Radionuclide-fluorescence Reporter Gene Imaging to Track Tumor Progression in Rodent Tumor Models
Summary
We describe a protocol for preclinical in vivo tracking of cancer metastasis. It is based on a radionuclide-fluorescence reporter combining the sodium iodide symporter, detected by non-invasive [18F]tetrafluoroborate-PET, and a fluorescent protein for streamlined ex vivo confirmation. The method is applicable for preclinical in vivo cell tracking beyond tumor biology.
Abstract
Metastasis is responsible for most cancer deaths. Despite extensive research, the mechanistic understanding of the complex processes governing metastasis remains incomplete. In vivo models are paramount for metastasis research, but require refinement. Tracking spontaneous metastasis by non-invasive in vivo imaging is now possible, but remains challenging as it requires long-time observation and high sensitivity. We describe a longitudinal combined radionuclide and fluorescence whole-body in vivo imaging approach for tracking tumor progression and spontaneous metastasis. This reporter gene methodology employs the sodium iodide symporter (NIS) fused to a fluorescent protein (FP). Cancer cells are engineered to stably express NIS-FP followed by selection based on fluorescence-activated cell sorting. Corresponding tumor models are established in mice. NIS-FP expressing cancer cells are tracked non-invasively in vivo at the whole-body level by positron emission tomography (PET) using the NIS radiotracer [18F]BF4–. PET is currently the most sensitive in vivo imaging technology available at this scale and enables reliable and absolute quantification. Current methods either rely on large cohorts of animals that are euthanized for metastasis assessment at varying time points, or rely on barely quantifiable 2D imaging. The advantages of the described method are: (i) highly sensitive non-invasive in vivo 3D PET imaging and quantification, (ii) automated PET tracer production, (iii) a significant reduction in required animal numbers due to repeat imaging options, (iv) the acquisition of paired data from subsequent imaging sessions providing better statistical data, and (v) the intrinsic option for ex vivo confirmation of cancer cells in tissues by fluorescence microscopy or cytometry. In this protocol, we describe all steps required for routine NIS-FP-afforded non-invasive in vivo cancer cell tracking using PET/CT and ex vivo confirmation of in vivo results. This protocol has applications beyond cancer research whenever in vivo localization, expansion and long-time monitoring of a cell population is of interest.
Introduction
Metastatic disease is the cause for most cancer-related deaths 1. Despite extensive research into metastatic processes, reliable monitoring of cancer metastasis in animal model systems is difficult to achieve. Recent advances in whole-body imaging technologies and multi-modal imaging approaches have enabled non-invasive in vivo cell tracking2,3,4,5. The latter can be used as a tool to monitor the presence, distribution, quantity, and viability of cells, non-invasively and repeatedly in a live animal or a human.
The purpose of the method described here is to longitudinally and non-invasively track cancer cells in 3D in living rodent tumor models. Using this method, researchers will be able to accurately quantify tumor progression including metastatic spread in 3D. Compared to traditional non-imaging-based techniques, this method offers the acquisition of quantitative data with largely reduced animal numbers. Another feature of this method is that it allows correlation of in vivo imaging with streamlined downstream ex vivo analysis of the tracked cells in harvested tissues by histology or cytometry3,6.
The rationale for the development of this method was to provide an in vivo tool for the monitoring and quantification of the whole metastatic process in rodent tumor models. Importantly, it was designed to minimize animal use while at the same time reducing inter-animal variability. Longitudinal non-invasive whole-body imaging is excellently suited to inform on metastatic outgrowth, for which per se it is difficult to predict accurately the time and location of its occurrence. Whole-body 3D imaging has therefore been at the center of method development. To close the scale gap between whole-body in vivo imaging and potential downstream ex vivo histological confirmation, a multi-scale imaging approach based on a dual-mode radionuclide-fluorescence reporter was adopted3,6.
Positron emission tomography (PET) is the most sensitive 3D whole-body imaging technology currently available offering excellent depth penetration and absolute quantification7 with a resolution of <1 mm8,9. Currently, preclinical cell tracking on the whole-body level by radionuclide imaging can reliably detect cells at densities of ~1,000 cells per volume of a million cells3,6 with resolutions in the sub-millimeter region. Unlike intravital microscopy, it cannot detect single spreading cancer cells, but it does not require surgical procedures (e.g. window chambers), is not limited to a small field of view, and not by low tissue penetration and scatter. Bioluminescence imaging provides an inexpensive alternative, but is associated with scatter and light absorption issues as well as poor depth penetration, and consequently severely limited in quantification2. Fluorescence whole-body imaging has been used for the acquisition of 3D images, but it is much less sensitive compared to bioluminescence or radionuclide technologies2. Nonetheless, fluorescence offers the opportunity to perform ex vivo downstream tissue analysis by cytometry or microscopy. The latter closes the scale-gap between macroscopic whole-body imaging (mm resolution) and fluorescence microscopic tissue analysis (µm resolution)3. Therefore, radionuclide and fluorescence modalities complement each other, ranging from the whole-body level to the (sub-)cellular scale.
Reporter gene imaging is ideally suited for prolonged cell tracking as required in metastasis research. In this application it is superior to direct cell labeling as it is (i) not affected by label dilution and thus not limited in tracking time, and (ii) better reflects live cell numbers. Consequently, whole-body cell tracking is particularly useful for applications in which traceable cells proliferate or expand in vivo, for example in cancer research3,6, for the detection of teratoma formation in stem cell research, or for the quantification of immune cell expansion5.
Various radionuclide-based reporter genes are available2. These include enzymes such as the herpes simplex virus HSV11 thymidine kinase (HSV1-tk), transporters such as the sodium iodide symporter (NIS) or the norepinephrine transporter (NET), as well as cell surface receptors such as the dopamine D2 receptor (D2R). NIS is a glycosylated trans-membrane protein that actively mediates iodide uptake, for example in follicular cells in the thyroid gland for the subsequent synthesis of thyroid hormones10. This process is driven by the symport of Na+ and relies on the cellular sodium gradient, which is maintained by the Na+/K+-ATPase11. Consequently, NIS better reflects living cell numbers than other reporters as iodide/radiotracer uptake is linked to an active Na+/K+ gradient rather than the mere presence of the transporter. Traditionally, radioiodide has been used for NIS imaging. For cell tracking, alternative NIS radiotracers that are not metabolically entrapped in the thyroid have been reported to be superior6. This recently developed PET radiotracer [18F]tetrafluoroborate ([18F]BF4–)12,13 shows superior pharmacokinetics as compared to radioiodide6 while being available at high specific activities14 without the need for complex radiochemistry facilities. [18F]BF4– can be synthesized via two different ways. The first method is based on isotope exchange of non-radioactive 19F in BF4– with radioactive 18F12. The second method is through addition of 18F to non-radioactive boron trifluoride14. The latter method was reported to yield higher specific activities14 and is the method of choice for preclinical imaging.
NIS is highly expressed in the thyroid tissues. It is also expressed in the salivary, lachrymal and lactating mammary glands as well as the stomach, but at lower levels as compared to the thyroid gland10. Therefore, excellent contrast imaging in other body regions can be achieved using NIS. It is also highly homologous between human, rat and mouse10. Moreover, there are no reports of toxicity upon ectopic NIS expression in non-thyroidal cells. Importantly, NIS has also not been associated with host immune responses, neither in humans nor in rodents. NIS has been used as a reporter gene to measure promoter activity15,16,17 and gene expression18,19,20,21,22,23 in several different contexts. It has also been used for non-invasive imaging of gene therapy vectors24,25, and to track cells in cardiac4, hematopoietic26, inflammation5, and neural studies27. Recently, NIS has also been used as a reporter gene to track cancer metastasis in vivo3,6.
In summary, the main advantages of this method over previous techniques are: (i) highly sensitive non-invasive 3D in vivo localization and quantification of metastatic spread, (ii) automated production of [18F]BF4– at high molar activities, (iii) a significant reduction in required animals through longitudinal imaging, (iv) the acquisition of paired data from subsequent imaging sessions resulting in improved statistical data, which in turn further reduces animal use, and (v) the intrinsic option for ex vivo confirmation of cancer cells in tissues by cytometry or fluorescence microscopy.
Protocol
This protocol meets all requirements set by United Kingdom (UK) legislation and the local Ethical Review panel. When following this protocol, ensure the procedures also meet all requirements dictated by national legislation and local Ethical Review panel. Ensure every experiment involving radioactivity is compliant with legislation and local rules and performed safely.
1. Engineering and characterization of cancer cells to express the radionuclide-fluorescence fusion reporter NIS-FP
NOTE: For simplicity, mEGFP A206K is abbreviated as "GFP", and mCherry as "RFP" in the subsequent sections of this protocol.
- Generation of lentiviral particles
- To produce lentivirus particles, co-transfect 293T cells with the following four plasmids using a suitable transfection method: (i) the reporter gene encoding plasmid (pLNT SFFV NIS-GFP or pLNT SFFV NIS-RFP (see Supplementary Information), (ii) the third-generation lentiviral packaging plasmids pRRE and (iii) pRSV-Rev, and (iv) a virus envelope containing plasmid, e.g, pMD2.G. Pre-mix plasmids before adding them to the transfection mix. Perform transfection in a cell culture hood.
NOTE: Additional transfection information is provided in Supplementary Information. - Assess transfection success after 48 h by standard wide-field fluorescence microscopy with filter settings appropriate for the chosen fusion reporter (NIS-GFP or NIS-RFP).
NOTE: Fluorescence signals are indicative of reporter gene transfection and therefore only a surrogate for successful co-transfection, not an indicator of successful virus production. - Harvest the virus particle-containing supernatant using a syringe and remove floating cells and cell debris by filtering through a 0.45 µm sterile polyethersulfone (PES) filter. Transfer to a sterile 1.5 mL polypropylene reaction tube. Perform virus work in a cell culture hood and ensure no live virus leaves the contained environment.
