Engineering Artificial Factors to Specifically Manipulate Alternative Splicing in Human Cells
Summary
This report describes a bioengineering method to design and construct novel Artificial Splicing Factors (ASFs) that specifically modulate the splicing of target genes in mammalian cells. This method can be further expanded to engineer various artificial factors to manipulate other aspects of RNA metabolism.
Abstract
The processing of most eukaryotic RNAs is mediated by RNA Binding Proteins (RBPs) with modular configurations, including an RNA recognition module, which specifically binds the pre-mRNA target and an effector domain. Previously, we have taken advantage of the unique RNA binding mode of the PUF domain in human Pumilio 1 to generate a programmable RNA binding scaffold, which was used to engineer various artificial RBPs to manipulate RNA metabolism. Here, a detailed protocol is described to construct Engineered Splicing Factors (ESFs) that are specifically designed to modulate the alternative splicing of target genes. The protocol includes how to design and construct a customized PUF scaffold for a specific RNA target, how to construct an ESF expression plasmid by fusing a designer PUF domain and an effector domain, and how to use ESFs to manipulate the splicing of target genes. In the representative results of this method, we have also described the common assays of ESF activities using splicing reporters, the application of ESF in cultured human cells, and the subsequent effect of splicing changes. By following the detailed protocols in this report, it is possible to design and generate ESFs for the regulation of different types of Alternative Splicing (AS), providing a new strategy to study splicing regulation and the function of different splicing isoforms. Moreover, by fusing different functional domains with a designed PUF domain, researchers can engineer artificial factors that target specific RNAs to manipulate various steps of RNA processing.
Introduction
Most human genes undergo Alternative Splicing (AS) to produce multiple isoforms with distinct activities, which has greatly increased the coding complexity of the genome1,2. AS provides a major mechanism to regulate gene function, and it is tightly regulated through diverse pathways in different cellular and developmental stages3,4. Because splicing misregulation is a common cause of human disease5,6,7,8, targeting splicing regulation is becoming an attractive therapeutic route.
According to a simplified model of splicing regulation, AS is mainly controlled by Splicing Regulatory cis-Elements (SREs) in pre-mRNA that function as splicing enhancers or silencers of alternative exons. These SREs specifically recruit various trans-acting protein factors (i.e. splicing factors) that promote or suppress the splicing reaction3,9. Most trans-acting splicing factors have separate sequence-specific RNA binding domains to recognize their targets and effector domains to control splicing. The best-known examples are the members of the serine/arginine-rich (SR) protein family that contain N-terminal RNA Recognition Motifs (RRMs), which bind exonic splicing enhancers, and C-terminal RS domains, which promote exon inclusion10. Conversely, hnRNP A1 binds to exonic splicing silencers through the RRM domains and inhibits exon inclusion through a C-terminal glycine-rich domain11. Using such modular configurations, researchers should be able to engineer artificial splicing factors by combining a specific RNA-Binding Domain (RBD) with different effector domains that activate or inhibit splicing.
The key of such a design is to use an RBD that recognizes given targets with programmable RNA binding specificity, which is analogous to the DNA-binding mode of the TALE domain. However, most native splicing factors contain RRM or K Homology (KH) domains, which recognize short RNA elements with weak affinity and thus lack a predictive RNA-protein recognition "code"12. The RBD of PUF repeat proteins (i.e. the PUF domain) has a unique RNA recognition mode, allowing for the redesign of PUF domains to specifically recognize different RNA targets13,14. The canonical PUF domain contains eight repeats of three α-helices, each recognizing a single base in an 8-nt RNA target. The side chains of amino acids at certain positions of the second α-helix form specific hydrogen bonds with the Watson-Crick edge of the RNA base, which determines the RNA binding specificity of each repeat (Figure 1A). The code for RNA base recognition of the PUF repeat is surprisingly simple (Figure 1A), allowing for the generation of PUF domains that recognize any possible 8-base combination (reviewed by Wei and Wang15).
This modular design principle allows for the generation of an Engineered Splicing Factor (ESF) that consists of a customized PUF domain and a splicing modulation domain (i.e. an SR domain or a Gly-rich domain). These ESFs can function as either splicing activators or as inhibitors to control various types of splicing events, and they have proven useful as tools to manipulate the splicing of endogenous genes related to human disease16,17. As an example, we have constructed PUF-Gly-type ESFs to specifically alter the splicing of the Bcl-x gene, converting the anti-apoptotic long isoform (Bcl-xL) to the pro-apoptotic short isoform (Bcl-xS). Shifting the ratio of the Bcl-x isoform was sufficient to sensitize several cancer cells to multiple anti-cancer chemotherapy drugs16, suggesting that these artificial factors may be useful as potential therapeutic reagents.
In addition to controlling splicing with known splicing effector domains (e.g., an RS or Gly-rich domain), the engineered PUF factors can also be used to examine the activities of new splicing factors. For example, using this approach, we have demonstrated that the C-terminal domain of several SR proteins can activate or inhibit splicing when binding to different pre-mRNA regions18, that the alanine-rich motif of RBM4 can inhibit splicing19, and that the proline-rich motif of DAZAP1 can enhance splicing20,21. These new functional domains can be used to construct additional types of artificial factors to fine-tune splicing.
Protocol
1. Construction of a PUF Scaffold with Customized RNA-binding Specificity by Overlapping PCR
- Design a series of PCR primers containing the PUF sequences that specifically recognize different RNA nucleotides in each position12 (see Table 1 for the primer sequences and see Figure 1A for the RNA:PUF recognition code). For each PUF repeat, design four different primers to recognize a different base on each position.
NOTE: These primers will be used in a series of four rounds of PCR reactions to generate PUF fragments that are joined together (Figure 1B). - Select the recognition site of the target gene near the alternative splice site.
NOTE: This site can be decided by the user, and it usually is chosen within 10 – 50 nt from the alternative splice sites. - Round 1: Generate coding sequences for 4 PUF repeats, universal bridge fragments, and cap fragments.
