positions of the scanning area; then repeat the measurement at five sample locations to check reproducibility, replacing the AFM cantilever if the image quality deteriorates.
6. Representative Results
Chemical shift analysis of the CPMAS 13C NMR spectra (Fig. 3) identified the major functional groups present in the exhaustively dewaxed tomato cuticle (cutin). The key carbon moieties in the cutin biopolyester were found to be long-chain aliphatics (0-45 ppm), oxygenated aliphatics (45-110 ppm), multiply-bonded and aromatics (110-160 ppm), and carbonyls (160-220 ppm). The oxygenated alkyl moieties (CHO + CH2O) play a crucial role in establishing covalent connections between the monomeric units of the cutin biopolymer, thereby forming the molecular architecture of the cutin matrix. Differences in relative peak areas observed in the spectral region between 45 and 100 ppm suggest that the mutant cutin has a relatively large proportion of cross-link forming CHO structural moieties compared to the wild-type cutin; careful quantitative measurements using direct polarization (DPMAS) NMR5 methods support this inference (data not shown).
CPMAS 13C NMR spectra also showed a progressively diminishing wax peak at 31 ppm (Fig. 4), indicating the sequential removal of epi- and intracuticular waxes from the cutin-wax composite while retaining the principal chemical architecture of the cutin biopolymer. Parallel AFM image analysis (Fig. 5) revealed surface irregularities due to the stepwise extraction of epi- and intracuticular waxes from the fruit cuticle, signifying alterations in the organization of the cuticular assembly.
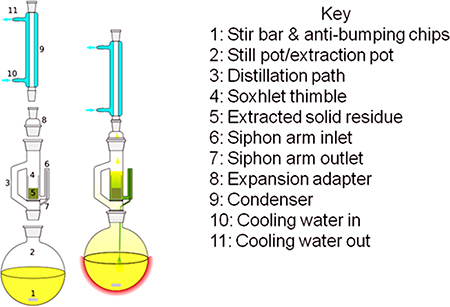
Figure 1. Soxhlet extractor (image source: Wikipedia).
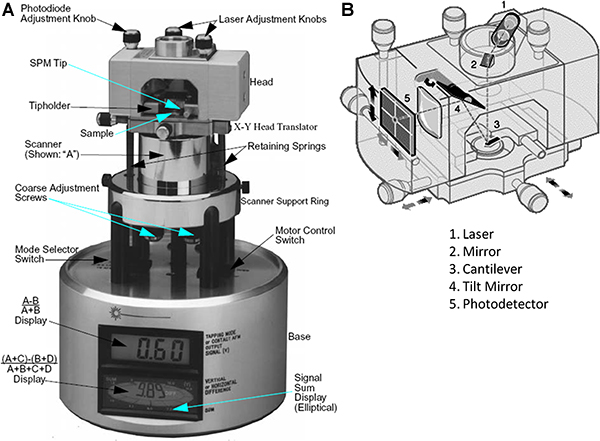
Figure 2. A) Scanning probe microscope. B) SPM head (adapted from the AFM training manual provided by Digital Instruments).
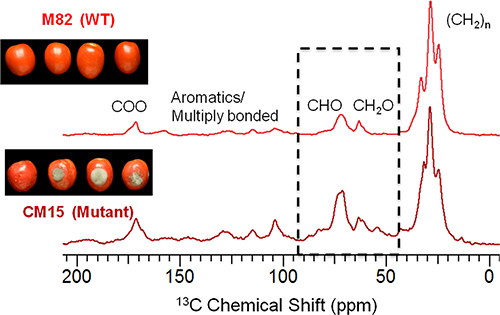
Figure 3. 150 MHz CPMAS 13C NMR spectra of exhaustively dewaxed red ripe tomato fruit cuticles (cutins) from wild-type (M82) and mutant (CM15) show resonances with chemical shifts corresponding to the functional groups of a cross-linked hydroxyfatty acid-based polyester and also some multiply bonded moieties. All spectra were acquired with a 10 kHz spinning frequency.