- To produce lentivirus particles, co-transfect 293T cells with the following four plasmids using a suitable transfection method: (i) the reporter gene encoding plasmid (pLNT SFFV NIS-GFP or pLNT SFFV NIS-RFP (see Supplementary Information), (ii) the third-generation lentiviral packaging plasmids pRRE and (iii) pRSV-Rev, and (iv) a virus envelope containing plasmid, e.g, pMD2.G. Pre-mix plasmids before adding them to the transfection mix. Perform transfection in a cell culture hood.
- Transduction and selection of NIS-FP expressing cancer cell lines
- Use pure fresh virus from step 1.1.3 mixed 1:1 (v/v) with the optimal growth medium of each cancer cell line (DMEM for MDA-MB-231 cells and RPMI 1640 for 4T1 cells; see Materials Table for media composition). Perform transduction in a cell culture hood.
NOTE: A more general protocol is referred to in Supplementary Information. - Transduce cancer cells with virus-containing medium in an incubator with humidified atmosphere containing 5% (v/v) CO2 at 37 °C for 72 h (work on 6-well or 12-well scale using 1 mL or 0.4 mL of the virus mixture from step 1.2.1).
- Monitor target cell NIS-FP expression by fluorescence microscopy.
- Expand successfully transduced cells to a scale of 3-10 million cells using standard culture conditions (see ATCC for MDA-MB-231 and 4T1 cell lines).
- Use fluorescence-activated cell sorting (FACS) to purify NIS-FP expressing cells from non-transduced cells.
NOTE: FACS can be a source of mycoplasma infections; it is recommended to check for mycoplasma before virus production and transduction but also after FACS and before step 1.3.
- Use pure fresh virus from step 1.1.3 mixed 1:1 (v/v) with the optimal growth medium of each cancer cell line (DMEM for MDA-MB-231 cells and RPMI 1640 for 4T1 cells; see Materials Table for media composition). Perform transduction in a cell culture hood.
- Characterization NIS-FP expressing cancer cell lines
- Confirm reporter expression by standard flow cytometry as described elsewhere28.
- Confirm reporter integrity by standard immunoblotting as described elsewhere29.
- Analyze intracellular fusion reporter localization by confocal fluorescence microscopy.
NOTE: Staining with or co-expression of a plasma membrane marker6 and subsequent co-localization analysis30 will facilitate this step. - Analyze radiotracer uptake in NIS-FP expressing cell lines.
NOTE: Any radioactive NIS substrate compatible with existing equipment is suitable, e.g. the iodide isotopes (123I–, 124I–, 125I–, 131I–), 99mTcO4–, 188ReO4–, [18F]SO3F– or [18F]BF4–.- Seed 106 purified cells in 6-well plates in their optimal growth medium (see Table of Materials) one day prior to the experiment. Prepare all samples in triplicate and include control samples: (i) "specificity controls", i.e. NIS-FP expressing cells pre-incubated with a competitive NIS substrate to test uptake specificity; (ii) "parental cell controls", i.e. cells not expressing NIS-FP but receiving radiotracer to test basal uptake in parental cells.
- On the next morning, wash cells once with serum-free growth medium.
- Incubate the cells in serum-free growth medium in the presence of 50 kBq 99mTcO4– or [18F]BF4– for 30 min at 37 °C (1 mL total volume). For specificity controls, pre-incubate the cells for 30 min with the competitive substrate NaClO4– (12.5 µM final concentration). Keep the competitive substrate concentration constant throughout the experiment.
- Collect the supernatant and transfer 100 µL to a prepared collection tube labeled "supernatant".
- Wash the cells twice with 1 mL ice-cold phosphate-buffered saline (PBS) containing Ca2+/Mg2+. Collect each wash solution and transfer 100 µL of each into a prepared collection tube labeled "wash1" or "wash2", respectively.
- Lift the cells by adding 500 µL PBS containing 0.25 % (w/v) trypsin and 0.53 mM EDTA, and incubating at 37 °C until the cells detach (check visually using a microscope). Transfer the suspension into a prepared collection tube labeled "cells". Wash the wells with 500 µL ice-cold PBS containing Ca2+/Mg2+ and add to the tube "cells". Pellet the cells by centrifugation (250 x g, 4 min, 4 °C).
- Count all four sample types from each well using a gamma counter appropriately set for the radioisotope of choice (here: 99mTc or 18F).
NOTE: Due to the large number of samples in this assay, it is recommended to use an automated γ-counter capable of automatic decay correction. - Analyze data by summing obtained gamma counts of samples from each well to determine a total radioactivity count per well.
NOTE: take aliquoting in steps 1.3.4.4 and 1.3.4.5 into account by multiplying numbers by 10 for "supernatant", and "wash" fractions. - Express cellular radiotracer uptake as %Uptake as indicated in Equ.1. Calculate averages and standard deviations from triplicate experiments.
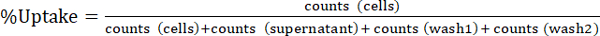 (Equ.1)
(Equ.1)
NOTE: Here, reporter validation experiments are described, but non-reporter-related cellular functions (e.g. proliferation, cancer cell invasion, gene expression etc.) are ultimately application-specific and the responsibility of each user.
2. Establishment of in vivo tumor models
- Use only fully characterized and validated cells for in vivo experiments. Check fluorescence of cells prior to administration by any suitable technique (e.g. fluorescence microscopy, flow cytometry) on every occasion.
- Establish the tumor model in 5-6 weeks old young-adult female mice. Use BALB/cAnNCrl or BALB/cAnN.Cg-Foxn1nu (BALB/c Nude) mice for the 4T1 tumor model and NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ (NSG) mice for the MDA-MB-231-based tumor model. Shave animals locally and use aseptic technique. Directly inject 50 µL of a suspension containing 106 NIS-FP expressing cancer cells/mL into the mammary fat pad between the fourth and the fifth nipple31.
NOTE: To improve injection accuracy, it is recommended performing the injection under general anesthesia using an inhalable anesthetic such as isoflurane (1-2 % (v/v) in O2).
NOTE: Surgical implantation of tumor pieces from NIS-FP expressing tumors is an alternative approach to tumor model establishment31. - Check fluorescence of cells at injection sites after administration and in the early days post injection. Use a fluorescence torch and filter glasses suitable for the FP of choice.
- Monitor tumor growth and check for any clinical signs (particularly at later time points).
NOTE: For superficial tumors, i.e. the orthotopic breast tumors in this protocol, use calipers and the formula for the mean tumor diameter (MTD) = ½·(L+W)32.
3. Production of [18 F]BF 4 – using an automated radiotracer synthesis (ARS) platform.
NOTE: Here, the automated [18F]BF4– synthesis based on the method of 18F addition to boron trifluoride is described. Users of a more widely available ARS platform (see Table of Materials), can download the corresponding Extensible Markup Language (XML) file required to run the automated sequence on this platform (Supplementary File). A detailed explanation of the cassette layout shown in Figure 1 is provided in (Table 1) as well as a detailed description of each step in the XML sequence file (Table 2) to support translation to any other automated platform.
- Set up the ARS platform as described in Table 1 and ensure it is operational with the correct XML file loaded onto the control computer. Ensure the ARS platform is placed in a chemical hood suitable for safe work with GBq amounts of radioactivity.
- Aspirate the cyclotron-produced [18F]F– (typically 1.5-2 GBq in 2-7 mL [18O]H2O) via the inlet reservoir (V6).
- Trap the radioactivity on an anion exchange resin (e.g. quaternary ammonium anion exchange cartridge; V5 to V4), recover the [18O]H2O in a separate vial (V1).
- Elute the [18F]F– (reverse elution, V5 to V4) into the reactor (V8) with 750 µL of 0.9 % (w/v) saline solution (V2), followed by 1.5 mL of acetonitrile (V16).
- Remove the water by azeotropic evaporation under vacuum and nitrogen flow by heating at 105 °C then 120 °C for 5 min.
- Reduce the reactor temperature to 80 °C.
- Add 800 µL of 15-crown-5 in anhydrous acetonitrile (46 mg, 0.21 mmol) to the dry [18F]F– through the central port of the reactor (V8).
- Add 850 µL of BF3.OEt2 in anhydrous acetonitrile (0.16 mg, 1.13 µmol) to the reactor.
- React for 5 min while returning to room temperature.
- Pass the reaction mixture through an aluminum oxide cartridge (V17 to V18) to trap the trifluoroborate.
- Return the reaction mixture to syringe S2 and dilute with water (approx. 1.6 mL).
- Pass the reaction mixture through a second anion exchange cartridge (V19 to V20) to trap the [18F]BF4– product.
- Wash the reactor with water (approx. 5.5 mL).
- Pass the resulting solution through the alumina and second anion exchange cartridges.
- Rinse syringes S2 and S3 with water.
- Wash the second anion exchange cartridge with water and dry it with nitrogen gas.
- Elute the product (V19 to V20) with 1 mL of 0.9 % NaCl (V14) into syringe S3.
- Transfer the product (400-500 µL) via the outlet line (V21) to a 1 mL glass collection vial.
NOTE: Molar activity is an important aspect of every radiotracer. However, its routine determination is not only time-consuming but also requires significant amounts of the freshly synthesized [18F]BF4–, such that it becomes a limiting factor for the number of animals that can be imaged. To test reproducibility and molar activities of the resultant [18F]BF4–, it is recommended to schedule a few dedicated test-runs for this purpose. For more details about molar activities, please refer to the Discussion section.
4. In vivo imaging of NIS-FP expressing cells by nanoPET/CT
- Animal preparation
- Anesthetize mice with 1.5-2.0 % (v/v) isoflurane in O2 at a flow rate of 1.0-1.5 L/min in an induction chamber. To check for sufficient anesthesia look for absence of the pedal reflex.
- Ensure to apply vet ointment on animal eyes to prevent dryness while under anesthesia
- Move the mouse onto a heating pad with the nose in an anesthetic supply mask and warm the tail (e.g. by dipping it into 37 °C water or using an infrared light lamp).