- Use the target site to define the recognition code for each repeat of the customized PUF. Select the set of PCR primers for PUF repeats 1 – 8. Conduct four consecutive rounds of PCR to produce a customized PUF domain, as described below (Figure 1B).
- In the first round of PCR, set up standard PCR reaction mixtures (containing 2.5 µL of 10x buffer, 0.5 µL of 10 mM dNTPs, 0.5 µL of 10 µM forward and reverse primers, 50 ng of the DNA template (human cDNA or expression vector of WT PUF domain), and 0.5 U high-fidelity DNA polymerase in a 25 µL final volume).
NOTE: These reactions will produce DNA sequences corresponding to each PUF repeat, to universal bridge fragments, and to two cap fragments with different restriction sites (Figure 1B). A brief list of templates, primers, and generated PCR products used in this step are shown in Table 2. - Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 15 s at 72 °C; and 5 min of incubation at 72 °C. Use high-fidelity DNA polymerase to minimize point mutations during PCR.
- Round 2: Generate coding sequences for R1/R2/R3/R4 (R1 – 4) and R5/R6/R7/R8 (R5 – 8).
- Separate the PCR products obtained in step 1.3.3 using electrophoresis with a 1.5% agarose gel (25 min at a constant voltage of 120 V). Gel-purify all products of the expected size using a gel purification kit. Use a different mixture of these products as the template in the following rounds of PCR.
NOTE: Mix the three PCR products in roughly a 1:1:1 ratio. They usually do not need to be quantified, as long as the intensity of each product looks similar in the gel. - Mix about 5% of the purified PCR products as a template in the next round of PCR. Alternatively, quantify the purified PCR products using a UV-Vis spectrophotometer and use 15 ng of each PCR product in the template mixture.
- Use purified PCR products R1/R2, R3/R4, and Bridge 2/3 as mixed templates and R1-F and R4-R as primers. Set up the PCR reaction as described in step 1.3 to obtain overlapping PCR product R1 – 4.
- Use purified PCR products R5/R6, R7/R8, and Bridge 6-7 as mixed templates and R5-F and R8-R as primers to obtain overlapping PCR product R5-8 (Figure 1B). Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C; and 5 min at 72 °C. Gel-purify R1-4 and R5-8.
- Separate the PCR products obtained in step 1.3.3 using electrophoresis with a 1.5% agarose gel (25 min at a constant voltage of 120 V). Gel-purify all products of the expected size using a gel purification kit. Use a different mixture of these products as the template in the following rounds of PCR.
- Round 3: Generate coding sequences for R1/R2/R3/R4/R5/R6/R7/R8 (R1 – 8 without caps).
- In the third round of PCR, using the purified R1-4, R5-8, and Bridge 4/5 as mixed templates and R1-F and R8-R as primers, set up the PCR reaction as described in step 1.3 to obtain overlapping PCR product R1-8 without caps.
- Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 55 s at 72 °C; and 5 min at 72 °C. Gel-purify the R1-8 without caps.
- Round 4: Generate coding sequences for complete PUF domains.
- In the last round of PCR, using the purified R1-8 without caps and 5'-end and 3'-end caps as mixed templates and Cap-F and Cap-R as primers, set up the PCR reaction as described in step 1.3 to obtain the final mutated PUF domains.
- Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C; and 5 min at 72 °C.
- Gel-purify the PCR products of the final reprogrammed PUF domains. Use these products for the construction of the ESF expression plasmid in the subsequent steps. Sequence the final construct to verify the reprogrammed sequences of PUF domains.
NOTE: Cap-F encodes NLS (PPKKKRKV) between BamHI and XbaI sites and Cap-R encodes a stop codon and the SalI site (restriction sites were designed for constructing expression vectors of ESFs, Figure 1C).
2. Construction of a Functional Module of ESFs
- Two strategies are commonly used to clone functional modules of ESFs (RS domains or Gly-rich domains): to amplify these domains by PCR using total human cDNA as a template (step 2.2) or to directly synthesize the DNA fragments that code for different RS or Gly-rich domains (step 2.3).
- Use PCR to clone RS domains from residues 123 – 238 of 9G8 (NP001026854), residues 180-272 of SRP40 (NP008856), or residues 117 – 221 of SC35 (NP003007). Clone Gly-rich domains from residues 195 – 320 of hnRNP A1 (NP_002127), residues 203 – 353 of hnRNP A2 (NP112533), or residues 211 – 378 of hnRNP A3 (NP919223).
- Use the NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) or Primer3 (http://frodo.wi.mit.edu/primer3/) to design primers for cloning RS domains or Gly-rich domains (functional module of ESFs).
NOTE: The forward primer contains an N-terminal FLAG tag after the NheI site. The reverse primer contains a BamHI site for cloning into expression vectors (Figure 1C). - Set up the standard PCR reaction, as described in step 1.3, to amplify the RS or Gly-rich domains. Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C; and 5 min at 72 °C.
- Use the NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) or Primer3 (http://frodo.wi.mit.edu/primer3/) to design primers for cloning RS domains or Gly-rich domains (functional module of ESFs).
- Clone RS domains or Gly-rich domains through direct DNA synthesis. Instead of generating PCR products that encode for the effector domains in step 2.2, synthesize an oligonucleotide that encodes a short-fragment RS domain (RSRSRSRSRSRS) or a Gly-rich domain (GYGGGGPGYGNQGGGYGGG) using a commercial source of DNA oligonucleotides.
3. Construction of ESF Expression Plasmids
- Construct ESF expression plasmids using any expression vector.
NOTE: An expression construct pGL-Gly-MS2 (a gift from Dr. R. Breathnach from the Institute de Biologie-CHR11) was originally used, which encodes from the N- to the C-terminal, a FLAG epitope, a Gly-rich domain of hnRNP A1, and the MS2 coat protein. Therefore, this expression construct is used as an example in the following step.