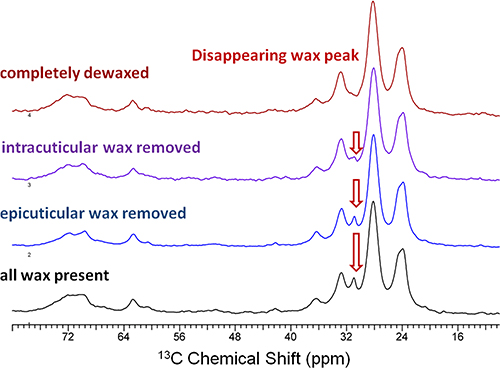
Figure 4. 150 MHz 13CCPMAS NMR spectra of commercial red ripe tomato fruit cuticles exhibit compositional changes upon sequential removal of epicuticular and intracuticular waxes by gum Arabic mechanical extraction and a two-minute chloroform dip, respectively. All spectra were acquired with a 15 kHz spinning frequency.
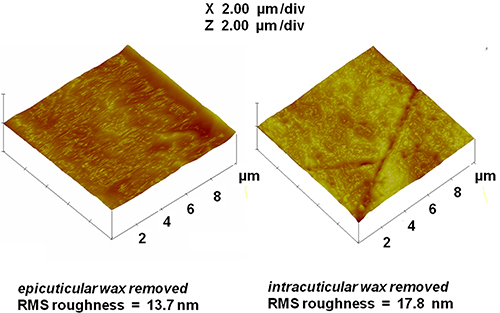
Figure 5. AFM images and roughness estimates for the partially dewaxed commercial tomato cuticles described in Figure 4 after the stepwise removal of epicuticular (left) and intracuticular (right) waxes.
Discussion
The protocols described herein allow for detailed molecular and microscale characterization of a complex intractable plant material without the need for destructive chemical breakdown. To investigate the blending of the cutin biopolyester with various lipids (waxes) that control the structural organization of the cuticular assembly,10 we conducted and monitored procedures for selective removal of epicuticular and intracuticular waxes from the heterogeneous cuticular blend. Solid-state 13C NMR was used to gauge extraction of wax molecular components, and atomic force microscopy served to examine concomitant changes in surface roughness.6,11 To compare the cross-linking capabilities of cutins from cultivated wild-type and single-gene mutant tomato fruits, solid-state 13C NMR was also used to estimate the relative numbers of CHO and CH2O chemical moieties.
A number of design features of this protocol are notable. As the wax materials encompass a wide range of lipids, treating the fruit cuticle with a series of solvents having divergent polarities is essential to achieve exhaustive dewaxing. In addition, dewaxing time can vary from 8 hours to 24 hours depending on the nature of the cuticle samples. To extract epicuticular waxes consistently from the intact fruit cuticle, it is imperative to apply the adhesive coating uniformly to the surface.
Solid-state CPMAS 13C NMR12 is a rapid qualitative method for identifying various structural components of highly heterogeneous and insoluble plant biopolymers while preserving their native physical characteristics;13 traditional solution-state NMR can also be used to characterize the extracted wax mixtures. If quantitative estimation of functional groups is desired for the intact plant polymers,5 high-fidelity direct-polarization magic-angle spinning (DPMAS) 13C NMR5,14 should be used as a complementary method. Accurate quantitation of the functional groups requires careful optimization of recycle times, excitation pulse lengths, and the strength of heteronuclear decoupling.15 The heteronuclear decoupling can be set for a 1H field strength ranging from 50 kHz to 185 kHz by using the TPPM16 or SPINAL7 methodologies. In addition to these parameters, the sensitivity of CPMAS measurements depends on the spin-lock time and Hartmann-Hahn matching condition.15 In place of traditional CPMAS, a ramped-amplitude CP (RAMP-CP) technique can be implemented to maximize the cross-polarization efficiency by varying the 1H amplitude linearly (~20-50 %) or tangentially while keeping the amplitude of 13C field strength constant during the spin-lock period (or vice versa).17,18 Carrying out the CPMAS measurements at a minimum of two different rotor-spinning frequencies is imperative to distinguish spinning sidebands from the main spectral peaks.