- Dilute the freshly prepared sterile-filtered [18F]BF4– solution to 5 MBq per 50 µL with 0.9 % sterile saline.
- Using a syringe connected to a hypodermic needle (gauge 29-31), draw 100 µL of the [18F]BF4– solution, measure the radioactivity in the syringe, and note the value and the time of the measurement.
- Intravenously administer 50 µL of the [18F]BF4– solution into the pre-warmed tail vein.
- Measure the remaining radioactivity in the syringe and note the value and the time of the measurement. The difference between values measured at steps 4.1.7 and 4.1.5 is the injected dose (ID).
- Set a timer to count down from 45 min (start time of PET imaging = 0 min).
- Place the mouse onto the bed of the nanoPET/CT scanner and ensure the anesthetic supply is correctly re-attached.
- Check anesthesia remains complete by testing for absence of the pedal reflex.
- Ensure the mouse is positioned on the bed in the desired way, e.g. the 'sphinx'-like position.
- Install animal monitoring devices according to the manufacturer's recommendations, e.g. a rectal temperature probe, a probe measuring animal breathing, or electrodes for recording electrocardiograms. Check the proper function of all instruments.
NOTE: For in vivo specificity tests with the NIS radiotracer [18F]BF4–, animals are imaged as described above, and then rested awake until the radioactivity has decayed sufficiently to be regarded as negligible, e.g. 48 h later when only 1.3·10−6 % residual 18F radioactivity will be present in the animal. In the subsequent imaging session, the competitive substrate ClO4– is administered at a dose of 200 mg/kg 30 min prior to radiotracer administration, and imaging is performed as described above.
- Imaging by nanoPET/CT
- Set the desired CT imaging parameters, e.g. using the nanoPET/CT 55 kVp tube voltage, set the exposure time to 1200 ms with one-degree angular stepping and 180-degree projections.
- Set the parameters for PET image acquisition. Use static scan PET parameters with a duration of 30 min, 1:5 coincidence mode and 400-600 keVp energy window.
- At countdown time = 15 min start CT image acquisition.
- At countdown time = 0 min start PET image acquisition.
- If serial animal imaging is required, let animals fully recover from anesthesia, i.e. regain consciousness under supervision. Subsequently, transfer them to a maintenance unit.
- If this is the terminal imaging session, proceed to animal euthanasia by either anesthetic overdose, rising concentration of carbon dioxide, or dislocation of the neck.
5. In vivo data analysis
- Reconstruct the PET/CT data using a Monte Carlo-based full 3D iterative algorithm. Ensure that corrections for attenuation, dead time, and radioisotope decay are considered. For details refer to the manufacturer's instructions of the PET/CT instrument in use.
- Check CT and PET images are correctly co-registered and save the data in a suitable exchange format such as 'Digital Imaging and Communications in Medicine' (DICOM).
- Analyze images
- Load the reconstructed DICOM files into a suitable image analysis software that enables the recognition and delineation of regions of interest (ROIs) and subsequent PET signal quantification in these ROIs.
- Segment the ROIs using manual or adaptive thresholding to define ROIs33,34 using a suitable software package. Anatomical image information from the CT scan helps guide ROI assignment, e.g. superficial tumors or lung volumes.
- Use the analysis software as per manufacturer's instructions and ensure data are calibrated to the injected radioactivity dose and corrected for attenuation and radioactive decay.
- Draw graphs showing data from this in vivo quantification. Express data as either percent injected dose/volume (%ID/mL) or standard uptake value (SUV), which is an alternative measure considering the radioactivity in the whole body of the subject.
- Calculate %ID/g values assuming the tissue density to be like water, i.e. ~1 g/L. It is noteworthy that this assumption can be invalid for organs with significantly different densities, such as lung or bone.
- Calculate different SUVs to estimate the true SUV (e.g. SUVmean, SUVmax); SUVmax is more reliable for small objects and is more frequently used than SUVmean35.
6. Ex vivo analyses
Perform the listed downstream analyses: (i) fluorescence imaging of organs containing fluorescent cancer cells (primary tumor and metastases) during animal dissection, (ii) measurement of radiotracer tissue distribution, and (iii) histologic or (iv) cytometric assessment of cancerous organs.
- Measurement of radiotracer distribution by γ-counting (ex vivo biodistribution) and ex vivo fluorescence imaging of cancerous tissues.
- Measure radioactivity of the whole dead animal and note the value and the time.
- Dissect the animals and harvest the following tissues: lung, heart, blood (using 20 mm glass capillaries), liver, stomach, kidneys, spleen, small and large intestines, thyroid and salivary glands, a piece of muscle from the leg, and bone of the rear femurs, and relevant and dissectible lymph nodes and cancerous tissues.
- Measure the radioactivity of the remaining carcass first including then excluding the tail and note the values and the times of measurement.
NOTE: Radioactivity in the tail can be considered stemming from radiotracer that was mis-injected and thus did not reach circulation; hence, this amount of radiotracer was not contributing to the injected dose. The tail radioactivity serves also as a retrospective parameter of injection quality. - Weigh all tissues (use pre-weighed tubes).
- Take photographs of cancerous organs in daylight and under fluorescence light.
NOTE: Use a camera stand to keep the distance between camera lens and organ constant (or use a dedicated commercial instrument for this purpose). - Embed organs/tissues intended for downstream histology into OCT or immerse them in formalin for fixation. For other downstream applications sample preparation can differ.
- Prepare radiotracer calibration standards in duplicate, e.g. 0 to 1000 kBq [18F]BF4–.
NOTE: Calibration standards are required to (i) relate the measured counts per min to radioactivity values (in kBq), and (ii) simplify decay correction; 18F– can replace [18F]BF4–. - Count the radioactivity of all harvested tissues using a γ-counter together with radioactivity calibration standards from step 6.1.7. Note the time of measurement. If count rates are too high (i.e. outside of linearity of calibration standard or indicated by too high detector dead times), re-count the samples two radiotracer half-lives later.
- Present data either as %ID/g or standard uptake values (SUV) (Equ.2).
SUV =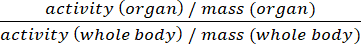 (Equ.2)
(Equ.2) - Discard all harvested tissues that are not required for further downstream analyses according to local waste management rules.
- Analyze cancerous tissues by cytometry or histology according to the user's preferences and standard protocols (as described elsewhere3,6,28).
Representative Results
The first step requires genetic engineering of the cancer cells of interest. Here, the results of lentiviral transduction of metastatic murine inflammatory 4T1 breast cancer cells and human metastatic MDA-MB-231 cells with lentivirus particles carrying DNA encoding either NIS-GFP or NIS-RFP are shown. Transduction efficiencies varied between cancer cell lines (Figure 2A, left column). However, all resultant transduced cancer cell lines were selected by FACS to purity (Figure 2A, right). Confocal fluorescence microscopy (Figure 2B) demonstrated correct plasma membrane localization of NIS-FPs. NIS-FP function was quantified using NIS-afforded radiotracer uptake (Figure 2C-2E) and demonstrated NIS function and specificity. Notably, no significant differences between 4T1.NIS-GFP and 4T1.NIS-RFP expressing cell lines with similar NIS expression levels were found (Figure 2C).
Following full in vitro cell line characterization, tumor models were set up with the newly generated traceable cancer cell lines. As an example, the 4T1.NIS-GFP tumor model, a model for inflammatory breast cancer, is shown here (Figure 3). In tumor-bearing animals longitudinal whole-body PET imaging then informed on tumor progression including metastatic spread (Figure 3B). The PET radiotracer [18F]BF4– was necessary for imaging and freshly produced in the morning of every PET imaging session. Synthesis of [18F]BF4– was performed using the described ARS method. Typically, ~1.6 GBq 18F– was used as input and obtained ~244 MBq [18F]BF4– in 40.5±3.9 min (N=17). The product was analyzed by radio thin layer chromatography or ion chromatography and showed a radiochemical purity of 94.7±1.4 %. The radiochemical yield was 19.4±4.0 % (decay-corrected).
On day 19 after tumor inoculation, the primary tumor was clearly identified using PET, but found no metastases. Ten days later (day 29), the same tumor-bearing mice were re-imaged and distant metastasis at various locations in all animals (lung metastasis, metastasis to various inguinal and/or axillary lymph nodes) were identified. The example in Figure 3 showed extensive lung metastasis with several clearly identifiable and quantifiable nodules in the lung (Figure 3B-3E). Moreover, the animal presented with regional spread of the tumor into the peritoneal wall as well as metastasis to the inguinal and both axillary lymph nodes. %ID values of individual metastases in the lung (Figure 3E) differed widely, but so did the occupied volumes of the underlying metastatic nodules. In contrast, volume-normalized %ID/mL values (Figure 3E) were much more uniform. This was comprehensible for different metastases at similar development stages (i.e. developed between days 19 and 29; Figure 3B). In contrast, the normalized %ID/mL value for the primary tumor was lower than those for the lung metastases, which is in line with a tumor mass that had more time to progress and remodel including the influx of other cell types (stromal cells, immune cells), particularly in this model of inflammatory breast cancer.
Guided by in vivo images and the fluorescence of cancer cells (visible during animal dissection under fluorescence light), small deep-seated organs such as lymph nodes were reliably harvested and, at the same time, assessed for cancerous nodule content (Figure 4A). While the fluorescence signal during animal dissection was indicative of tumor cell presence, it was important to ensure this classification was accompanied by ex vivo radioactivity measurements of the harvested tissues. Figure 4B shows the standard uptake values (SUV) obtained for the various tissues across a cohort of three animals, all of which presented with metastasis. Endogenously NIS-expressing organs such as the thyroid and salivary glands (harvested combined) or the stomach also showed the expected high radiotracer uptake. Furthermore, this NIS-FP approach allowed straightforward cancer cell identification during histology (Figure 4C). This immunofluorescence histology example data showed tumor vascularization in the 4T1.NIS-GFP tumor model. This data also showed that the NIS-GFP reporter resided predominantly in the plasma membranes of the tumor cells also in vivo (Figure 4C), thereby validating the uptake results.