NOTE: Other expression vectors with multiple cloning sites can also be used, and the standard cloning steps will be applied to join the fragments together. - Digest 1.5 µg of the expression plasmids (pGL-Gly-MS2) mentioned in step 3.1 to remove the MS2 coat protein fragment. Use the restriction endonucleases BamHI and SalI for 1 h at 37 °C. Digest the recognition module of ESFs (encoding an NLS and reprogrammed PUF domains, generated in step 1.5) with BamHI and SalI in same condition.
- Add 5x loading dye to the restriction digest reactions and slowly electrophorese the total volume with a 1.5% agarose gel containing a DNA gel stain for 40 min at 120 V. Gel-purify the digested products.
- Prepare 10 µL ligation reactions with a purified recognition module of ESF inserts and expression vector DNA in a 3:1 ratio, 1 µL of DNA ligase, and 1 µL of 10x ligation buffer. Incubate the ligation reactions overnight at 4 °C; the resulting construct expresses a Gly-PUF-type ESF (pGL-Gly-PUF) under the control of a CMV promoter (Figure 1C).
- Remove the fragment encoding the FLAG/Gly-rich domain with NheI and BamHI digestion for 1 h at 37 °C. Replace it with a fragment that encodes the RS domain (generated in step 2.2, also digested by NheI and BamHI); the resulting construct expresses an RS-PUF-type ESF (Figure 1C).
4. Construction of the Splicing Reporter
- Synthesize oligonucleotides containing the candidate sequences (i.e. target sequences of PUF domains) flanked by XhoI and ApaI sites or XhoI and EcoRI sites. Anneal the oligonucleotides for 5 min at 55 °C to obtain double-stranded inserts containing the recognition sites of PUF domains flanked by the cohesive ends of XhoI and ApaI or XhoI and EcoRI sites.
- Start from a previously described modular splicing reporter22, pGZ3, which contains two GFP exons separated by a test exon (exon 12 of the human IGF2BP1, Ensembl ID ENSG00000159217) and its flanking introns.
NOTE: The test exon was engineered with XhoI and ApaI sites, which can be used to insert synthesized oligonucleotides containing the candidate sequences (target sequences of PUF domains). This modular splicing reporter (pGZ3) is provided through the plasmid repository Add-gene. - Digest the base reporter pGZ3 (prepared in step 4.2) with the XhoI and ApaI restriction endonucleases for 1 h at 37 °C.
- Gel-purify the digested products as described in step 3.3.
- Prepare 10 µL of ligation reactions with the double-stranded inserts generated at step 4.1 and the digested pGZ3 vector generated at step 4.4 in a 3:1 molar ratio, 1 µL of DNA ligase, and 1 µL of 10x ligation buffer. Incubate the ligation reactions overnight at 4 °C to obtain the constructing splicing reporter vector pGZ3.
- Insert the double-stranded inserts generated in step 4.1 into the previously published reporters, pEZ-1B (using XhoI and EcoRI sites) and pEZ-2F (using XhoI and ApaI sites)23, to obtain the competing 5' ss reporter and the competing 3' ss reporter, as described in steps 4.3-4.5.
NOTE: Both types of splicing reporters will be available through Add-gene.
5. Construction of Lentiviral Expression Vectors for ESF
- Set up the standard PCR reaction as described in step 1.3 to amplify the full-length ESFs from the original expression vectors (pGL-Gly-PUF or pGL-RS-PUF) with primers containing MluI/SpeI sites. Set the PCR program as follows: 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 30 s at 55 °C, and 1.5 min at 72 °C; and 5 min at 72 °C.
- Through a digestion reaction, gel purification, and ligation reaction, integrate the ESFs into the lentiviral expression vector, pWPXLd, between the MluI/SpeI sites, as described in steps 3.2-3.4.
- Use the standard calcium phosphate precipitation method, as previously reported24, to generate lentiviruses by cotransfecting HEK293T cells by packaging the vectors, psPAX2 and pMD2.G, with either pWPXLd-Gly-PUF(Bcl-x), pWPXLd-Gly-PUF(wt) (as a specificity control), or pWPXLd-GFP (mock).
- Seed 5×106 HEK293T cells into 10 cm dishes and grow the cells overnight in 10 mL of Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) per dish in a humidified incubator at 37 °C and 5% CO2. Perform the transfection when the cells are 70 – 90% confluent.
- Prepare the plasmid mixture by adding the three plasmids (7.5 µg of the packaging vector, psPAX2; 2.5 µg of pMD2.G; and 10 µg of either pWPXLd-Gly-PUF(Bcl-x), pWPXLd-Gly-PUF(wt), or pWPXLd-GFP) to a 2 mL tube.
- Add 560 µL of 0.25 M CaCl2 and 560 µL of 2x BBS solution to the 2-mL tube. Mix gently several times and incubate it for 15 min at RT to prepare the transfection mixture.
- Add all the transfection mixtures to the 10-cm dishes. Swirl the dishes gently and incubate them overnight at 3% CO2 and 37 °C.
- Remove the medium, add 10 mL of fresh DMEM with 2% FBS to each dish, and incubate them at 10% CO2 and 37 °C O/N.
- Collect the first supernatant from the dishes. Add 10 mL of fresh DMEM with 2% FBS to each dish. Incubate the dishes O/N at 10% CO2 and 37 °C. Store the supernatant at 4 °C.
- Collect the second supernatant from the dishes. Pool the supernatant from the first and second harvests. Clear the supernatant of cell debris by filtering it through a 0.4-µm filter. Use the cleared supernatant directly or store it at -80 °C.
- Determine the titer of the lentivirus by infecting HEK293T cells with serial dilutions of the virus preparation, as previously reported25.
6. Specifically Modulating Exon Inclusion and the Alternative Use of Splice Sites with ESFs
- Seed 2 x105 HEK293T cells into each well of a 24-well plate. Grow the cells overnight in 500 µL of DMEM supplemented with 10% FBS in a humidified incubator at 37 °C and 5% CO2.
