Laser-guided Neuronal Tracing In Brain Explants
Summary
We describe a technique to label neurons and their processes via anterograde or retrograde tracer injections into brain nuclei using an in vitro preparation. We modified an existing method of in vitro tracer electroporation by taking advantage of fluorescently labeled mouse mutants and basic optical equipment in order to increase labeling accuracy.
Abstract
We present a technique which combines an in vitro tracer injection protocol, which uses a series of electrical and pressure pulses to increase dye uptake through electroporation in brain explants with targeted laser illumination and matching filter goggles during the procedure. The described technique of in vitro electroporation by itself yields relatively good visual control for targetting certain areas of the brain. By combining it with laser excitation of fluorescent genetic markers and their read-out through band-passing filter goggles, which can pick up the emissions of the genetically labeled cells/nuclei and the fluorescent tracing dye, a researcher can substantially increase the accuracy of injections by finding the area of interest and controlling for the dye-spread/uptake in the injection area much more efficiently. In addition, the laser illumination technique allows to study the functionality of a given neurocircuit by providing information about the type of neurons projecting to a certain area in cases where the GFP expression is linked to the type of transmitter expressed by a subpopulation of neurons.
Introduction
In order to define a certain neuronal (micro)circuit, one must start by finding the various participants of said circuit, and their connection pattern. Ever since Waller's publication about neurofiber tracing through lesioning1 a large variety of neuroanatomical tracing techniques has been established. Some of these techniques can be applied in fixed tissue post mortem2-4, others rely on the active transport of the dye in live neurons, as discovered in 19715-6. The latter can be further subdivided in two groups discriminating between methods taking advantage of active retrograde (from the injected area to the source of a given projection, i.e. the somata of neurons that are projecting to said area) and active anterograde (from the injected area to the target of a given projection, i.e. the axonal projections and the axonal terminals of the labeled neurons) transport. Also, in some cases tracer material is injected into live animals which then survive the injection by several days or weeks (in vivo tracer injections), while in other cases explanted brains are injected and incubate for several hours after the injection in artificial cerebrospinal fluid (in vitro tracer injections).
In this protocol we modified an existing in vitro electroporation technique7-8 to label neuronal somata and processes in anterograde and retrograde tracing experiments using choleratoxin subunit-b and tetramethylrhodamine dextran as the tracing substances. The overall goal of this protocol is to provide neuroscientists with an efficient tool to trace neuronal connectivity patterns between different brain nuclei, while taking advantage of available transgenic mouse lines and basic optical equipment in order to increase targeting accuracy during tracer injections. Although the method of anterograde and retrograde tracing using choleratoxin and dextran amine and their respective fluorescently labeled conjugates is not new9-13 (as is the method of electroporation, e.g. Haas et al.14), the combination of tracer injections with electroporation in an in vitro preparation involving blocks of brain tissue is a more recent development7. Its main advantage over neuronal tracing techniques using the same type of tracer dyes in live animals is the increased labeling intensity, because of the higher efficiency with which the electroporated dye is being taken up by neurons. An additional advantage is the shortened incubation period (required for the dye transport) and its increased target accuracy during the tracer injection, because the experimenter has visual control over the target area of the injection. The latter also means that no expensive stereotactic equipment is required to find the nucleus or brain area of interest.
To additionally increase targeting accuracy, we took advantage of a transgenic mouse line, which expresses GFP in its glycinergic subpopulation of neurons15 and basic optical equipment consisting of a hand-held laser pointer of 405 nm wavelength and matching band-pass filtering goggles (450 – 700 nm). Thus, we achieved a significant further increase in targeting accuracy by identifying the injection area through its fluorescent signal and by providing a finer way to control for the dye spread within the injection area through the observation of the interaction between the indigenous GFP signal and the tracer fluorescence. Our technique also allows to uncover the functionality of a circuit along with its connectivity by identifying GFP-positive inhibitory neurons (or excitatory in other mouse lines) that were filled with the tracer.
In summary, we further enhanced a powerful neuroscientific tool to study the connectome of the vertebrate brain and assess the different neuroanatomical features of a given neurocircuit. By using transgenic mice along with inexpensive and widely available optical equipment we were able to significantly increase the targeting accuracy of our injections. Furthermore, the transgenic mice allowed us to identify the type of the traced connections, which helped uncovering the functionality of an inhibitory microcircuit in the auditory brainstem.
Protocol
1. Optical Genotyping
1. Optical Genotyping of Mouse Pups
- Check for expression of the respective fluorescent marker using a laser pointer of the appropriate excitation wavelength (405 nm in the experiments described here) and corresponding filter goggles blocking the excitation wavelength but passing the emission wavelength (450 – 700 nm in the experiments described here). Point the laser pointer at the back of the head or the spinal cord of the mouse pup (see Figure 1). Avoid shining the laser into the eyes and lengthy exposure of the skin to laser light.