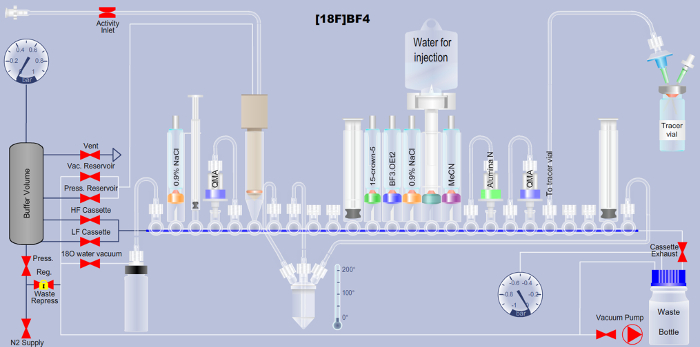
Figure 1. Scheme detailing the set-up of the automated radiotracer synthesis platform for the production of [18F]BF4– via the fluorine-18-to-boron trifluoride addition method. Reagent names are printed onto the respective tubes in the scheme. QMA is the abbreviation for quaternary ammonium anion exchange, and indicates the used solid-phase chromatographic separation material. Additional details are available in Tables 1 and 2. Please click here to view a larger version of this figure.
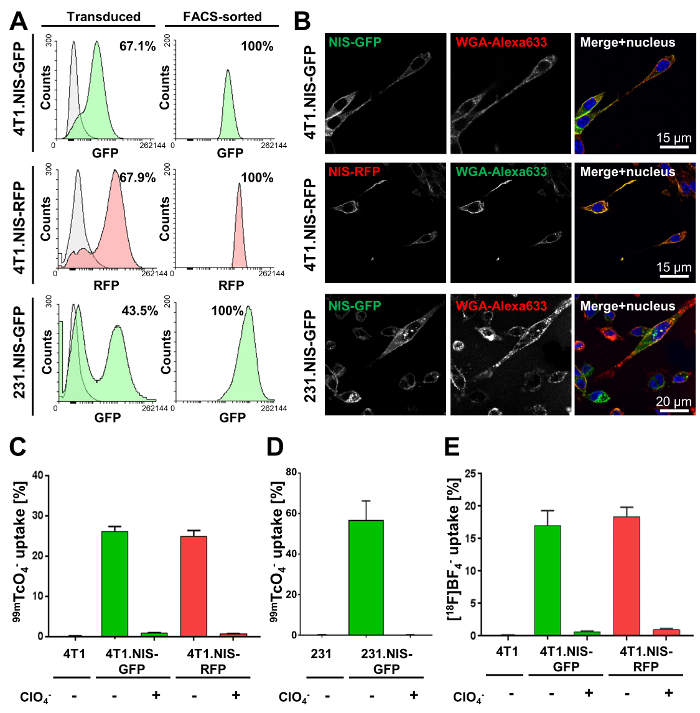
Figure 2. Typical characterization results of cancer cell lines stably expressing NIS-GFP or NIS-RFP. (A) The indicated cell lines were made using lentiviruses transferring either NIS-GFP or NIS-RFP. The left column shows the transduced population (green or red fluorescent) as compared to the respective parental cells (grey; 4T1 and MDA-MB-231 cells, respectively). Percentages show transduction efficiencies as determined by flow cytometry. The right column shows the results of flow cytometric analyses after FACS purification of the mixed populations in the left column. All cell lines were found to be >99 % pure for indicated NIS-expressing cells (by flow cytometry). (B) Confocal fluorescence microscopy of purified cell lines shows plasma membrane localization of NIS-GFP or NIS-RFP in the respective cell lines. WGA-Alexa633 was used as a plasma membrane marker. (C, D) Functional validation of NIS-FP protein expressed in the indicated newly generated cancer cell lines. NIS function was measured using the radiotracer 99mTcO4– (50 kBq per million cells). As controls, parental cells were used as well as fusion reporter expressing cells that were treated with the NIS co-substrate perchlorate before and during the assay (specificity control). Results clearly demonstrate NIS-FP function and specificity in all cell lines. (E) Functional validation of 4T1.NIS-FP cell lines using [18F]BF4– as a radiotracer for NIS. All other conditions were identical to (C). Importantly, very similar relative uptake results were obtained for both 4T1-derived cell lines with both radiotracers (Figure 2C and E), thereby justifying the interchangeable use of both for in vitro functional characterization of NIS-FP expressing cell lines. Please click here to view a larger version of this figure.
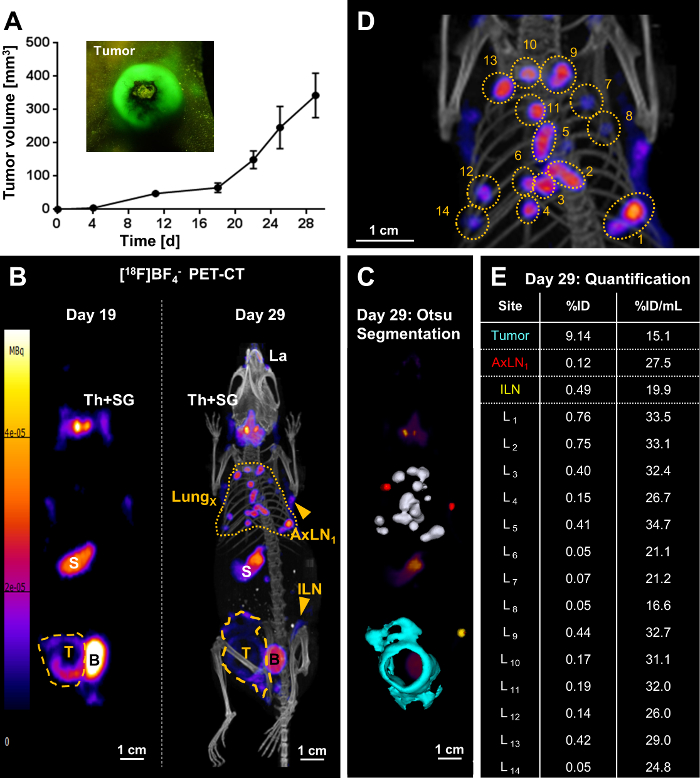
Figure 3. Representative result of metastasis tracking by [18F]BF4–-PET/CT imaging in a mouse bearing a 4T1.NIS-GFP tumor. (A) One million 4T1.NIS-GFP cells were injected into the mammary fat pads of 5-6 weeks old BALB/c CanN.Cg-Foxn1nu/Crl mice and tumor growth was followed over time using calipers. Owing to the GFP fluorescence of the cancer cells, crude visual identification/growth assessment was also possible using a fluorescence torch and suitable filter glasses (see inset). (B/left) On day 19 post tumor inoculation, the primary tumor (yellow dashed line) was clearly identified but no metastasis. The image presented is a maximum intensity projection (MIP) of the PET image. Endogenous NIS signals (white descriptors) were also recorded, i.e. the thyroid and salivary glands (Th+SG), the stomach (S), and, at very low levels, some parts of the mammary and lachrymal glands. The bladder (B) signal stems from tracer excretion. (B/right) On day 29 post tumor inoculation, metastasis was clearly identified: multiple metastases in the lung (yellow dotted line) as well as metastatic lymph nodes (ILN, AxLN; yellow arrowheads). The image presented is a MIP of the PET/CT image. The primary tumor (yellow dashed line) grew not only in a globular shape at this time point, but also had invaded into the peritoneal wall. (C) A 3D implementation of the Otsu thresholding technique enabled 3D surface rendering of the cancerous tissues; these are superimposed onto a PET MIP. Lung metastases are shown in white, metastatic axillary lymph nodes in red, the metastatic inguinal lymph node in yellow, and the primary tumor that invaded into the peritoneal wall in turquoise. (D) A blow-up image of the PET/CT MIP in (B/right) to indicate individual lung metastases. (E) Radiotracer uptake into cancerous tissues was quantified from 3D images (%ID) and normalized by their respective volumes (%ID/mL). Individual lung metastases correspond to the numbering in (D). Please click here to view a larger version of this figure.