Concurrent AFM measurements conducted in contact mode enable direct imaging of the cuticle surface condition with high scanning speeds and high resolution,19 for instance during sequential removal of waxy constituents. Operating AFM in tapping (non-contact) mode can be used as an alternative for surface characterization of delicate “soft” plant materials, avoiding possible damage due to lateral (shear) forces and scraping of the sample surface.5,20 In either case, sequential acquisition of multiple images of the same spot on the surface serves to identify any surface damage due to “probe-surface interactions” in AFM measurements.6,21 For optimal reproducibility, AFM probes with spring constants suitable for soft cuticular surfaces should be used, and constancy of temperature and humidity should be maintained.6,15,20 Whereas solid-state NMR offers a molecular profile of ensemble average (bulk) properties in tomato fruit cuticles, atomic force imaging provides a complementary noninvasive probe 22,23 for tracking the surface topography of these exquisitely complex macromolecular assemblies.1,2
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by U.S. National Science Foundation grants # MCB-0741914 and MCB-0843627; additional infrastructural support was provided at The City College of New York by National Institutes of Health 2 G12 RR03060-26 from the National Center for Research Resources. We gratefully acknowledge the J.K.C. Rose group in the Cornell University Plant Biology Department for providing M82 (wild type) and CM15 (mutant) tomato cuticles. We thank Dr. Spyros Monastiriotis from the CCNY Chemical Engineering group of Prof. Alexander Couzis for his generous help with the AFM experiments. We thank Ms. Lauren Gohara for graphical design support.
Materials
| Name of the reagent | Company | Catalog no. | Comments |
| Sodium acetate trihydrate | Sigma-Aldrich | S8625-500G | |
| Pectinase | TCI America | P0026 | EC 3.2.1.15; 10 U ml-1, store in refrigerator |
| Cellulase | Sigma-Aldrich | C1184-100KU | EC232.734.4; 1.3 units/mg, store in refrigerator |
| Glacial Acetic acid | Sigma-Aldrich | A9967 | |
| Sodium azide | Sigma-Aldrich | S2002-100G | Extremely hazardous |
| Incubator/shaker | New Brunswick Scientific Co. | Model No.G24 | MFG No.M1036-000G |
| Vacuum Oven | Precision Scientific | 31566 | |
| Variac Controller | |||
| Sintered glass thimble (85 mm/25mm) | VWR | 89056 | |
| Disposable extraction thimble ( 80 mm/ 25 mm) | VWR | 28320 | |
| Methanol | VWR | EMD-MX0485-7 | |
| Glass wool | VWR | RK20789 | |
| Aluminum foil | Fisher | 01-213-100 | |
| Tweezers | VWR | 82027-452 | |
| Chloroform | VWR | EM-CX1050-1 | |
| Hexane | Fisher Scientific | H302-4 | |
| Nitrogen gas | |||
| Parafilm | VWR | 52858 | |
| Paper towels | VWR | 89002-984 | |
| Kim wipes | VWR | 21905-026 | |
| Gum arabic | Sigma | G9752 | |
| 1.6 mm fastMAS zirconia rotor | Varian (Agilent) | ||
| NMR spectrometer | Varian 600 NMRS | standard bore magnet | |
| Glycine | Sigma-Aldrich | 50046 | Model compound for CPMAS |
| Glutamine | Sigma-Aldrich | 49419 | Model compound for CPMAS |
| Adamantane | Sigma-Aldrich | 100277 | To calibrate 90° pulse in NMR |
| Multimode Scanning Probe Microscope (Nanoscope IIIA) | Digital Instruments (Bruker AXS) | ||
| Nanoscope software | Digital Instruments (Bruker AXS) | Version 5.30r3sr3 (2005) | |
| AFM probe (Nonconductive silicon nitride tip) | Veeco (Bruker AXS) | Model NP-20 | |
| Light microscope | Digital Instruments | ||
| Magnetic puck | Digital Instruments | ||
| Double sided tape | VWR | ||
| Fruit Peeler | |||
| Büchner funnel | VWR | 89038 |
References
- Dom#237;nguez, E., Heredia-Guerrero, J. A., Heredia, A. The biophysical design of plant cuticles: an overview. New Phytologist. 189, 938-949 (2011).