- Mix the liposomal transfection reagent by gently inverting the bottles before use. Dilute 2 µL of liposomal transfection reagent in 50 µL of reduced serum medium. Mix gently and incubate them for 5 min at RT.
- Dilute 0.04 µg of pGL-Gly-PUF expression vectors and 0.2 µg of pGZ3 reporter plasmids in 50 µL of reduced serum medium in a sterile tube. Dilute 0.4 µg of pGL-RS-PUF expression vectors and 0.2 µg of pGZ3 reporter plasmids in 50 µL of reduced serum medium in a sterile tube. Dilute 0.4 µg of pGL-RS-PUF expression vectors and 0.2 µg of pEZ-1B or pEZ-2F reporter plasmids in 50 µL of reduced serum medium in a sterile tube.
- After 5 min of incubation at RT, gently mix the diluted liposomal transfection reagent prepared in step 6.2 with the diluted plasmids prepared in step 6.3. Incubate the mixture for 20 min at RT.
- Add the entire transfection mixtures containing the expression vectors and transfection reagent prepared in step 6.4 to each well and incubate for at least 12 h in a humidified incubator at 37 °C and 5% CO2.
- After 12 h in a humidified incubator at 37 °C and 5% CO2, discard the medium from each well and wash them with 500 µL of Phosphate-buffered Saline solution (PBS)/well.
- Discard the PBS and add 200 µL of trypsin to each well. Incubate the plate in a humidified incubator at 37 °C and 5% CO2 for 5 min. Add 1 mL of the medium to stop the digestion and transfer the cells to a sterile 1.5 mL tube.
- Centrifuge the tubes for 3 min at 5,000 x g and discard the medium. Add 0.5 mL of RNA extraction buffer per tube to lyse the cells by repetitive pipetting. Incubate the homogenized samples for 5 min.
- For each RNA extraction buffer-treated sample, add 0.1 mL of chloroform per 0.5 mL of RNA extraction buffer. Invert the tubes for 15 s and incubate them for 3 min at RT.
- Centrifuge the tubes for 15 min at 12,000 x g and 4 °C. Transfer the aqueous phase to a fresh tube. Add 0.25 mL of isopropanol per 0.5 mL of RNA extraction buffer used for the initial homogenization.
- Mix by vortexing and incubate them at RT for 10 min. Centrifuge the tubes at 12,000 x g for 10 min at 4 °C. After centrifugation, the RNA precipitate is usually visible on the bottom of the tube.
- Discard the supernatant and wash the RNA pellet with 0.5 mL of 75% ethanol per 0.5 mL of RNA extraction buffer.
- Vortex vigorously and centrifuge it at 7,500 x g for 5 min at 4 °C. Remove the supernatant and dry the RNA pellet. Dissolve the RNA in 50 µL of RNase-free water.
- Add 2 µL of 5U/µL DNase I, 7 µL of 10x buffer, and 11 µL of H2O to each 50-µL RNA solution. Incubate the tubes at 37 °C for 1 h. Heat them at 70 °C for 15 min to inactivate the DNase.
- For each sample, add the following components to a 0.2 mL nuclease-free tube: 1 µL of 50 µM oligo dT, 5 µL of 400 ng/µL RNA (2 µg), 1 µL of 10 mM dNTP, and 3 µL of H2O. Perform the reverse PCR.
- Heat the mixture to 65 °C for 5 min and incubate it on ice for at least 2 min to prevent the reformation of the secondary structure. Add the following components to the same tube: 4 µL of 5x first-strand buffer, 1 µL of 0.1 M DTT, 1 µL of 200 U/µL reverse transcriptase, and 4 µL of H2O. Mix by pipetting gently up and down and incubate at 50 °C for 60 min. Stop the reaction by heating it at 70 °C for 15 min.
- For each sample, add the following components to a PCR tube in order to complete the body-labeled PCR: 2.5 µL of 10x PCR buffer, 0.5 µL of 10 mM dNTP mix, 1 µL of 10 µM forward primer, 1 µL of 10 µM reverse primer, 0.25 µL of 5 U/µL Taq DNA polymerase, 0.5 µL of 25 nM Cy5-dCTP, and 2 µL of the cDNA prepared in step 6.15.
- Heat the reaction to 94 °C for 2 min to denature the molecules, perform 25 cycles of PCR (94 °C for 30 s, 60 °C 30 s, and 72 °C 30 s), keep the reaction at 72 °C for 7 min, and then leave it at a 4 °C hold.
- Resolve the PCR products by electrophoresis through a 10% polyacrylamide gel with a 1x Tris base, Boric acid, and EDTA (TBE) buffer. Perform a scan using a fluorescence scanner. Measure the amount of each splicing isoform using a densitometry software.
7. Use ESF to Modulate Endogenous Bcl-x Splicing and Measure Its Effects on Apoptosis
- Plate 2 x 105 HeLa cells to each well of a 24-well plate. Grow the cells overnight in 500 µL of DMEM supplemented with 10% FBS in an incubator at 37 °C and 5% CO2.
- After 12 h, transfect the cells with 2 µg of pGL-Gly-PUF(WT) or 0.2 µg, 1 µg, and 2 µg of pGL-Gly-PUF(531) (a reprogrammed PUF domain that recognizes Bcl-x pre-mRNA with high affinity, see Figure 2A) .
- 24 h later, harvest the cells. 1/3 of the cells are for the RNA isolation and PCR analysis (steps 6.8 – 6.17) and 2/3 are for the protein isolation.
- For the Western blot analysis, boil the total cell pellets in 2X Sodium Dodecyl Sulfate-PolyAcrylamide Gel Electrophoresis (SDS-PAGE) loading buffer for 10 min, and then resolve the proteins in a 12% SDS-PAGE gel. Transfer the proteins onto a nitrocellulose membrane.
