Hairy Root Transformation and Regeneration in Arabidopsis thaliana and Brassica napus
Summary
The protocol describes hairy root induction using Arabidopsis primary inflorescence stems and Brassica napus hypocotyls. The hairy roots can be cultured and used as explants to regenerate transgenic plants.
Abstract
Hairy root transformation represents a versatile tool for plant biotechnology in various species. Infection by an Agrobacterium strain carrying a Root-inducing (Ri) plasmid induces the formation of hairy roots at the wounding site after the transfer of T-DNA from the Ri plasmid into the plant genome. The protocol describes in detail the procedure of the injection-based hairy root induction in Brassica napus DH12075 and Arabidopsis thaliana Col-0. The hairy roots may be used to analyze a transgene of interest or processed for the generation of transgenic plants. Regeneration medium containing cytokinin 6-benzylaminopurine (5 mg/L) and auxin 1-naphthaleneacetic acid (8 mg/L) successfully elicits shoot formation in both species. The protocol covers the genotyping and selection of regenerants and T1 plants to obtain plants carrying a transgene of interest and free of T-DNA from the Ri plasmid. An alternative process leading to the formation of a composite plant is also depicted. In this case, hairy roots are kept on the shoot (instead of the natural roots), which enables the study of a transgene in hairy root cultures in the context of the whole plant.
Introduction
Plant transformation is the bottleneck of any genetic study in plant biology. A soil-borne bacterium, Agrobacterium tumefaciens, is widely used as a means for gene delivery by floral dip or tissue culture to generate transformants. A. tumefaciens infects plants at a wounding site and causes tumors due to the transfer and integration of T-DNA from a Tumor-inducing (Ti) plasmid into the host plant genome. Engineered A. tumefaciens strains with modified Ti plasmid without the wild-type T-DNA and a binary vector with artificial T-DNA and cloning sites for inserting a gene of interest are commonly used as an efficient plant transformation system1. However, many model species and crops are recalcitrant to floral dip or in vitro plant regeneration or have long growth cycles, impacting the efficiency of this transformation system.
Agrobacterium rhizogenes induces the formation of adventitious roots, or hairy roots, at the wounding site after infecting a host plant. Similar to A. tumefaciens, A. rhizogenes transfers a T-DNA from a Root-inducing (Ri) plasmid to the host plant genome, causing the development of transgenic hairy roots. This process is controlled mainly by the root oncogenic loci (rol) genes2,3. Using agrobacterial strains carrying both the Ri plasmid and an artificial binary vector encoding a gene of interest, hairy root cultures have been used to produce recombinant proteins, analyze the function of promoters or genes, or edit genomes using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs)/CRISPR-associated protein 9 (Cas9)4,5,6.
Our protocol uses the transconjugant strain Ti-less A. tumefaciens C58C1 carrying the Ri plasmid pRiA4b7. The T-DNA of the Ri plasmid consists of two regions, right and left T-DNA (TR-DNA and TL-DNA, respectively), that can independently integrate into the plant genome8. Exploiting this system, the lengthy explant transformation process in Brassica napus DH12075 cultivar was optimized9. The protocol detailed below allows for the regeneration of selected hairy root lines and obtaining T1 plants carrying the transgene of interest and free of rol genes in roughly 1 year. The injection-based hairy root transformation can be used in other Brassicaceae species, as shown by transforming Arabidopsis thaliana Col-0. While hypocotyl is used to transform B. napus, A. thaliana is injected into the primary inflorescence stem.
Protocol
1. Preparation of media and solutions
- Prepare the hormone stock solutions.
- For preparation of 50 mL of 1-naphthaleneacetic acid (NAA), 6-benzylaminopurine (BAP), and indole-3-butyric acid (IBA) stock solution with a concentration of 5 mg/mL, dissolve 250 mg of the powdered hormone in 2 mL of 1 M sodium hydroxide (NaOH) and adjust the volume to 50 mL with ultrapure water.
- To prepare 50 mL of gibberellic acid (GA3) with a concentration of 1 mg/mL, dissolve 50 mg of the powdered hormone in 2 mL of ethanol and adjust the volume to 50 mL of ultrapure water.
- Filter the solution using a sterile syringe filter with 0.22 µm pore size and distribute it into sterile 2 mL tubes for storage. Store the solution at -20 °C or 4 °C, depending on the manufacturer's recommendation.