2. Optical Genotyping of Older Animals
- Deeply anesthetize the mouse (aged p14 to p138 in the experiments described here) with an overdose of pentobarbital through an intraperitoneal injection (120 mg/kg bodyweight). Confirm proper anesthesia by checking the animal's reflexes (briefly pinch the tip of the ear or one of the hind legs to elicit the retraction reflex of the ear or leg).
- Note: Despite the use of an overdose of pentobarbital (120 mg/kg bodyweight) we refer to the injection procedure as "deep anesthesia" instead of euthanasia, because the animal is ideally still alive (i.e. its heart is still beating), when the surgery and the perfusion begin. This ensures a more efficient perfusion of the inner organs including the brain.
- Once the animal does not show any reflexes, carefully remove the skin overlying the skull (Figure 2A) by making an incision in the skin covering the back of the head and cutting to the midsection of the skull. Expose the uncovered skull to laser light and observe the fluorescence through the filter goggles as described above. If a positive GFP signal is observed, proceed to step 2, if not, sacrifice the animal through decapitation (second/ensuring form of euthanasia following our IACUC regulations).
Note: In even older mice ( >1 month) the fluorescence can be observed through the eyes of the animal.
2. Transcardial Perfusion and Brain Preparation
- Perfuse transcardially with ice-cold phosphate buffered saline (PBS; NaCl: 137 mM, KCl: 2.7 mM, KH2PO4: 1.76 mM, Na2HPO4: 10 mM) using a 30 G syringe needle to largely remove the blood from the animal.
- First, carefully open the rib cage with sharp scissors and then push the needle into the left ventricle of the heart. Start pumping the PBS into the heart and the aorta and immediately following this step open the right atrium of the heart with the scissors to allow for the blood to exit the body. Note: Perfusion takes 5 – 10 min depending on the size of the animal.
- After exsanguination through perfusion, decapitate the animal, secure its head with pin needles (or 30 G syringe needles) by piercing them through the eye sockets in a preparation dish and remove the brain from the skull.
- First, use sharp scissors to cut off the skin overlaying the skull: make a small incision in the back of the head, then cut along the midline on the dorsal side of the head, cut to the sides just caudally of the eyes of the animal and lastly cut in the caudal direction to completely remove the flaps of skin at the back of the head.
- Next, repeat the same cutting pattern for the skull itself, make sure to remove both cochleas in the process, because this will significantly simplify the removal of the brain stem in the end. After this procedure the caudal half of the brain should lie completely exposed.
- In the next step, use a sharp razor blade or a scalpel to cut through the whole forebrain at an angle of about 45° and about 2 mm rostral from the cerebellum. Scoop out the separated front half of the brain with a bent spatula.
- Use the spatula to elevate the caudal half of the brain at its rostral end and subsequently cut through the cranial nerves that are still connected to the brain stem on its ventral side. As an end result, the caudal half of the brain including the brainstem should fall freely off the rest of the prepared animal head.
- Flip the removed caudal half of the brain to expose its ventral side and cut along the coronal plane just rostral (for the anterograde injections) or just caudal (for the retrograde injections) of the ventrally located "bulbs" containing the trapezoid body (see step 2.3).
- Always use a sharp razor blade or a scalpel for the cutting procedure to minimize cell death caused by excessive mechanical stress on the tissue. As an end result, a brain explant spanning about 1 cm in length and containing the complete superior olivary complex along other brain regions should have been obtained.
Note: This protocol demonstrates the procedure for a tracer injection into the trapezoid body located in the auditory brainstem. Adapt the location and orientation of the cutting planes and injection sites as needed. If the fluorescing brain regions of interest lie close enough to the brain surface, so that they can be identified and injected without cutting the brain (see Figure 2B), then the coronal cutting step can be omitted.
- Identify the trapezoid body as two ventral prominent "bulbs" containing the ventral nucleus of the trapezoid body (VNTB).
Note: For further anatomical reference regarding the approximate stereotactic location of these cutting planes, see Franklin & Paxinos, 2008. Panel 78 of the atlas, Bregma ~ -5.7 mm shows the medial nucleus of the trapezoid body (=MNTB) labeled as "Tz". Panel 69 shows the ventral nucleus of the trapezoid body (VNTB) labeled as "MVPO" (medioventral periolivary nucleus)19.
3. Anterograde/Retrograde Tracing
Note: Perform all following procedures at RT (25° C).
- After cutting, place the brainstem explant in a preparation dish containing oxygenated (95% O2, 5% CO2) dissecting solution (NaCl: 125 mM, KCl: 2.5 mM, MgCl2: 1 mM, CaCl2: 0.1 mM, glucose: 25 mM, NaH2PO4: 1.25 mM, NaHCO3: 25 mM, ascorbic acid: 0.4 mM, myo-inositol: 3 mM, pyruvic acid: 2 mM), securing it with 30 G syringe needles. Ensure that the area of injection is facing upwards.