- Block the membrane with 5% milk for 1 h at RT and incubate the membrane O/N with Caspase-3 (1:1,000), PARP (1:1,000), or beta-actin (1:5,000) primary antibodies (diluted in 5% milk) at 4 °C.
- Wash the membrane with PBS containing 0.1% Tween 20 (PBS-T) for 5 min at RT 3x on a rocking shaker. Incubate the membrane with HRP-linked antibodies (1:5,000, diluted in 5% milk) for 1 h at RT.
- Wash the membrane with PBS-T 3x at RT, and then develop the membrane using ECL Western blotting detection reagents.
- Add 250 µL of poly-L-Lysine (PLL) onto coverslips, incubate them for 15 min at RT, and siphon off the liquid. Wash the PLL-coated glass coverslips with PBS 3x for 5 min each. For the immunofluorescence assay measuring apoptosis, seed 5 x 105 HeLa cells onto poly-lysine-coated glass coverslips in a 6-well plate. Transfect pGL-Gly-PUF(WT) or pGL-Gly-PUF(531) plasmids into the HeLa cells using a liposomal transfection reagent (see steps 6.1 – 6.5).
- 24 h after the transfection, fix the cells on the coverslips with 1 mL of 4% paraformaldehyde (PFA) in 1x PBS for 20 min at RT. CAUTION: PFA is toxic; handle it with care in a fume hood.
- Gently wash the cells on the coverslips by adding 2 mL of 1x PBS. Incubate them for 5 min. Remove the PBS with pipettes. Repeat the wash 3x.
- Permeabilize the cells with 0.2% Triton X-100 in 1x PBS for 10 min, and then wash them 3x with 1x PBS.
- Block the cells with 3% Bovine Serum Albumin (BSA) in 1x PBS for 10 min and wash them 3x with 1x PBS.
- Dilute the FLAG antibody 1:1,000 in 3% BSA/PBS and pipet 30 µL of the diluted FLAG antibody onto a parafilm sheet.
- Take out the coverslip with the cells, carefully dry the excess buffer with lab wipes, and put it upside-down (cell-side down) on the 30 µL anti-FLAG solution. Incubate it for 1 h at RT.
- Add about 500 µL of 1x PBS to the side of the coverslip incubated with the primary antibody until the coverslip floats on top of the solution. Return it to the 6-well plate with 1x PBS in the wells.
- Wash it 3x with 1x PBS for 5 min each.
- Dilute the anti-mouse secondary antibody (1:500) in 3% BSA/PBS; again, use 30 µL for each coverslip. Put the coverslip upside-down on the secondary antibody solution and incubate it for 15 min at RT.
- Remove the coverslip from the parafilm, as described in step 7.15. Put it back into the 6-well plate and wash it with 1x PBS 3x for 5 min each.
- Mount the coverslips with mounting medium (with DAPI), remove the excess medium, and seal the edge with nail polish.
- Visualize the cells using a fluorescence microscope (at 100X magnification) and photograph them using a digital camera.
NOTE: The expression of ESF is visualized by the fluorescence-labeled secondary antibody against the FLAG tag, and the nuclear fragmentation is visualized using DAPI staining.
8. Measure the Apoptosis of Different Cancer Cells Expressing ESF
- Split 2 x 106 HeLa cells, MDA-MB-231 cells, and A549 cells into 60-mm dishes with 4 mL of DMEM supplemented with 10% FBS in an incubator at 37 °C and 5% CO2 to detect apoptosis with Propidium Iodide (PI) staining.
- 24 h later, refresh the medium and prepare the lentivirus, as previously reported24.
- Dilute 10 x 106 lentivirus pWPXld-Gly-PUF(WT), pWPXld-Gly-PUF(531), or pWPXld-GFP stocks into 4 mL of fresh medium to make the ratio of virus to cell number equal to 5.
- Change the medium in the plates to 4 mL of the virus-containing medium prepared in step 8.3 and incubate the plates in an incubator at 37 °C and 5% CO2. 12 h after the infection, change the medium.
- After 24 h of infection, collect and stain the cells for 5 min in a PBS solution containing a final concentration of 2 µg/mL PI.
- Analyze the PI-stained cells with a flow cytometer, as described previously16.
Representative Results
This report describes the complete protocol for the design and construction of ESFs and splicing reporters. It also outlines the further application of ESFs in manipulating the AS of endogenous genes16. To illustrate typical results of ESF-mediated splicing changes, we use the data from our previous work as an example. The ESFs with different functional domains can be used to promote or inhibit the inclusion of the target cassette exon (Figure 1D & E). ESFs can also affect the usage of alternative 5' and 3' splice sites in the reporter system (Figure 1F & G).
The alternative splicing of the endogenous gene can also be specifically regulated with designer ESFs. We have demonstrated this application by specifically targeting Bcl-x, which can be spliced into two antagonistic isoforms with alternative 5' splice sites. We designed an ESF, Gly-PUF(531), that recognizes an 8-nt RNA element between the alternative 5' splice sites. This Gly-PUF(531) specifically shifted the splicing towards the production of Bcl-xS (Figure 2A). After transfecting the Gly-PUF(531) into HeLa cells, the level of Bcl-xS isoforms and Bcl-xS proteins increased in a dose-dependent manner, whereas the control ESF, Gly-PUF(WT), did not affect the ratio of Bcl-xS to Bcl-xL (Figure 2B & C). In addition, the designer ESF can induce the cleavage of caspase 3 and poly (ADP-ribose) polymerase (PARP), two well-known molecular markers of apoptosis (Figure 2D). As expected, the designer ESFs are predominantly localized in the nuclei of transfected cells, as demonstrated by immunofluorescence microscopy (Figure 2E). Consistently, the splicing shift by Gly-PUF(531) caused the fragmentation of nuclear DNA, indicating that these cells are undergoing apoptosis (Figure 2E). The increase of apoptotic cells was further confirmed by examining more than 200 cells from randomly selected fields and by quantifying the percent of cells with fragmented nuclear DNA (Figure 2F).