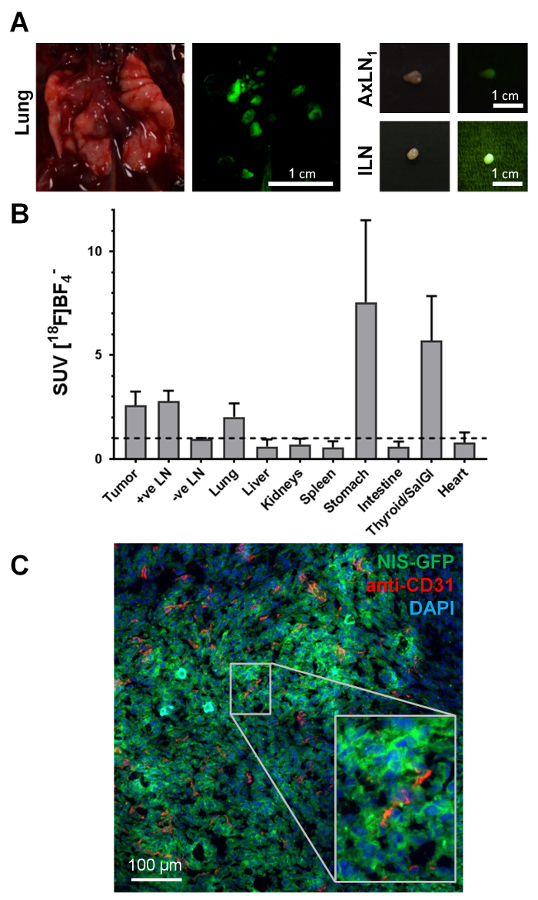
Figure 4. Typical examples of ex vivo data accessible from NIS-FP tumor-bearing mice. (A) During tissue harvesting for downstream analyses, the fluorescent properties of the NIS-FP expressing tumor cells served as an indicator guiding animal dissection. As exemplars, tissues from the animal in Figure 3, i.e. the lung with several metastatic lesions and two positive lymph nodes are shown. Daylight photography as well as fluorescence images are shown. The fluorescence images were taken with the same camera as the daylight images but under blue light excitation (450±10 nm bandpass filter) with a green emission filter (530±30 nm bandpass filter) placed in front of the camera lens. (B) Distribution of radiotracer in different organs ('biodistribution') of animals with 4T1.NIS-GFP tumors (N=3; day 29 post tumor inoculation; 5 MBq [18F]BF4–). Standard uptake values (SUV) were calculated and values >1 indicate specific accumulation of radiotracer in the respective organs. The data show specific radiotracer uptake in cancerous tissues, i.e. primary tumor, metastatic lymph nodes (as identified by imaging and dissection under fluorescence light), lung (was dissected as a whole without separating individual metastases), as well as organs endogenously expressing NIS, i.e. thyroid and salivary glands and stomach. (C) Immunofluorescence histology of the primary tumor from the same mouse as shown in Figure 3. The primary tumor was harvested, embedded in OCT and frozen before being sectioned (10 µm) and processed for staining. NIS-GFP expressing cancer cells were directly identified without the need for antibody staining. Blood vessels were stained with a rabbit antibody against mouse PECAM-1/CD31 (2 µg/mL) and a Cy5-conjugated goat anti-rabbit secondary antibody. Nuclei were stained with 2'-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5'-Bi-1H-benzimidazole (1 µg/mL) and the sample mounted in poly(vinyl alcohol – vinyl acetate) containing 2.5 % (w/v) Dabco as an antifade. Confocal images were obtained using a confocal microscope with settings appropriate for 2'-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5'-Bi-1H-benzimidazole, GFP and Cy5. These example data clearly show that the 4T1.NIS-GFP tumor is vascularized but also that vascularization differs in its extent (cf. top left with bottom middle). It also shows that the NIS-GFP reporter predominantly resides in the plasma membranes of the tumor cells in vivo (inset), thereby validating the in vitro uptake results. Please click here to view a larger version of this figure.
Supplemental Information. Please click here to download this file.
Supplemental File. Please click here to download this file.
| FASTlab manifold valve | Reagent, solvent, cartridge or tubing* | Details | ||
| V1 | Silicone tubing to [18O]H2O waste bottle | 14 cm | ||
| V2 | 0.9% NaCl solution, 750 µL | 11 mm vial | ||
| V3 | Syringe S1 | 1 mL | ||
| V4 | anion exchange cartridge C1, pre-conditioned with 1M NaCl (10 mL) and H2O (10 mL) | e.g. Sep-Pak Accell Plus QMA Plus Light (Waters, cat. No. WAT023525) | ||
| V5 | Silicone tubing to anion exchange cartridge C1 | 14 cm | ||
| V6 | [18O]H2O/18F inlet reservoir | Max 5 mL | ||
| V7 | Silicone tubing to reactor vessel (left side; gas inlet) | 14 cm | ||
| V8 | Silicone tubing to reactor vessel (central port; liquid inlet/outlet) | 14 cm | ||
| V9 | Closed | |||
| V10 | Closed | |||
| V11 | Syringe S2 | 5 mL | ||
| V12 | 15-crown-5, 46 mg in 800 µL MeCN | 11 mm vial | ||
| V13 | Trifluoroborate diethyl etherate, 0.14 µL in 850 µL MeCN (dilute 14 µL of BF3.OEt2 with 1 mL MeCN. Dilute 10 µL of this solution to 850 µL with MeCN). | 13 mm vial | ||
| V14 | 0.9% NaCl solution, 1 mL | 13 mm vial | ||
| V15 | Water bag spike | |||
| V16 | Acetonitrile (MeCN), 1.5 mL | 13 mm vial | ||
| V17 | Silicone tubing to Alumina neutral cartridge C2 | 14 cm | ||
| V18 | Alumina neutral cartridge C2, pre-conditioned with H2O (10 mL), acetone (10 mL) and air (20 mL) | e.g. Sep-Pak Alumina N Plus Light (Waters, cat. No. WAT023561) | ||
| V19 | Silicone tubing to anion exchange cartridge C3 | 14 cm | ||
| V20 | anion exchange cartridge C3, pre-conditioned with 1 M NaCl (10 mL) and H2O (10 mL) | e.g. Sep-Pak Accell Plus QMA Plus Light (Waters, cat. No. WAT023525) | ||
| V21 | Silicone tubing to collection vial | 40 cm | ||
| V22 | Closed | |||
| V23 | Closed | |||
| V24 | Syringe S3 | 5 mL | ||
| V25 | Silicone tubing to reactor vessel (right side; vacuum port) | 40 cm | ||
| *NOTE: Because of the plastic spikes, the dead volume for 11 mm vials and 13 mm vials is approximately 0.35 mL and 0.4 mL, respectively. Therefore, the actual amounts of reagents transferred to the reactor are slightly different. All quantities indicated in this method refer to the actual amounts introduced in each reagent vial. | ||||
Table 1. Description of the cassette layout for the automated [18F]BF4– synthesis via the fluorine-18-to-boron trifluoride addition method (cf. Figure 1).
| Sequence steps | Comment | ||||
| [1-2] | Pressurize the system and flush the manifold with N2 | ||||
| [3-15] | Rinse syringe S3 twice with H2O (V15), flush the manifold with N2 | ||||
| [16-23] | Pressurize reagent vials in positions V16, V14, V13 and V12, flushing the manifold with N2 between each vial | ||||
| [24-26] | Open the activity inlet (V6) | ||||
| Connect the vial containing 18F. If the total volume is > 5 mL, only insert the needle halfway into the vial before continuing. | |||||
| [27-39] | Close the activity inlet (V6), trap 18F in QMA cartridge C1 (V5), collect the [18O]H2O in the waste bottle (V1). If the total volume is > 5 mL, pause the sequence at step 37, return to step 26, fully insert the needle into the vial containing 18F, and resume the process. | ||||
| [40] | Close the [18O]H2O waste bottle (V1), flush the manifold with N2 | ||||
| [41] | Pressurize the eluent vial in position V2 | ||||
| [42-44] | Open reactor valve V8, aspirate eluent from V2 into syringe S1 | ||||
| [45-50] | Elute QMA cartridge C1 into reactor (V8) using saline from syringe S1, set the reactor temperature to 90 °C | ||||
| [51] | Flush QMA cartridge C1 with N2 and increase the reactor temperature to 105 °C | ||||
| [52-53] | Draw acetonitrile from V16 into syringe S2 | ||||
| [54-57] | Transfer acetonitrile from syringe S2 to the reactor (V8) | ||||
| [58-60] | Heat the reactor at 120 °C for 5 min. Evaporate the solvent with a flow of N2 to the reactor (V7). | ||||
| [61-65] | Set the temperature to 105 °C, dry syringe S1 with N2 | ||||
| [66-69] | Draw the 15-crown-5 solution from V13 into syringe S2, increase the reactor temperature to 120 °C | ||||
| [70-71] | Reduce the temperature to 105 °C, flush the manifold with N2 | ||||
| [72] | Cool down the reactor (set the temperature to 40 °C) for 5 min | ||||
| [73-78] | Set the reactor temperature to 80 °C, transfer the 15-crown-5 solution from syringe S2 to the reactor (V8) | ||||
| [79-81] | Draw the BF3.OEt2 solution from V14 into syringe S2 | ||||
| [82-87] | Transfer the BF3.OEt2 solution from syringe S2 to the reactor (V8), flush the reactor line with N2 | ||||
| [88] | Flush the manifold with N2 | ||||
| [89] | React for 5 min, let the temperature return to RT | ||||
| [90-95] | Transfer the reaction mixture (V8) to syringe S2 | ||||
| [96-104] | Pass the reaction mixture through Alumina N cartridge C2, into syringe S3 | ||||
| [105] | Flush the manifold with N2 | ||||
| [106-109] | Return the reaction mixture to syringe S2 | ||||
| [110-112] | Empty syringe S3, draw H2O (V15) into syringe S2 to dilute the reaction mixture | ||||
| [113-115] | Load the reaction mixture onto QMA cartridge C3 | ||||
| [116-118] | Draw H2O (V15) into syringe S2 | ||||
| [119-124] | Rinse the reactor (V8) with H2O from syringe S2, aspirate the washings into syringe S2 | ||||
| [125-128] | Pass the washings through cartridges C2 and C3 | ||||
| [129-130] | Dry the cartridges and the manifold with N2 | ||||
| [131-136] | Wash syringe S1 with H2O (V15) | ||||
| [137-142] | Wash syringe S2 with H2O (V15) | ||||
| [143] | Flush the manifold with N2 | ||||
| [144-147] | Draw H2O (V15) into syringe S2 | ||||
| [148-151] | Flush QMA cartridge C3 with H2O from syringe S2 | ||||
| [152-153] | Dry QMA cartridge C3 with N2 and flush the manifold with N2 | ||||
| [154-157] | Elute QMA cartridge C3 with 0.9% NaCl (V14) into syringe S3 | ||||
| [158-161] | Transfer the product from syringe S3 to the collection vial (V21) | ||||
| [162-163] | Flush QMA cartridge C3 with N2 to the collection vial (V21) | ||||
| [164-166] | Flush the manifold with N2 | ||||
| [167-170] | Flush cartridges C2 and C3 (to waste bottle) and the manifold with N2 | ||||
| [171] | Flush the collection tubing (V21) with N2 | ||||
Table 2. Description of steps in the XML sequence file.