- Pull injection pipettes from borosilicate glass and fill them with one of the following three solutions: a) a 1.0 mg/ml solution of choleratoxin subunit-b conjugated to an Alexa fluorophore 555 in PBS12-13 (mainly used for retrograde transport), b) a 1%-dilution in 0.9% saline of dextran tetramethylrhodamine 555 (3,000 MW, mainly used for retrograde transport) or c) a 1%-dilution in 0.9% saline of 10,000 MW dextran-TRITC 555 (mainly used for anterograde transport).
- Ensure that injection pipette resistances range from 2.5 to 3.5 MOhm and that they are manufactured in 3 – 4 pulling cycles. For the experiments described here, achieve this by selecting a pre-programmed pulling algorithm on the pipette puller.
- Illuminate the brain explant using a statically mounted laser pointer of the appropriate wavelength (item No. 2 in Figure 3A; 405 nm wavelength in the experiments described here). Use the laser pointer as a guide for the injections, and aim the laser beam at the fluorescently labeled brain nucleus or cells of interest that will be injected.
- Find and observe the illuminated target area through a binocular microscope while wearing the compatible filter goggles (450 – 700 nm band-pass filter in the experiments described here). Insert the injection electrode via the micromanipulator into the area of interest.
- Inject the tracer substance. The injections consist of 2 – 10 pressure pulses at 15 psi for 50 msec each, using a pressure injection device, directed perpendicular into the region of interest (ROI) within the brain section. Administer each pressure pulse at intervals of 10 – 15 sec to allow for the dye to spread.
- In the case of the tetramethylrhodamine (TRITC) injections, additionally stimulate electrically to enhance the dye uptake through electroporation with the electrode placed in the brain area of interest (MNTB and VNTB in the experiments described here).
- Use 8 TTL pulses of 8 Volts for 50 msec with 50 msec interpulse intervals, driven by a stimulator and amplified to 55 V with a stimulation isolation unit (items No. 7 and 9 in Figure 3B). Use 10 – 20 repetitions of this protocol, administered over several minutes7,8. Make sure to ground the electrode properly – the grounding wire should be connected to the bath solution!
- Check for success of the injection. Since a fluorescent tracer with a longer emission wavelength is also emitting a fluorescent signal upon laser excitation with shorter wavelengths, visually check for the dye uptake/spread in the injected target area while illuminating the area with the laser and observing through the filter goggles.
4. Incubation
- After the injection, incubate the brainstems in oxygenated artificial cerebrospinal fluid (ACSF; NaCl: 125 mM, KCl: 2.5 mM, MgCl2: 1 mM, CaCl2: 2 mM, glucose: 25 mM, NaH2PO4: 1.25 mM, NaHCO3: 25 mM, ascorbic acid: 0.4 mM, myo-inositol: 3 mM, pyruvic acid: 2 mM, bubbled with 95% O2 – 5% CO2) at room temperature (RT, 25° C) for 1 – 4 hr, while the dye is being transported. Bubble the solution during the entire incubation period.
- Subsequently, transfer the brainstems in 4% paraformaldehyde (PFA) dissolved in PBS (approx. 100 ml; pH 7 for the PFA/PBS mix) and incubate them at 4° C to allow post-fixation overnight (O/N).
5. Tissue Slicing and Mounting
- The next day, wash the brainstem three times in PBS (5 min for each washing step), cover it in 4% agar and then cut into 50 – 80 μm thick slices on a vibratome. Mount slices using a mounting medium on a glass slide, and coverslip.
- Optionally, label the sections with fluorescent Nissl for 25 min (for example blue Nissl at a 1:100 dilution in AB media).
- To prepare AB media, mix 250 ml of 0.2 M phosphate buffer (PB; 51 mM KH2PO4, 150 mM Na2HPO4), 15 ml 5 M NaCl, 15 ml 10% Triton-X, 220 ml ddH2O and 5 g bovine serum albumine.
- Precede and succeed the Nissl labeling step by 3 washing steps in PBS (10 min each).
Note: In cases where thicker sections need to be mounted and analyzed (in experiments described here such slices were 100 – 500 microns thick to preserve axonal connections over bigger distances), the technique can be combined with tissue clearing. We found the ClearT212 to be successful20. When a tissue clearing step is included, perform it before mounting the sections.
Representative Results
Figures 1 and 2 show how the laser pointer and laser goggles can be used to quickly and inexpensively genotype GFP positive animals from a litter. In cases of young mouse pups, the technique can be used to noninvasively identify GFP label in the animal's brains through the skull and overlying skin (Figure 1A – D). Emitted fluorescence can be seen through the skin and the skull of mouse pups at least up to postnatal day 3 (Figure 1C). The procedure is shown in Figure 1 for our green-fluorescent protein (GFP) labeled mouse line expressing GFP in its glycinergic subpopulation of neurons (GlyT2-GFP). In older animals with thicker skulls and pigmented skin, the GFP label shows less well through skin and bone, and therefore the skin on the head and/or neck needs to be removed before the optical genotyping (Figure 2A).