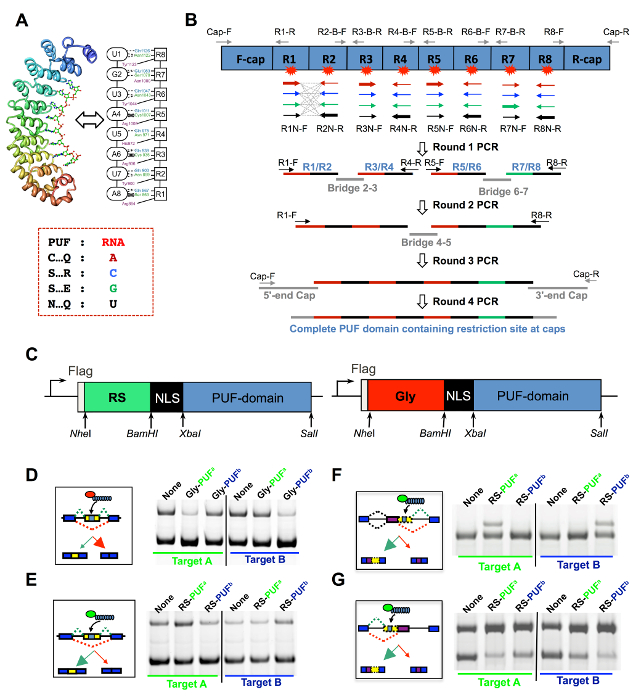
Figure 1: Design of ESFs and Their Activity in Modulating Exon Skipping. (A) Specific binding between the PUF domain and RNA targets is illustrated with the RNA-PUF structure and a schematic diagram. The PUF binding code for each of the four RNA bases, shown on the right with different colors, is used to design PUF mutations. (B) Flow chart to obtain a customized PUF domain. The PUF that recognizes "UGUAUAUA" was used as an example. A 4-round PCR strategy is used to assemble a PUF scaffold with customized RNA-binding specificity (color coded similarly to panel A). In the first round, a series of PCR primers that incorporate the desired RNA-recognition codes for two adjacent PUF repeats are used to generate four fragments that include the eight RNA-recognition codes of a full PUF protein (R1/R2, R3/R4, R5/R6, and R7/R8). Cap fragments encoding an N-terminal nuclear localization signal, a C-terminal stop codon, and bridge fragments are also produced separately (5'-end and 3'-end cap, bridge 2/3, bridge 4/5, and bridge 6/7). In the second round, new templates are generated by mixing overlapping fragments encoding adjacent repeats with the appropriate bridge (e.g., mixing R1/R2, R3/R4, and bridge 2/3 generates the template for R1 – 4) and then extending with DNA polymerase to fill the gaps. Similarly, the third round joins R1-4, R5-8, and bridge 4/5. Finally, the fourth round adds the 5'-end and 3'-end caps of the PUF domain together with the cloning sites for subsequent cloning to expression vectors. (C) Modular domain organization of ESFs. ESFs are driven by CMV promoters (arrow) and encode, from the N- to the C-terminal: a FLAG epitope (for the detection of ESFs), a functional module (a Gly-rich domain or an RS domain), an NLS (facilitating the nuclear localization of ESFs), and an RNA-recognition domain (a PUF domain). NheI and BamHI are designed to insert a functional module, while XbaI and SalI are designed to insert an RNA-recognition domain. (D) Gly-PUF ESFs are co-expressed with exon skipping reporters, and the splicing pattern is assayed by RT-PCR. The modified PUFa and PUFb specifically bind to 8-mer targets A and B, respectively (in the same colors). All combinations are used, so the PUF-target pairs of different color serve as the controls. The effects of RS-PUF on exon skipping (E), the competing 5' splice site (F), or the competing 3' splice site reporter (G) were assayed by methods similar to panel D. The data of the RT-PCR are from Wang et al.16. Please click here to view a larger version of this figure.
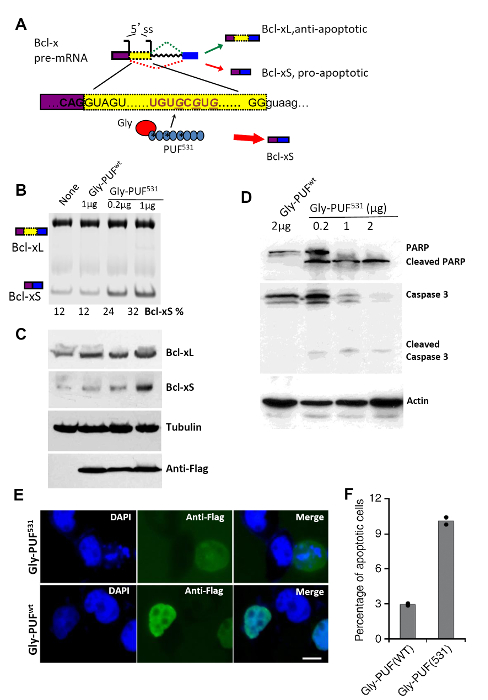
Figure 2: Regulation of Endogenous Bcl-x pre-mRNA Splicing with ESFs. (A) Schematic of the alternative splicing of endogenous Bcl-x pre-mRNA. Two alternative 5' splice site in exon 2 of Bcl-x are used to generate two isoforms of different sizes, Bcl-xL and Bcl-xS. The sequence UGUGCGUG between the two 5' splice sites is selected as the ESF target, and WT PUF repeats 1, 3, and 5 (Q867E/Q939E/C935S/Q1011E/C1007S) are reprogrammed (asterisks) to recognize this target sequence. The resulting ESF containing a Gly-rich domain inhibits the use of the downstream 5' ss (indicated by the red arrow). (B) Modulation of Bcl-x 5' ss usage. Different amounts of the Gly-PUF(531) expression construct are transfected into HeLa cells. Gly-PUF(WT) is used as a control. Two isoforms of Bcl-x are detected with RT-PCR using primers corresponding to exons 1 and 3 of the Bcl-x gene. The percentage of the Bcl-xS isoform is quantified and shown at the bottom. (C) ESFs affect the expression levels of Bcl-xL and Bcl-xS. Samples are loaded in the same order as in panel B, and all proteins are detected by Western blots. The expression of ESFs is detected by the anti-FLAG antibody, and the tubulin level is used as a control. (D) Different amounts of ESF expression constructs are transfected into HeLa cells, resulting in the cleavage of PARP and caspase 3. Samples are detected by Western blot 24 h after transfection. The actin level is detected as a control. (E) The subcellular localization of ESFs in transfected HeLa cells is detected by immunofluorescence microscopy with the anti-FLAG antibody. The cells are co-stained with DAPI to show the nuclei. Some nuclei, especially in cells transfected with Gly-PUF(531), are fragmented due to apoptosis. Scale bar: 5 µm. (F) Percentage of apoptotic cells (i.e. cells with fragmented nuclear DNA) are measured from randomly chosen fields of fluorescence microscopy images. The bars indicate the mean, while the dots indicate the data from the two experiments. The figures are modified from our earlier report by Wang et al.16 in accordance with the policy of Nature Publishing Group. Please click here to view a larger version of this figure.