Discussion
The first step to render cancer cells traceable in vivo by this method requires engineering them to express the NIS-FP fusion reporter. The choice of the fluorescent protein in the fusion reporter is critical as oligomerizing fluorescent proteins can lead to artificial reporter clustering, thereby negatively affecting its function. We have had success with proven monomeric fluorescent proteins such as mEGFP (with the monomerizing mutation A206K36,37), mTagRFP, or mCherry. NIS can either be of human or mouse origin (hNIS or msNIS) depending on the purpose of the experiment and the cancer model. Transduction efficiencies generally vary between different cancer cell lines. However, generated cancer cell lines are subsequently purified by FACS in this protocol, thereby reducing the need for optimizing transduction conditions. Transduction with high multiplicity of infection is not always advisable as multiple construct integration into the genome is likely to result not only in higher construct expression but also in more unwanted/unregulated genome modification. Therefore, it is important to let polyclonal transduced cells grow to stability of expression (monitored by flow cytometry) and avoid sorting the brightest clones only by FACS. It also renders functional validation of non-reporter features crucial before these cells should be used for in vivo experiments. A recently developed alternative to viral gene delivery is gene editing technology38, which offers more specific control over viral integration sites. Expression analysis by flow cytometry and immunoblotting is important. Flow cytometry allows acquisition of population-based single cell data, for example to examine whether there is any drift in reporter expression levels over time. It relies on the FP moiety only, unless cells are also stained with an antibody directed against surface or total NIS. Flow cytometry does not inform on fusion reporter integrity. In contrast, immunoblotting reports on the integrity of the fusion reporter. The molecular weight of NIS and the FP must be added to determine the expected molecular weight of the chosen NIS-FP. Confocal fluorescence microscopy demonstrated fusion reporter colocalization with the plasma-membrane marker wheat germ agglutinin in all newly made cell lines. This was the expected cellular location for most of the protein and indicated a go-ahead milestone for subsequent functional validation. If minimal/no NIS-FP was found on the plasma membrane (e.g. only in internal cellular compartments), this would indicate a cell biological issue with the fusion reporter in this cell line, or a potential mutation of the fusion reporter affecting its intracellular trafficking. It is noteworthy that we have not observed such a case in any of the cancer cells we tested so far, which included: A375P, A375M2, SK-Mel28, WM983A/B (human melanoma); MCF-7, MDA-MB-231, MDA-MB-436 (human breast cancer); NCI-H1975 (human lung cancer); SK-Hep1 (human liver cancer); 4T1, 4T1.2, 66cl4, 67NR, FARN168 (murine inflammatory breast cancer); B16F0, B16F3, B16F10 (murine melanoma); MTLn3 (rat breast adenocarcinoma).
NIS function must be measured using uptake assays with radioactive NIS substrates. Due to the SPECT radiotracer 99mTcO4– being generator-produced and therefore widely available in hospitals without the need for any radiotracer synthesis as well as having a more convenient longer half-life (6.01 h for 99mTc as compared to 110 min for 18F), we used this NIS substrate for routine functional validation of new NIS-FP expressing cell lines. Pre-blocking of NIS-expressing cells with the NIS co-substrate sodium perchlorate resulted in the expected reduction/abolishment of radiotracer uptake, thereby demonstrating specificity of radiotracer uptake. This NIS specificity test is a critical validation step. If a NIS specificity experiment would not result in reduced radiotracer uptake comparable to the respective parental cells, either a technical error during the experiment has occurred, or the radiotracer uptake was not due to NIS. It is also possible that sodium perchlorate pre-blocking reduces radiotracer uptake in a parental cell line; this would identify cell lines with endogenous functional NIS expression (e.g. stimulated thyroid cells6).
A crucial advantage of this imaging protocol is that information is collected in 3D and over time. This allows the comparison of images from the same animal over time, thereby providing paired data and thus overcoming the issues caused by inter-animal variability. This contrasts with most non-imaging related metastasis assessment methods that are based on sacrificing different animals at different time points. In Figure 3B it is evident how metastatic spread and outgrowth progressed over time in an individual animal. The signals detected by PET/CT imaging are fundamentally caused by NIS expression. This includes all signals from exogenously NIS-expressing cancer cells as well as all organs endogenously expressing NIS. Typical endogenous NIS signals are found in thyroid and salivary glands, the stomach, and, at low levels in some parts of the mammary and lachrymal glands. In addition to endogenous NIS expression, the NIS radiotracer [18F]BF4– is also excreted via the kidneys, thereby explaining radiotracer uptake in urine-filled bladders. Kidney uptake is no longer detectable at the imaging time point recommended in this protocol (45 min post radiotracer injection6). If signals from urine-filled bladders should lead to signal-to-background issues, the bladder can be mechanically emptied under anesthesia before imaging. Importantly, the endogenous signals can vary between animal strains. It is also noteworthy that endogenous NIS expression in the mammary glands can be higher under lactating conditions10. In the presented case and in the cases of those metastatic cell lines successfully characterized before (cf. list above), we did not find endogenous NIS expression to significantly interfere with metastasis detection. It is noteworthy, that [18F]BF4– remains more available for uptake into cancerous tissues as compared to iodide, because iodide is metabolized into thyroid hormones6. This phenomenon might also contribute to larger amounts of radioiodide in the blood stream as compared to [18F]BF4– 6. For different applications (cancer cell tracking in other cancers or non-cancer cell tracking applications), this might differ, and it is therefore recommended to assess whether endogenous NIS expression is likely to cause signal-to-background issues through preliminary experiments. An important aspect in preclinical imaging is the molar activity of the radiotracer. The method described here uses ~1.5 GBq 18F– as starting material14 and has been shown to produce molar activities significantly above the previously reported substitution method12. [18F]BF4– produced at molar activities ≤1 GBq/µmol12 can lead to reduced uptake in NIS-expressing tissues. This is of particular importance when the injected amount of radioactivity per kilogram is high, i.e. when small animals such as mice are imaged39; it is less important in the human setting40. High molar activities are therefore imperative for high-quality preclinical PET imaging. Molar activities obtained by the boron trifluoride addition method14, which is shown in its automated form in this protocol, overcome this issue. Furthermore, it is noteworthy that the presented protocol for [18F]BF4– synthesis is not compliant with good manufacturing practice (GMP) and therefore unsuitable for use in human clinical trials in this form. A GMP protocol (via the 18F substitution method to radiolabel BF4–) is available elsewhere40.
PET/CT imaging allows the visualization of radiotracer uptake, which is indicative of NIS-mediated radiotracer uptake stemming from NIS-FP expressing cancer cells. More importantly, the associated PET signals can be quantified. It is necessary to apply reliable thresholding procedures to ensure a consistent and unbiased differentiation of relevant signals from any potential background. As the background varies in different locations in vivo, it is important to consider local/regional thresholding and segmentation. One such method was developed by and named after Otsu34, and its 3D implementation is employed for 3D rendering of the primary tumor and metastases in this protocol. Generally, the image seen by the observer visually corresponds best to the quantified %injected dose (%ID) values. As for image-based quantification, it is also important to normalize the measured radioactivity values of the different tissues to their volumes. There are two predominantly used ways of expressing normalized results, (i) %ID per volume (e.g. %ID/mL), and (ii) standard uptake value (SUV35). They differ in that %ID/mL takes into account the individual volume only, while SUV is a measure that is relative to the average radioactivity across the whole animal. It is also important to note that NIS imaging renders the live tumor volume (LTV) accessible, because dead/dying cells not synthesizing ATP can no longer import radiotracer10. This explains the large low-signal area within the primary tumor ("donut shaped" tumor) indicating areas of tumor cell death/necrosis. Importantly, LTV was a much more reliable measure of tumor burden as compared to the crude tumor volume accessible by caliper measurements (which does not take into account viability and assesses only superficial tumor regions).
A major advantage of this dual-mode tracking strategy is evident when harvesting tissues after animal culling. Guided by in vivo images and assisted by fluorescent cancer cells during animal dissection, small and deep-seated organs/metastases can also be reliably harvested. Frozen tissue preservation/sectioning methodology enables the direct fluorescence imaging of GFP without the need for staining with an anti-GFP antibody, but at the expense of reduced structural tissue preservation as compared to formalin-fixed paraffin-embedded methodology (FFPE). The latter critically requires also anti-FP staining, because the FFPE method is incompatible with intact preservation of fluorescent proteins (due to fixation/dehydration/rehydration). While the fluorescence signal is indicative of tumor cell presence, it is important to ensure this classification is confirmed by ex vivo radioactivity measurements of the harvested tissues ('biodistribution'). Ex vivo radioactivity measurements are more sensitive than visual detection of fluorescence, hence can allow the identification of cancer cell-dependent signals that would otherwise remain undetected. In the case of a terminal imaging session, it is critical to accurately note the injected radiotracer amounts as well as the times of radiotracer radioactivity measurements, animal injection, animal culling, and calibrated scintillation counter measurements of harvested tissues. This is crucial to ensure correction for radiotracer decay and thereby enable reliable biodistribution analysis.
PET/CT imaging enables repeated non-invasive 3D quantification of tumor progression including the assessment of metastatic spread on a whole-body level. This feature is a significant advantage over conventional methods, which often rely on large cohorts of animals that are euthanized for the assessment of tumor progression at varying time points. The advantages of this imaging-based approach are: (i) highly sensitive non-invasive 3D in vivo quantification, (ii) a significant reduction in animal numbers due to the possibility of repeat imaging, (iii) the acquisition of longitudinal paired data from subsequent imaging sessions improving statistics by excluding inter-animal variability, which in turn further reduces animal numbers, (iv) automated production of [18F]BF4– at high specific activities, and (v) the intrinsic option for ex vivo confirmation in tissues by fluorescence methodologies such as microscopy or cytometry.