After perfusing the animal the brain is taken out and illuminated again to find the fluorescently labeled nuclei of interest. The bright structures that light up close to the ventral surface of the brainstem in Figure 2B are the two ventral nuclei of the trapezoid body, which we used as both a guide and a target for our tracer injections. Figures 4 and 5 show representative results of an anterograde injection into the ventral nucleus of the trapezoid body (VNTB) using tetramethylrhodamine dextrane (TRITC; Figure. 4) and two retrograde injections into the medial nucleus of the trapezoid body (MNTB) using choleratoxin subunit-b (CTB; Figures 5A and B) and TRITC (Figures 5C and D). Bright axonal labeling indicating (in this case inhibitory) projections from VNTB to MNTB can be seen following an anterograde injection into VNTB (Figure 4). Figures 5A and 5C show an overview over the site of injection (MNTB) with the nucleus containing the retrogradely labeled cells (VNTB) next to it. Figures 5B and 5D show magnified images of retrogradely labeled somata of glycinergic VNTB cells in the same sections as displayed in Figure 5A and 5C. Note how the retrograde tracing with CTB results in a more punctate pattern12-13 (Figure 5B), whereas VNTB cells retrogradely labeled with TRITC display a very dense Golgi-like labeling21 (Figure 5D).
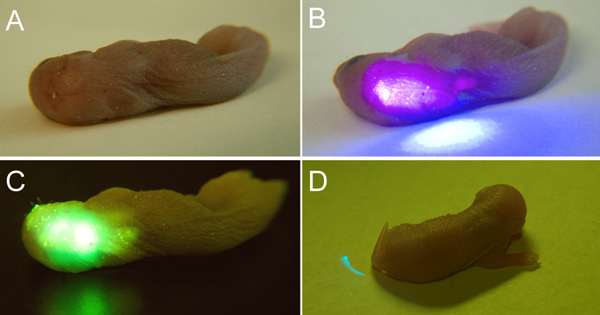
Figure 1: Optical Genotyping of GlyT2-GFP-positive and GlyT2-GFP-negative Mice. The GFP-positive mice were photographed under (A) normal lighting conditions, (B) upon laser illumination with a 405 nm laser pointer without the green-filtering goggles and (C) upon laser illumination with the laser and UV safety goggles. A strong GFP signal is visible in the area above the cerebellum and the brainstem. (D) By contrast, GlyT2-GFP-negative pups from the same mouse line and age (p2) do not show any signs of fluorescence in the same areas (back of the head and spinal cord). The blue laser light is effectively filtered by the goggles.
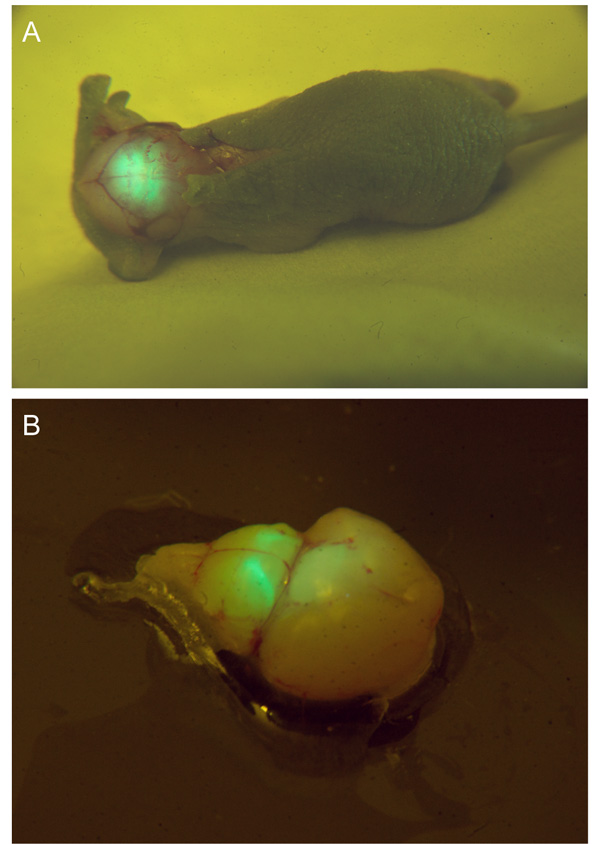
Figure 2: Optical Genotyping and a Prepared Brain Explant. (A) A p4 GlyT2-GFP mouse pup's skull is exposed to laser illumination and viewed through band-pass filtering goggles. The fluorescing GFP can be observed through the skull. (B) A brain explant from the same pup in a preparation dish upon laser illumination, as seen through the filtering goggles. Glycinergic axons and brain nuclei, such as the ventral nucleus of the trapezoid body (VNTB) can be seen as very bright structures close to the brain surface.