Discussion
This report provides a detailed description for the design and construction of artificial splicing factors that can specifically manipulate the alternative splicing of a target gene. This method takes advantage of the unique RNA binding mode of PUF repeats to produce an RNA-binding scaffold with customized specificity. It can be used to either activate or repress splicing.
The critical step in this protocol is the generation of the reprogramed PUF domain that defines the specificity of ESFs. A PCR stitching protocol has been developed and optimized for the rapid generation of the PUF scaffold. The key for its success is to adjust the ratio of different overlapping templates to 1:1:1. The purification of the PCR products after each round is also critical, because unpurified products may have primer contamination from the last round. Another important step is to assay the splicing ratio using semi-quantitative RT-PCR. Generally, too many amplification cycles should be avoided, as they may saturate the PCR reaction. In our experiments with the co-expression of a splicing reporter, 20 – 25 cycles were routinely used, but this may vary depending on the abundance of the mRNA when measuring the splicing of endogenous genes. When quantifying the splicing isoforms using a new pair of primers, we suggest calibrating the PCR experiment each time, as previously described16.
A potential limitation with designer ESFs is their off-target effects, because the specificity is determined by the number of repeats in the PUF scaffold. Wildtype PUF recognizes an 8-nt site, which is comparable to the specificity of an siRNA that recognizes its target through "seed match." However, since any 8-nt sequence could occur once by chance in a transcript 65,000 nt long (48 = 65,536), there will be other off-target transcripts recognized by the designer PUF. The off-target effect can be reduced by using PUFs with additional repeats; however, it is still useful to evaluate the specificity and off-target effects of ESFs. To minimize potential off-target effects, the expression of a combination of multiple designer ESFs at a lower level may also be carried out. In such a case, the off-targeted genes may not be affected by the low amount of ESFs, whereas the splicing of the real target will be affected by the multiple ESFs that function synergistically. This solution is similar to what researchers used in gene silencing with RNAi, where the pooled siRNAs (each at a reduced concentration) that target multiple sites of a single mRNA can decrease the off-target effects.
The other main method to manipulate AS is to use antisense oligonucleotides that pair with certain regions of the pre-mRNA. Compared to this existing method, the ESFs can cause prolonged effects in stably transfected cells. In addition, the in vivo delivery of ESF can take advantage of the increasing arsenal of gene therapy vectors, whereas the in vivo delivery of antisense oligonucleotides is very hard to control. In addition, this method can avoid complicated and costly modifications of oligonucleotides. Using various inducible promoters, a more precise control of the ESF expression in the correct cell types and at the correct times may also be achieved. The main disadvantage of this method is the relatively low specificity (an 8-nt recognition site versus a 16- to 20-nt recognition site in typical antisense oligonucleotides).
The dysregulation of alternative splicing causes many diseases, including cancer26,27. Genome-wide studies have revealed more than 15,000 tumor-associated splice variants in various types of cancers28,29,30. For instance, intronic splice-site mutations of tumor-suppressor genes often cause exon-skipping events and produce aberrant proteins that may contribute to tumor genesis26,31,32. Moreover, some splicing factors are found to be overexpressed in many cancer types, which contributes to cell transformation33,34, indicating that splicing factors can also play important roles in cancer biogenesis. Therefore, in addition to providing a useful tool to modulate gene function, manipulation of splicing with designer ESFs may restore misregulated splicing events in cancer, thus providing a potential therapeutic tool. In addition, by fusing a designer PUF domain with different functional domains, multiple artificial factors that manipulate various RNA metabolism processes can be designed. For example, the fusion of a translational activator (GLD2) or a translational repressor (CAF1) with the PUF domain produced novel factors that can activate or inhibit mRNA translation27. Using the same design principal, we combined a non-specific RNA endonuclease domain (PIN) with a series of designer PUF domains to generate artificial site-specific RNA endonucleases (ASREs) that function analogously to DNA restriction enzymes17.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH grant R01-CA158283 and NSFC grant 31400726 to Z.W. Y.W. is funded by the Young Thousand Talents Program and the National Natural Science Foundation of China (grants 31471235 and 81422038). X.Y. is funded by the postdoctoral science foundation of China (2015M571612).