In vivo cell tracking is a growing field. It has been fueled by recent advancements in imaging technology, which resulted in enhanced resolution, detection limits and multiplex capability (via multi-modal imaging). In this protocol, we apply this concept to track tumor progression including spontaneous cancer cell metastasis in 3D by repeat imaging. Applications include studies aimed at unraveling the mechanisms of spontaneous cancer cell metastasis. For example, traceable tumor cells could be used to study the impact of different immune cell components (as present/functional in animal strains of different levels of immunocompromisation) on the metastatic process. Similarly, the impact of individual genes, either in the animal strain or the cancer cell line, could be studied. Furthermore, the presented protocol could be used to assess/validate the efficacy of specific drugs or therapeutic concepts on tumor progression. Importantly, this reporter gene:radiotracer pair for PET imaging (NIS:[18F]BF4–) could also be used for different cell tracking applications. For example, several cell therapies are currently emerging as promising therapeutic approaches. This includes cellular therapeutics for cancer treatment41 but also in transplantation42 and regenerative medicine43,44 settings. Whole-body in vivo cell tracking is becoming increasingly important for the development and clinical translation of cellular therapeutics, for example, for evaluating safety and for therapy monitoring.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The research was supported by the King's College London and UCL Comprehensive Cancer Imaging Centre, funded by Cancer Research UK and EPSRC in association with the MRC and DoH (England); the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London; the Centre of Excellence in Medical Engineering funded by the Wellcome Trust and EPSRC under grant number WT 088641/Z/09/Z; a Cancer Research UK Multidisciplinary Project Award to GOF and PJB, and a King's Health Partners grant to GOF. The nanoPET/CT and nanoSPECT/CT scanners were purchased and maintained by an equipment grant from the Wellcome Trust. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the DoH.
Materials
| Step 1) Engineering and characterization of cancer cells to express the radionuclide-fluorescnece fusion reporter NIS-FP. | |||
| 2'-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5'-Bi-1H-benzimidazole | Thermo Scientific | H3570 | Trivial name: Hoechst 33342; CAS number: 23491-52-3; Hoechst 33342, Trihydrochloride, Trihydrate – 10 mg/mL Solution in Water. |
| 4T1 murine breast cancer cell line | ATCC | CRl-2539 | for details see ATCC website |
| Automatic Cell Counter, e.g. CASYCounter | Roche Diagnostics GmbH | 5651697001 | CASY Model TT Cell Counter and Analyzer |
| CASYclean Cleaning Reagent | Sedna Scientific | 2501036 | |
| CASYton Isotonic Diluent | Sedna Scientific | 2501037 | |
| Confocal Fluorescence Microscope, e.g. Leica TCS SP5 | Leica, Wetzlar, Germany | Equipped with Plan-Neofluor 25×0.5NA and Plan-Apochromat 63×1.4NA oil UV objectives and Diode (405 nm), Argon-ion (458, 477, 488, 496, 514 nm) and HeNe (543 and 633 nm) lasers; A Leica LAS AF Lite Software 4.0.11706 (Leica Microsystems CMS GmbH) was used for image acquisition and and anaysis | |
| Cover slips No. 1.5 thickness | VWR International | 631-0150 | |
| Dabco | Sigma | 290734 | Stock 125 mg/mL |
| DMEM | Sigma | D5546 | Supplement with 10 % (v/v) FBS and L-glutamine (2 mM) to make up the optimal growth medium for MDA-MB-231 cells. |
| FACS sorter, e.g. BD FACSAria III | BD Biosciences | Equipped with a BD FACS DIVA Software, a 6 Laser System (375/405/488/561/633 nm lasers) – cells sorted with a 100 μm nozzle under 20 psi flow pressure, window extension of 2.0 μm, 2.0 Neutral Density Filter and 3 kV plate voltage | |
| Fetal Bovine Serum (FBS) | Sigma | F9665 | Heat inactivated at 56 °C for 30 min |
| Automated Gamma Counter, e.g. 1282 Compugamma | LKB Wallac Laboratory | 99mTc-pertechnetate energy window 110-155 keV 18F energy window 175-220 keV | |
| Hoechst 33342 solution | Life Technologies | H1399 | 1-3 µg/mL; DAPI (from various supplieres) can be used instead. |
| L-glutamine | Sigma | G7513 | Solution 200 mM concentrated, sterile-filtered |
| Linear polyethylenimine (PEI) | Polyscience | 23966-2 | Linear, 25 kDa; transfection reagent for 293T cell line. |
| MDA-MB-231 human breast cancer cells | ATCC | HTB-26 | for details see ATCC website |
| Mowiol 4-88 | Sigma | 81381 | |
| pLNT SFFV NIS-mEGFP | request from our lab | n/a | For details (generation and maps) see Supplementary Information |
| pLNT SFFV NIS-mCherry | request from our lab | n/a | For details (generation and maps) see Supplementary Information |
| pMD2.G | Addgene | #12259 | plasmids encoding for the VSV-G envelope |
| pRRE | Addgene | #12251 | packaging plasmid |
| pRSV-Rev | Addgene | #12253 | packaging plasmid |
| Paraformaldehyde solution 4 % (w/v) in PBS | Santa Cruz Biotechnology | sc-281692 | |
| Penicillin-Streptomycin | Sigma | P43330 | Containing penicillin (10,000 units/mL) and streptomycin (10 mg/mL), sterile-filtered |
| Phosphate Buffered Saline (PBS) | Sigma | D8537 | pH 7.4, sterile-filtered and without calcium chloride and magnesium chloride |
| Poly(vinyl alcohol – vinyl acetate) | Polysciences | 17951 | Trivial name: Mowiol 4-88; CAS number: 9002-89-5 |
| Puromycin dihydrochloride | Sigma | P8833 | From Streptomyces alboniger, reconstituted in sterile water |
| Benchtop centrifuge, e.g. Rotina 380 R Benchtop centrigfuge | Hettich Lab Technology | ||
| RPMI 1640 | Sigma | R0883 | Supplement with 10% (v/v) FBS and L-glutamine (2mM) to make up the optimal growth medium for 4T1 cells. |
| SFCA Syringe filter 0.45 μm | Corning | ||
| Syringes 10 mL | BD Emerald | Disposable non-sterile syringes | |
| Tissue culture fluorescence microscope, e.g. EVOS-FL | Life Technologies | Cell Imaging System equipped with a 10× objective (PlanFL PH2, 10×/0.25, ∞/1.2) and a colour camera | |
| Trypsin-EDTA solution 10X | Sigma | 59418C | (0.5 % (w/v) trypsin, 0.2 % (w/v) EDTA) gamma irradiated by SER-TAIN process and without phenol red |
| Wheat Germ Agglutinin Alexa Fluor 633 Conjugate | Life Technologies | W21404 | Used at 1:1000 (2 µg/mL) for cell immunofluorescence |
| Step 2) Establishment of in vivo tumor models. | |||
| Digital caliper | World Precision Instruments | 501601 | |
| Isoflurane 1000 mg/g | Isocare | For inhalation | |
| Fluorescence Torch, e.g. NightSea Fluorescence Torch DFP-1 | Electron Microscopy Sciences | SFA-LFS-RBS/GR | Equipped with GFP and RFP emission filters and NightSea filter goggles (DFP-1) |
| Syringes 0.3 mL U-100 insulin | Terumo | 29G × 1/2'' – 0.33 × 12 mm | |
| Standard materials/equipment for aseptic technique and animal maintenance | |||
| Step 3) Production of [18F]BF4– using an automated radiotracer synthesis platform. | |||
| 15-crown-5 | Sigma-Aldrich | 188832 | CAS 33100-27-5 |
| Acetonitrile (anhydrous) | Acros Organics | 326811000 | |
| Boron trifluoride diethyl etherate | Sigma-Aldrich | 216607 | BF3.OEt2, purified by redistillation, ≥46.5 % BF3 basis. CAS 109-63-7 |
| Automated Radiotracer Synthesis (ARS) platform, e.g. FASTLab | GE Healthcare | ||
| Disposable cassettes for ARS platform, e.g. FASTLab cassettes | GE Healthcare | FASTlab Developer pack | |
| Polygram Alox N/UV254 polyester sheets | Macherey-Nagel | 802021 | RadioTLC plates, 40×80 mm |
| Strong anion exchange cartridge, e.g. Sep-Pak Accell Plus QMA Plus Light | Waters | WAT023525 | Condition with 1M NaCl (10 mL) and H2O (10 mL) |
| Alumina neutral cartridge, e.g. Sep-Pak Alumina N Plus Light | Waters | WAT023561 | Condition with H2O (10 mL), acetone (10 mL) and air (20 mL) |
| Water for injection USP | GE Healthcare | ||
| Nitrogen filter | Millipore | SE2M049I05 | Sterile 0.2 µm FG Millex 13 mm |
| Step 4) In vivo imaging of NIS-FP expressing cells by nanoPET/CT. | |||
| Isoflurane 1000 mg/g | Isocare | For inhalation | |
| Preclinical PET/CT multimodal imaging instrument, e.g. nanoScan PET/CT | Mediso Medical Imaging System, Budapest, Hungary | ||
| Fluorescence Torch, e.g. NightSea Fluorescence Torch DFP-1 | Electron Microscopy Sciences | SFA-LFS-RBS/GR | Equipped with GFP and RFP emission filters and NightSea filter goggles (DFP-1) |
| Rodent anesthesia induction chamber | Vet-Tech | AN010R | With three-way valves (x2), tube mount connector for inlet, PVC tubing for gas inlet (2 m) and 22 mm scavenging tube (2 m) |
| Rodent anesthesia system | Vet-Tech | AN001B | Including animal face-mask suitably sized for animal of interest and isolflurane vaporizer |
| Sterile physiological saline | Thermo Scientific Oxoid | BO0334B | |
| Syringes 0.3 mL U-100 insulin | Terumo | 29G × 1/2'' – 0.33 × 12 mm, for intravenous injection of radiotracer | |
| Veterinary Scavenger | Vet-Tech | AN200 | VetScav filter weighing mechanism – 240 V with automatic temperature compensation and LED system |
| 5) In vivo data analysis. | |||
| Tera-Tomo Monte Carlo based full 3D iterative algorithm | Mediso Medical Imaging System, Budapest, Hungary | ||
| VivoQuant Software | Invicro LLC., Boston, USA | ||
| 6) Ex vivo analyses | |||
| 2-Methylbutane | Sigma | 59070-1L-D | Pre-cooled over liquid nitrogen to freeze OCT-embedded tissues |
| Bovine Serum Albumin (BSA) | Sigma | 85040C | |
| Cover slips 22×50 mm | VWR International | SMITMCQ211022X50 | |
| Cryostat, e.g. Cryostat MNT | SLEE Medical | two-piece modular histology embedding machine equipped with an embedding module, a tissue storage compartment and a cold plate | |
| Cy5 AffiniPure Goat anti-Rabbit IgG (H+L) | Jackson/Stratech | 111-175-144 | Used at 1:500 (2 µg/mL) |
| Dabco | Sigma | 290734 | Stock 125 mg/mL |
| Microtome blades, e.g. Feather S35 | CellPath | ||
| Fluorescence Microscope (wide-field or confocal), e.g. Nikon Eclipse Ti-E Inverted Fluorescence Microscope | Nikon | Equipped with 10×, 20× (air) and ideally 40× (oil) objectives and lasers/filters or filter cubes, respectively, that are suitable for Hoechst 33342, GFP and Cy5 | |
| Automated Gamma Counter, e.g. 1282 Compugamma | LKB Wallac Laboratory | 99mTc-pertechnetate energy window 110-155 keV, 18F energy window 175-220 keV | |
| Hoechst 33342 solution | Life Technologies | H1399 | |
| Fluorescence adapter for dissecting microscope, e.g. NightSea Adapter | Electron Microscopy Sciences | SFA-LFS-RBS/GR | Equipped with GFP and RFP emission filters |
| O.C.T. compound | VWR international | 361603E | |
| Wax pen, e.g. PAP-PEN | Dako UK Ltd | Wax pen to draw around tissue section to reduce required staining/washing solution volumes | |
| Paraformaldehyde solution 4 % (w/v) in PBS | Santa Cruz Biotechnology | sc-281692 | |
| Rabbit anti-CD31 | Abcam | ab28364 | Polyclonal anti-mouse used 1:50 (20 µg/mL) for tissues immunofluorescence |
| Microscope slides, e.g. Superfrost slides | VWR, Lutterworth, UK | ||
| Tris-buffered saline (TBS) | available from various suppliers. | Tris-buffered saline; 150 mM NaCl, 25 mM Tris/HCl at pH 7.4 | |
References
- Chaffer, C. L., Weinberg, R. A. A perspective on cancer cell metastasis. Science. 331 (6024), 1559-1564 (2011).