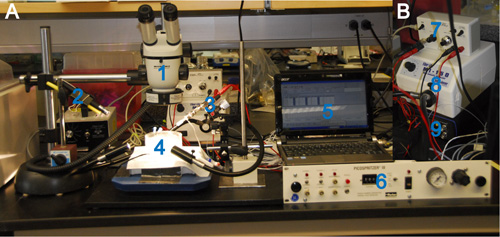
Figure 3: Overview Over an In Vitro Pressure Injection and Electroporation Setup. (A) 1: stereoscope, 2: mounted laser pointer (405 nm wavelength), 3: headstage with injection pipette mounted on a micromanipulator, 4: preparation dish containing the brain explant, 5: a PC with installed MC Stimulus software, 6: pressure injection device, connected to the injection pipette via tubing. (B) 7: stimulus isolation unit, 8: high-intensity illuminator, 9: stimulator. The stimulator is connected to the electrode in the pipette via the stimulation isolation unit and is driven by the PC.
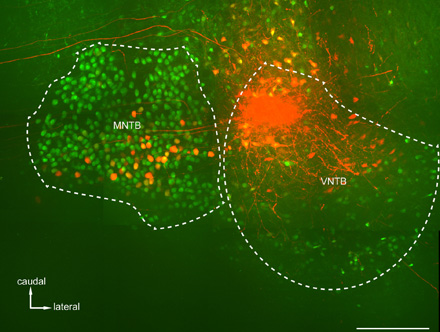
Figure 4: Anterograde Tracing of Inhibitory Connections from VNTB to the Medial Nucleus of the Trapezoid Body (MNTB). Shown is a maximum projection of a confocal stack taken in a horizontal slice spanning about 200 microns in depth in a cleared brain of a p14 GlyT2-GFP mouse. Following a tetramethylrhodamine dextrane (TRITC) injection into the caudo-medial portion of the VNTB, brightly labeled axonal connections (and in some cases their terminal endings) from VNTB to MNTB can be observed. Scale bar: 200 μm.
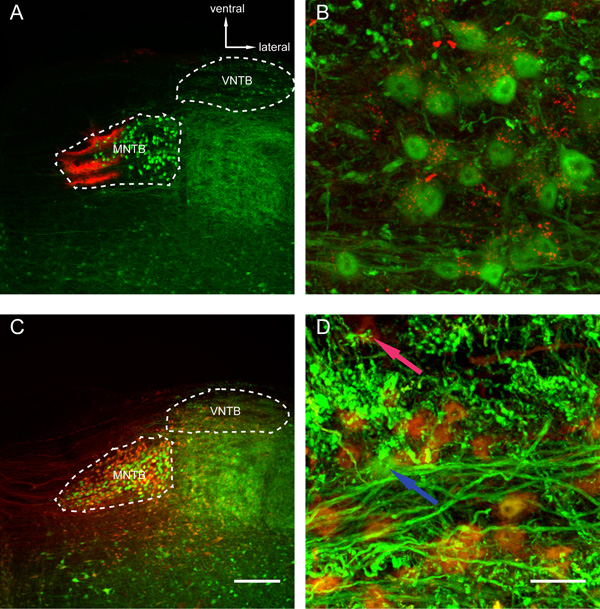
Figure 5: Retrograde Tracer Injections into MNTB Reveal Filled Glycinergic Cells in the VNTB. Shown are maximum projections (spanning over approx. 50 μm) of confocal stacks taken in coronal slices of a p88 (A, B) and a p80 (C, D) GlyT2-GFP mouse. (A) An overview of a choleratoxin subunit-b (CTB) injection into the medial portion of the MNTB. (B) Glycinergic cells in the VNTB are filled with CTB (red puncta inside the cell somata) as a result of the injection shown in Figure 5A. Some of the puncta appear outside of the cells, which could be indicative of other GFP-negative (non-glycinergic) cell types in the VNTB being filled, as well (because of injured fibers of passage in the area of injection, for example). (C) Another overview of a retrograde TRITC injection targeting the MNTB. Note, how a number of cells is filled with TRITC in the MNTB following electroporation (as opposed to the purely pressure-injected CTB in Figure 5A). (D) Retrograde labeling of glycinergic VNTB cells after a TRITC injection in the MNTB (same injection as shown in Figure 5C). Most of the cells display a dense yellow or orange labeling, because of the mixture of two fluorescent signals (the indigenous GFP expression and the filling with the red TRITC). Compare with a GFP-positive cell that for some reason did not take up the dye (blue arrow) and a tracer-labeled GFP-negative cell appearing deeply red and possibly filled, because of an axonal injury in the area of injection (red arrow). Note the different staining pattern of cell somata in retrograde tracer injections using CTB (Figure 5B) and TRITC (Figure 5D). Scale bars: Figures 5A and 5C: 200 μm, Figures 5B and 5D: 20 μm.