- Bargel, H., Koch, K., Cerman, Z., Neinhuis, C. Structure-function relationships of the plant cuticle and cuticular waxes – a smart material?. Functional Plant Biol. 33, 893-910 (2006).
- Jetter, R., Schäffer, S. Chemical Composition of the Prunus laurocerasus Leaf Surface. Dynamic Changes of the Epicuticular Wax Film during Leaf Development. Plant Physiol. 126, 1725-1737 (2001).
- Vogg, G. Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J. Exper. Botany. 55, 1401-1410 (2004).
- Isaacson, T. Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 60, 363-377 (2009).
- Round, A. N. The Influence of Water on the Nanomechanical Behavior of the Plant Biopolyester Cutin as Studied by AFM and Solid-State NMR. Biophysical J. 79, 2761-2767 (2000).
- Fung, B. M., Khitrin, A. K., Ermolaev, K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids. J. Magn. Reson. 142, 97-101 (2000).
- Morcombe, C. R., Zilm, K. W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162, 479-486 (2003).
- Fang, S. J., Haplepete, S., Chen, W., Helms, C. R., Edwards, H. Analyzing atomic force microscopy images using spectral methods. J. App. Phys. 82, 5891-5898 (1997).
- Pollard, M., Beisson, F., Li, Y., Ohlrogge, J. B. Building lipid barriers: biosynthesis of cutin and suberin. Trends. Plant Sci. 13, 236-246 (2008).
- Stark, R. E. NMR studies of structure and dynamics in fruit cuticle polyesters. Solid State Nucl. Mag. 16, 37-45 (2000).
- Schaefer, J., Stejskal, E. O. Carbon-13 nuclear magnetic resonance of polymers spinning at the magic angle. J. Amer. Chem. Soc. 98, 1031-1032 (1976).
- Sachleben, J. R., Chefetz, B., Deshmukh, A., Hatcher, P. G. Solid-State NMR Characterization of Pyrene-Cuticular Matter Interactions. Envir. Sci. & Tech. 38, 4369-4376 (2004).
- Zlotnik-Mazori, T., Stark, R. E. Nuclear magnetic resonance studies of cutin, an insoluble plant polyester. Macromolecules. 21, 2412-2417 (1988).
- Stark, R. E., Garbow, J. R. Nuclear magnetic resonance relaxation studies of plant polyester dynamics. 2. Suberized potato cell wall. Macromolecules. 25, 149-154 (1992).
- Bennett, A. E., Rienstra, C. M., Auger, M., Lakshmi, K. V., Griffin, R. G. Heteronuclear Decoupling in Rotating Solids. J. Chem. Phys. 103, 6951-6958 (1995).
- Metz, G., Wu, X. L., Smith, S. O. Ramped-Amplitude Cross-Polarization in Magic-Angle-Spinning NMR. J. Magn. Reson. Ser. A. 110, 219-227 (1994).
- Peersen, O. B., Wu, X. L., Smith, S. O. Enhancement of CP-MAS Signals by Variable-Amplitude Cross-Polarization – Compensation for Inhomogeneous B-1 Fields. J. Magn. Reson. Ser. A. 106, 127-131 (1994).
- lessandrini, A., Facci, P. AFM: a versatile tool in biophysics. Measurement Sci. & Tech. 16, 10-1088 (2005).
- Benítez, J. J., Matas, A. J., Heredia, A. Molecular characterization of the plant biopolyester cutin by AFM and spectroscopic techniques. J. Struct. Biol. 147, 179-184 (2004).
- Koch, K., Neinhuis, C., Ensikat, H. J., Barthlott, W. Self assembly of epicuticular waxes on living plant surfaces imaged by atomic force microscopy (AFM). J. Exper. Bot. 55, 711-718 (2004).
- Muller, D. J. AFM: A Nanotool in Membrane Biology. Biochemistry. 47, 7986-7998 (2008).
- Last, J. A., Russell, P., Nealey, P. F., Murphy, C. J. The Applications of Atomic Force Microscopy to Vision Science. Investigative Ophthalmology & Visual Science. 51, 6083-6094 (2010).