- Brader, P., Serganova, I., Blasberg, R. G. Noninvasive molecular imaging using reporter genes. J Nucl Med. 54 (2), 167-172 (2013).
- Fruhwirth, G. O., Diocou, S., Blower, P. J., Ng, T., Mullen, G. E. A whole-body dual-modality radionuclide optical strategy for preclinical imaging of metastasis and heterogeneous treatment response in different microenvironments. J Nucl Med. 55 (4), 686-694 (2014).
- Terrovitis, J., et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. J Am Coll Cardiol. 52 (20), 1652-1660 (2008).
- Seo, J. H., et al. Trafficking Macrophage Migration Using Reporter Gene Imaging with Human Sodium Iodide Symporter in Animal Models of Inflammation. J Nucl Med. 51 (10), 1637-1643 (2010).
- Diocou, S., et al. [18F]tetrafluoroborate-PET/CT enables sensitive tumor and metastasis in vivo imaging in a sodium iodide symporter-expressing tumor model. Scientific Reports. 7 (1), 946 (2017).
- Lajtos, I., et al. Cold wall effect eliminating method to determine the contrast recovery coefficient for small animal PET scanners using the NEMA NU-4 image quality phantom. Phys Med Biol. 59 (11), 2727-2746 (2014).
- Nagy, K., et al. Performance evaluation of the small-animal nanoScan PET/MRI system. J Nucl Med. 54 (10), 1825-1832 (2013).
- Deleye, S., et al. Performance evaluation of small-animal multipinhole muSPECT scanners for mouse imaging. Eur J Nucl Med Mol Imaging. 40 (5), 744-758 (2013).
- Portulano, C., Paroder-Belenitsky, M., Carrasco, N. The Na+/I- symporter (NIS): mechanism and medical impact. Endocr Rev. 35 (1), 106-149 (2014).
- Dohan, O., et al. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 24 (1), 48-77 (2003).
- Jauregui-Osoro, M., et al. Synthesis and biological evaluation of [F-18]tetrafluoroborate: a PET imaging agent for thyroid disease and reporter gene imaging of the sodium/iodide symporter. Eur J Nucl Med Mol Imaging. 37 (11), 2108-2116 (2010).
- Weeks, A. J., et al. Evaluation of [18F]-tetrafluoroborate as a potential PET imaging agent for the human sodium/iodide symporter in a new colon carcinoma cell line, HCT116, expressing hNIS. Nucl Med Commun. 32 (2), 98-105 (2011).
- Khoshnevisan, A., et al. [(18)F]tetrafluoroborate as a PET tracer for the sodium/iodide symporter: the importance of specific activity. EJNMMI Res. 6 (1), 34 (2016).
- Groot-Wassink, T., et al. Noninvasive imaging of the transcriptional activities of human telomerase promoter fragments in mice. Cancer Res. 64 (14), 4906-4911 (2004).
- Chen, L., et al. Radioiodine therapy of hepatoma using targeted transfer of the human sodium/iodide symporter gene. J Nucl Med. 47 (5), 854-862 (2006).
- Sieger, S., et al. Tumour-specific activation of the sodium/iodide symporter gene under control of the glucose transporter gene 1 promoter (GTI-1.3). Eur J Nucl Med Mol Imaging. 30 (5), 748-756 (2003).
- Chisholm, E. J., et al. Cancer-specific transgene expression mediated by systemic injection of nanoparticles. Cancer Res. 69 (6), 2655-2662 (2009).
- Klutz, K., et al. Targeted radioiodine therapy of neuroblastoma tumors following systemic nonviral delivery of the sodium iodide symporter gene. Clin Cancer Res. 15 (19), 6079-6086 (2009).
- Merron, A., et al. Assessment of the Na/I symporter as a reporter gene to visualize oncolytic adenovirus propagation in peritoneal tumours. Eur J Nucl Med Mol Imaging. 37 (7), 1377-1385 (2010).
- Merron, A., et al. SPECT/CT imaging of oncolytic adenovirus propagation in tumours in vivo using the Na/I symporter as a reporter gene. Gene Ther. 14 (24), 1731-1738 (2007).
- Dingli, D., et al. Combined I-124 positron emission tomography/computed tomography imaging of NIS gene expression in animal models of stably transfected and intravenously transfected tumor. Mol Imaging Biol. 8 (1), 16-23 (2006).
- Carlson, S. K., et al. Quantitative molecular imaging of viral therapy for pancreatic cancer using an engineered measles virus expressing the sodium-iodide symporter reporter gene. AJR Am J Roentgenol. 192 (1), 279-287 (2009).
- Barton, K. N., et al. GENIS: gene expression of sodium iodide symporter for noninvasive imaging of gene therapy vectors and quantification of gene expression in vivo. Mol Ther. 8 (3), 508-518 (2003).
- Siddiqui, F., et al. Design considerations for incorporating sodium iodide symporter reporter gene imaging into prostate cancer gene therapy trials. Hum Gene Ther. 18 (4), 312-322 (2007).
- Rad, A. M., et al. AC133+ progenitor cells as gene delivery vehicle and cellular probe in subcutaneous tumor models: a preliminary study. BMC Biotechnol. 9, 28 (2009).
- Hwang do, W., et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. Eur J Nucl Med Mol Imaging. 35 (1), 135-145 (2008).
- Edmonds, S., et al. Exploiting the Metal-Chelating Properties of the Drug Cargo for In Vivo Positron Emission Tomography Imaging of Liposomal Nanomedicines. ACS Nano. 10 (11), 10294-10307 (2016).
- Zhang, R., et al. Probing the Heterogeneity of Protein Kinase Activation in Cells by Super-resolution Microscopy. ACS Nano. 11 (1), 249-257 (2017).
- Dunn, K. W., Kamocka, M. M., McDonald, J. H. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 300 (4), C723-C742 (2011).
- Pulaski, B. A., Ostrand-Rosenberg, S. Mouse 4T1 breast tumor model. Curr Protoc Immunol. , (2001).
- Workman, P., et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 102 (11), 1555-1577 (2010).
- Foster, B., Bagci, U., Mansoor, A., Xu, Z., Mollura, D. J. A review on segmentation of positron emission tomography images. Comput Biol Med. 50, 76-96 (2014).
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics. 9 (1), 62-66 (1979).
- Kinahan, P. E., Fletcher, J. W. Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR. 31 (6), 496-505 (2010).
- Zacharias, D. A., Violin, J. D., Newton, A. C., Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296 (5569), 913-916 (2002).
- Snapp, E. L., et al. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 163 (2), 257-269 (2003).
- Yang, H., et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 154 (6), 1370-1379 (2013).
- Hume, S. P., Gunn, R. N., Jones, T. Pharmacological constraints associated with positron emission tomographic scanning of small laboratory animals. Eur J Nucl Med. 25 (2), 173-176 (1998).
- O’Doherty, J., et al. 18F-Tetrafluoroborate, a PET Probe for Imaging Sodium/Iodide Symporter Expression: Whole-Body Biodistribution, Safety, and Radiation Dosimetry in Thyroid Cancer Patients. J Nucl Med. 58 (10), 1666-1671 (2017).
- Couzin-Frankel, J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 342 (6165), 1432-1433 (2013).
- Boardman, D. A., et al. Expression of a Chimeric Antigen Receptor Specific for Donor HLA Class I Enhances the Potency of Human Regulatory T Cells in Preventing Human Skin Transplant Rejection. American Journal of Transplantation. , (2017).
- Rashid, T., Takebe, T., Nakauchi, H. Novel strategies for liver therapy using stem cells. Gut. 64 (1), 1-4 (2015).
- Ellison, G. M., et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 154 (4), 827-842 (2013).