Discussion
A general strength of in vitro tracer electroporation, as opposed to in vivo tracing studies, is that it gives researchers better access to the brain area of interest and hence, does not involve expensive stereotactic (and often electrophysiological) equipment. In addition, the survival period required for the brain explants spans only a few hours (1 – 4) instead of days or even weeks in the case of in vivo tracer injections (see a detailed review on the use of dextran amines and other tracers in in vivo injections by Lanciego and Wouterlood, 201122, but also Rodriguez-Contreras, et al., 200823), so that unsuccessful injections can be detected as early as during the injection procedure or at the latest, on the next day, so that the injection parameters can be adjusted accordingly. The short survival period for brain explants also means that there is usually a smaller difference between the effective (area, where the dye was taken up by neurons in anterograde or axonal terminals in retrograde injections) and the apparent (area where the dye spilled, without being taken up) site of injection, although this difference can never be ruled out completely and can only be quantified/controlled for post-hoc, which in some cases can be completely impossible (see Albrecht et al., 2014 for a more detailed discussion24).
It should be pointed out, that connections of virtually any length can be studied with this method, as long as the experimenter is able to preserve both the injected area and its target or source area of axonal projections in one single piece of brain tissue. The visualization of such connections can be achieved through either reconstruction of confocal image stacks acquired after slicing and mounting the brain (as described here) or whole-brain/brain slab imaging techniques after executing tissue clearing protocols (see section 5 in this protocol for an example).
The major advantage of our technique of laser-guided in vitro electroporation is the significant increase in targeting accuracy during tracer injections. The original method of in vitro electroporation relies on just visual control by targeting certain topographic markers or using them as approximate guides for the area of injection. The same is true for the dye spread in the target area: one can observe the dye spreading, but it is not easy to control the dye spread in a finer way. With the laser modification of this method, a researcher can easily find nuclei and cells of interest and observe the dye spread in the nucleus/cell population of interest through the interaction of the indigenous genetically driven fluorescence and the fluorescence of the dye. Thus, a researcher gains more control over his/her tracing experiment by minimizing false positives in antero- or retrograde fillings, because cells/axonal terminals outside the nucleus/area of interest were filled during the injection, as well. Obviously, this does not mean an absolute control over the effective site of injection, which, as mentioned earlier, can differ from the apparent site. A caveat of this technique is obviously the availability of genetically modified organisms. Mice are a widely used genetic model organism in mammalian studies, which is also true for the auditory field, but some mammalian models could be considered better, because they resemble the human audiogram more precisely. Thus, other techniques may be employed to label neuronal populations in wildtype animals of other species better suited for a certain type of research field (e.g. gerbils or chinchillas in studies of the auditory system25-28): this could include viral injections taking advantage of certain promoters targeting only certain compartments of the brain and expressing fluorescent markers in only a subset of brain cells. Naturally, extra care needs to be taken to ensure the validity of the expressed genetic constructs in the right subpopulation of neurons, no matter whether such constructs are expressed virally in wildtype animals or indigenously in knock-in mouse lines.
As pointed out earlier, the advantage of mouse mutants expressing fluorescent proteins in certain subpopulations of neurons also allows for the functional characterization of brain circuitry (in our case we were able to characterize inhibitory projections from one auditory brainstem nucleus to another24 by pure visualization of the results of our tracing experiments). Again, the validity of the expressed genetic constructs needs to be confirmed before any interpretation of the results of the tracing experiments can take place.
Another general strength of in vitro electroporation is its compatibility with other anatomical methods, such as (immuno)histochemistry and brain clearing. As demonstrated previously24, it is not affected by the application of additional staining techniques such as Nissl, or a brain clearing method called ClearT212 20,24. The major drawback that one has to keep in mind is that the penetration depth of Nissl and antibody stains will decrease with increasing tissue thickness, so one has to weigh the options of axonal process preservation in thicker slices versus signal strength of additional (immuno)histochemical staining in experiments where both anatomical labeling methods are needed. Of course, another factor to consider is the excitation/emission bandwidth separation of the different fluorescent labels, since the minimal number of labels starts at three markers at this point (indigenous fluorescent protein expression, tracer and Nissl/antibody labeling).
Obviously, our described method can be expanded to injections of any kind of fluorescent markers (including quantum beads) into brain explants or even brain slices of transgenic mice. Since the injection accuracy depends on both, a clear target detection (the observation of fluorescently labeled nuclei and cells) and a finer control over the spill of the fluorescent dye, only one of those two factors should suffice for increased injection accuracy, even when the other is absent. This means that different non-fluorescent markers (like viral constructs or other DNA/RNA vectors) can be injected with greater precision into brain nuclei of interest, as long as the target nuclei are clearly visible to the experimenter. This latter application is particularly interesting, because electroporation does allow for effective uptake of DNA material, as well29-30, making our method almost ideal for this type of injections. The only drawback that needs to be considered is that DNA constructs take time to express and the problem of live tissue preservation arises. This could be overcome by establishing organotypic brain slice cultures31-32 based on injected brain slices (or re-sliced brain tissue blocks), which allow for live tissue preservation on the order of weeks.