Materials
| High-fidelity DNA polymerase (Phusion High-Fidelity) with PCR buffer | New England Biolabs | M0530L | |
| DNA ligase (T4 DNA ligase) | New England Biolabs | M0202L | |
| Liposomal transfection reagent (Lipofectamine 2000) | Invitrogen | 11668-019 | |
| Reduced serum medium (Opti-MEM) | Gibco | 31985-062 | |
| RNA extraction buffer (TRIzol Reagent) | ambion | 15596018 | TRIzol reagent includes phenol, which can cause burns. Wear gloves when handling |
| BSA (Bovine Serum Albumin) | Sigma-Aldrich | A7638-5G | |
| PBS (1X) | Life Technologies | 10010-031 | |
| SuperScript III reverse transcriptase | Invitrogen | 18080044 | |
| Caspase-3 antibody | Cell Signaling Technology | 9668 | |
| PARP antibody | Cell Signaling Technology | 9542 | |
| Bcl-x antibody | BD Bioscience | 610211 | |
| beta-actin antibody | Sigma-Aldrich | A5441 | |
| alpha-tubulin antibody | Sigma-Aldrich | T5168 | |
| FLAG antibody | Sigma-Aldrich | F4042 | |
| Nitrocellulose membrane | Amersham-Pharmacia | RPN203D | |
| ECL Western Blotting detection reagents | Invitrogen | WP20005 | |
| Cy5-dCTP | GE Healthcare | PA55021 | |
| Fluorescence-activated cell sorter | BD Bioscience | FACSCalibur | |
| Dulbecco’s Modified Eagle’s Medium (DMEM) | GE Healthcare | SH30243.01 | |
| Fetal bovine serum | Invitrogen | 26140079 | |
| Propidium iodide (PI) | Sigma | P4170 | |
| Bovine Serum Albumin (BSA) | Sigma | A7638-5G | |
| Triton-X100 | Promega | H5142 | |
| Poly-lysine | Sigma | P-4832 | Filter sterilize and store at 4 °C |
| Vector pWPXLd | Addgene | 12258 | |
| Vector pMD2.G | Addgene | 12259 | |
| Vector psPAX2 | Addgene | 12260 | |
| DNase I (RNase-free) | New England Biolabs | M0303S | |
| Oligo(dT)18 Primer | Thermo Scientific | SO131 | |
| Anti-mouse secondary antibody (Anti-mouse IgG, HRP-linked Antibody) | Cell Signaling Technology | 7076S |
References
- Wang, E. T., et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 456 (7221), 470-476 (2008).
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J., Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 40 (12), 1413-1415 (2008).
- Wang, Z., Burge, C. B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 14 (5), 802-813 (2008).
- Matera, A. G., Wang, Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 15 (2), 108-121 (2014).
- Singh, R. K., Cooper, T. A. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 18 (8), 472-482 (2012).
- Wang, G. -. S., Cooper, T. A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 8 (10), 749-761 (2007).
- Li, Y. I., et al. RNA splicing is a primary link between genetic variation and disease. Science. 352 (6285), 600-604 (2016).
- Scotti, M. M., Swanson, M. S. RNA mis-splicing in disease. Nat Rev Genet. 17 (1), 19-32 (2016).
- Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 72 (1), 291-336 (2003).
- Graveley, B. R., Maniatis, T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol cell. 1 (5), 765-771 (1998).
- Del Gatto-Konczak, F., Olive, M., Gesnel, M. -. C., Breathnach, R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 19 (1), 251-260 (1999).
- Auweter, S. D., Oberstrass, F. C., Allain, F. H. Sequence-specific binding of single-stranded RNA: is there a code for recognition. Nucleic Acids Res. 34 (17), 4943-4959 (2006).
- Dong, S., et al. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J Biol Chem. 286 (30), 26732-26742 (2011).
- Filipovska, A., Razif, M. F., Nygård, K. K., Rackham, O. A universal code for RNA recognition by PUF proteins. Nat Chem Biol. 7 (7), 425-427 (2011).
- Wei, H., Wang, Z. Engineering RNA-binding proteins with diverse activities. Wiley Interdiscip Rev RNA. 6 (6), 597-613 (2015).
- Wang, Y., Cheong, C. -. G., Hall, T. M. T., Wang, Z. Engineering splicing factors with designed specificities. Nat Methods. 6 (11), 825-830 (2009).
- Choudhury, R., Tsai, Y. S., Dominguez, D., Wang, Y., Wang, Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 3, 1147 (2012).
- Wang, Y., et al. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 20 (1), 36-45 (2013).
- McCabe, B. C., Gollnick, P. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis. J Bacteriol. 186 (15), 5157-5159 (2004).
- Wang, Y., Ma, M., Xiao, X., Wang, Z. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol. 19 (10), 1044-1052 (2012).
- Choudhury, R., et al. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun. 5, (2014).
- Xiao, X., Wang, Z., Jang, M., Burge, C. B. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci U S A. 104 (47), 18583-18588 (2007).
- Wang, Z., Xiao, X., Van Nostrand, E., Burge, C. B. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 23 (1), 61-70 (2006).
- Li, M., Husic, N., Lin, Y., Snider, B. J. Production of lentiviral vectors for transducing cells from the central nervous system. J Vis Exp. (63), e4031 (2012).
- Tiscornia, G., Singer, O., Verma, I. M. Production and purification of lentiviral vectors. Nat Protoc. 1 (1), 241-245 (2006).
- Venables, J. P. Aberrant and alternative splicing in cancer. Cancer Res. 64 (21), 7647-7654 (2004).
- Cooke, A., Prigge, A., Opperman, L., Wickens, M. Targeted translational regulation using the PUF protein family scaffold. Proc Natl Acad Sci U S A. 108 (38), 15870-15875 (2011).
- He, C., Zhou, F., Zuo, Z., Cheng, H., Zhou, R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PloS one. 4 (3), 4732 (2009).
- Venables, J. P., et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 68 (22), 9525-9531 (2008).
- Shapiro, I., et al. An EMT-Driven Alternative Splicing Program Occurs in Human Breast Cancer. PLoS Genet. 7 (8), 1002218 (2011).
- David, C. J., Manley, J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24 (21), 2343-2364 (2010).
- Ladomery, M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell biol. 2013, (2013).
- Karni, R., et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 14 (3), 185-193 (2007).
- Anczukòw, O., et al. SRSF1-regulated alternative splicing in breast cancer. Mol cell. 60 (1), 105-117 (2015).