And of course other transgenic mouse mutants can be used to investigate the connectivity of other neuronal types like cholinergic neurons (e.g. in ChAT-GFP mice33), making this method a versatile tool to study connectivity and functionality of brain (micro)circuits.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Supported by NIH/NIDCD R01 DC 011582. Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core supported in part by NIH/NCRR Colorado CTSI Grant Number UL1 RR025780 and the Rocky Mountain Neurlogical Disorders Core Center Grant NIH P30NS048154. Dr. Sascha du Lac from the Salk Institute provided us with the GlyT2-GFP mice.
Materials
| Sodium chloride | Sigma-Aldrich | S7653 | All chemicals are from Sigma-Aldrich, unless noted otherwise. |
| Potassium chloride | P9333 | ||
| Potassium phosphate monobasic | P5655 | ||
| Sodium phosphate di-basic | S907 | ||
| Magnesium chloride | M2670 | ||
| Calcium chloride | C5080 | ||
| Glucose | G7528 | ||
| Sodium bicarbonate | S6297 | ||
| Ascorbic acid | A4544 | ||
| Myo-inositol | I5125 | ||
| Sodium pyruvate | P2256 | ||
| Bovine serum albumine | A2153 | optional, for additional (immuno)histochemistry | |
| Triton-X-100 | X100 | optional, for additional (immuno)histochemistry | |
| Poly(ethylene glycol), 8000 MW | P2139 | optional, for brain clearing | |
| Formamide | Fisher Scientific | F84 | optional, for brain clearing |
| Choleratoxin subunit-b | Molecular Probes | C-34776 (Alexa 555) | |
| Dextrane tetramethyl-rhodamine | Molecular Probes | D-7162 (Alexa 555) | |
| Fluorescent Nissl | Invitrogen | N-21479 (blue) | optional |
| Paraformaldehyde | Fisher Scientific | SF93 | |
| Agarose | Invitrogen | 16520 | |
| Fluoromount-G | Southern Biotech | 0100-01 | |
| Pentobarbi-tal | Vortech Pharmaceuticals | Fatal-Plus | |
| Borosilicate glass filaments | Harvard Apparatus | G150F-10 | |
| Pipette puller | Zeitz Instruments, Germany | DMZ Universal Puller | |
| Perfusion setup | Custom-made | ||
| Laser pointer | laserpointerpro.com, Hong Kong | HK-88007294 (405 nm) | |
| Filter/safety goggles | Dragon Lasers, China | LSG09 (band-pass 450-700 nm) | |
| Bionocular microscope | Wild Heerbrugg, Switzerland | Wild M3 | Equipped with high-intensity illuminator (MI-150; Dolan-Jenner Inc.) |
| Picospritzer | Parker Instruments | Picospritzer III | |
| PC with installed MC Stimulus software | Multi Channel Sys-tems, Germany (software) | ||
| 2-channel stimulator | Multi Channel Sys-tems, Germany | STG-1002 | |
| Stimulation isolation unit | A.M.P.I., Israel | Iso-Flex | |
| Micromanipulator | Narishige, Japan | YOU-1 | |
| Vibratome | Leica, Germany | VT1000S |
References
- Waller, A. Experiments on the sections of glossopharyngeal and hypoglossal nerves of the frog and observations of the alterations produced thereby in the structure of their primitive fibers. Philos. Trans. R. Soc. Lond. 140, 423-429 (1850).
- Fink, R. P., Heimer, L. Two methods for selective silver impregnation of degenerating axons and their synaptic endings in the central nervous system. Brain Res. 4, 369-374 (1967).
- Nauta, W. J. Selective silver impregnation of degenerating axons in the central nervous system. Stain Technol. 27, 175-179 (1952).
- Ramón y Cajal, S. Histologie du système nerveux de l’Homme et des vertébrés. , (1911).
- Kristensson, K., Olsson, Y. Uptake and retrograde transport of peroxidase in hypoglossal neurons. Electron microscopical localization in the neuronal perikaryon. Acta Neuropathol. 19, 1-9 (1971).
- Kristensson, K., Olsson, Y. Retrograde axonal transport of protein. Brain Res. 29, 363-365 (1971).
- Burger, R. M., Cramer, K. S., Pfeiffer, J. D., Rubel, E. W. Avian superior olivary nucleus provides divergent inhibitory input to parallel auditory pathways. J. Comp. Neurol. 481, 6-18 (2005).
- Ford, M. C., Grothe, B., Klug, A. Fenestration of the calyx of Held occurs sequentially along the tonotopic axis, is influenced by afferent activity, and facilitates glutamate clearance. J. Comp. Neurol. 514, 92-106 (2009).
- Glover, J. C., Petursdottir, G., Jansen, J. K. S. Fluorescent dextran amines used as axonal tracers in the nervous system of chicken embryo. J. Neurosci. Methods. 18, 243-254 (1986).
- Trojanowski, J. Q., Gonatas, J. O., Gonatas, N. K. Conjugates of horseradish peroxidase (HRP) with cholera toxin and wheat germ agglutinin are superior to free HRP as orthograde transported markers. Brain Res. 223, 381-385 (1981).
- Trojanowski, J. Q., Gonatas, J. O., Steiber, A., Gonatas, N. K. Horseradish peroxsidase (HRP) conjugates of cholera toxin and lectins are more sensitive retrograde transported markers than free HRP. Brain Res. 231, 33-50 (1982).
- Conte, W. L., Kamishina, H., Corvin, J. V., Reep, R. L. Topography in the projections of lateral posterior thalamus with cingulate and medial agranular cortex in relation to circuitry for directed attention and neglect. Brain Res. 1240, 87-95 (2008).
- Conte, W. L., Kamishina, H., Reep, R. L. Multiple neuroanatomical tract-tracing using fluorescent Alexa Fluor conjugates of cholera toxin subunit B in rats. Nat. Protoc. 4, 1157-1166 (2009).
- Haas, K., Jensen, K., Sin, W. C., Foa, L., Cline, H. T. Targeted electroporation in Xenopus tadpoles in vivo- from single cells to the entire brain. Differentiation. 70, 148-154 (2002).
- Zeilhofer, H. U., et al. Glycinergic neurons expressing enhanced green fluorescent protein in bacterial artificial chromosome transgenic mice. J. Comp. Neurol. 482, 123-141 (2005).
- Poyatos, I., Ponce, J., Aragön, C., Giménez, C., Zafra, F. The glycine transporter GLYT2 is a reliable marker for glycine-immunoreactive neurons. Brain. Res. Mol. Brain Res. 49, 63-67 (1997).
- Gong, S., et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 425, 917-925 (2003).
- Heintz, N. Bac to the future: the use of BAC transgenic mice for neuroscience research. Nat. Rev. Neurosci. 2, 861-870 (2001).
- Franklin, K. B. J., Paxinos, G. . The Mouse Brain in Stereotaxic Coordinates. , (2008).
- Kuwajima, T., Sitko, A. A., Bhansali, P., Jurgens, C., Guido, W., Mason, C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 140, 1364-1368 (2013).
- Wouterlood, F. G., Jorritsma-Byham, B. The anterograde neuroanatomical tracer biotinylated dextran amine: comparison with the tracer PHA-L in preparations for electron microscopy. J. Neurosci. Methods. 48, 75-87 (1993).
- Lanciego, J. L., Wouterlood, F. G. A half century of experimental neuroanatomical tracing. J. Chem. Neuroanat. 42, 157-183 (2011).
- Rodriguez-Contreras, A., Silvio, J., van Hoeve, S., Habets, L. P., Locher, H., Borst, J. G. G. Dynamic development of the calyx of Held synapse. PNAS. 105 (14), 5603-5608 (2008).
- Albrecht, O., Dondzillo, A., Mayer, F., Thompson, J. A., Klug, A. Inhibitory projections from the ventral nucleus of the trapezoid body to the medial nucleus of the trapezoid body in the mouse. Front. Neural Circuits. 8, 83 (2014).
- Benson, D. A., Teas, D. C. Single unit of binaural interaction in the auditory cortex of the chinchilla. Brain Res. 103, 313-338 (1976).
- Koka, K., Jones, H. G., Thornton, J. L., Lupo, J. E., Tollin, D. J. Sound pressure transformations by the head and pinnae of the adult Chinchilla (Chinchilla lanigera). Hear Res. 272 (1-2), 135-147 (2011).
- Ryan, A. Hearing sensitivity of the mongolian gerbil, Meriones unguiculatus. J. Acoust. Soc. Am. 59 (5), 1222-1226 (1976).
- Zwicker, E., Fastl, H. . Psychoacoustics Facts and Models. , (1990).
- Chu, G., Hayakawa, H., Berg, P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 15 (3), 1311-1326 (1987).
- Klenchin, V. A., Sukharev, S. I., Serov, S. M., Chernomordik, L. V., Chizmadzhev, Y. A. Electrically induced DNA uptake by cells is a fast process involving DNA electrophoresis. Biophys J. 60 (4), 804-811 (1991).
- Elias, L., Kriegstein, A. Organotypic Slice Culture of E18 Rat Brains. J. Vis. Exp. (6), e235 (2007).
- Gähwiler, B. H. Organotypic monolayer cultures of nervous tissue. J. Neurosci. Methods. 4 (4), 329-342 (1981).
- Tallini, Y. N., et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol Genomics. 27 (3), 391-397 (2006).