- Prepare antibiotic stock solutions. For 50 mL of cefotaxime and ticarcillin disodium stock solution with a concentration of 100 mg/mL, dissolve 0.5 g of the powdered antibiotic in 40 mL of ultrapure water and adjust to a final volume of 50 mL. Filter the solution using a sterile syringe filter with 0.22 µm pore size and distribute it into sterile 2 mL tubes for storage. Store the solution at -20 °C.
NOTE: Antibiotics and hormones are always added to the cooled medium. - Prepare 1 L of Luria Broth (LB) medium by adding 10 g of bacto-tryptone, 5 g of yeast extract, and 5 g of sodium chloride (NaCl) in a 1 L measuring cylinder and adjust the volume to 1 L with double distilled water. Adjust the pH to 7.0 with KOH with a pH meter (following the manufacturer's instructions). Transfer the solution to a 1 L bottle and add 15 g of bacteriological agar (1.5%), if a solid medium is prepared and autoclave the medium. If necessary, add the appropriate antibiotics (bacterial resistance is carried on the binary vector) to the cooled medium.
NOTE: Use an autoclave to sterilize the solution. Put the bottle into the basket, close the lid, and sterilize it for 20 min at 121 °C and 98.9 kPa. This protocol is always used for autoclaving solutions in further steps. - Prepare 1 L of Yeast Extract Beef (YEB) medium by mixing 5 g of beef extract, 1 g of yeast extract, 5 g of peptone, 5 g of sucrose, and 0.5 g of magnesium chloride (MgCl2) into double distilled water. Adjust the volume to 1 L. Transfer the solution to a 1 L bottle. Autoclave the medium.
- Prepare 1 L of medium for seed germination by mixing 2.2 g of Murashige and Skoog (MS) powder, 10 g of sucrose (1%), and 0.5 g of 2-(N-Morpholino)ethanesulfonic acid (MES) sodium salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH. Transfer the solution to a 1 L bottle and add 8 g of plant agar (0.8%). Autoclave the medium.
- Prepare 1 L of plant growth medium by mixing 2.2 g of MS powder, 5 g of sucrose (0.5%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH. Transfer the solution to a 1 L bottle and add 8 g of plant agar (0.8 %). Autoclave the medium.
- Prepare 1 L of hairy root growth medium by mixing 4.4 g of MS + B5 vitamins powder, 30 g of sucrose (3%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH, and transfer the solution to a 1 L bottle. Add 3 g of gelling agent (0.3%) and autoclave the medium. Add cefotaxime and ticarcillin disodium to a final concentration of 100 – 200 mg/L and 100 – 500 mg/L, respectively. If necessary, add the appropriate antibiotics (resistance is due to the T-DNA of a binary vector).
- Prepare 1 L of composite plant growth medium by mixing 2.2 g of MS + B5 vitamins powder, 10 g of sucrose (1%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH and transfer the solution to a 1 L bottle. Add 6 g of gelling agent (0.6%) and autoclave the medium. Add cefotaxime and ticarcillin disodium to a final concentration of 200 mg/L and 500 mg/L, respectively. If necessary, add the appropriate antibiotics (resistance is due to the T-DNA of a binary vector).
- Prepare 1 L of regeneration medium by mixing 4.4 g of MS + B5 vitamins powder, 30 g of sucrose (3%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH. Transfer the solution to a 1 L bottle and add 3 g of gelling agent (0.3 %). Autoclave the medium. Add NAA and BAP to a final concentration of 8 mg/L and 5 mg/L, respectively, and ticarcillin disodium to a final concentration of 100 mg/L.
- Prepare 1 L of shoot elongation medium by mixing 4.4 g of MS + B5 vitamins powder, 20 g of sucrose (2%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH. Transfer the solution to a 1 L bottle and add 3 g of plant agar (0.3%). Autoclave the medium. Add BAP and GA3 to a final concentration of 0.5 mg/L and 0.03 mg/L, respectively, and cefotaxime to a final concentration of 100 mg/L.
- Prepare 1 L of root induction medium by mixing 2.2 g of MS + B5 vitamins powder, 10 g of sucrose (1%), and 0.5 g of MES salts into double distilled water. Adjust the volume to 1 L, mix, and adjust the pH to 5.8 with KOH. Transfer the solution to a 1 L bottle and add 3 g of gelling agent (0.3%). Autoclave the medium. Add IBA and cefotaxime to a final concentration of 0.5 mg/L and 100 mg/L, respectively.
- Prepare 1 L of Cetyltrimethylammonium bromide (CTAB) buffer by adding 20 g of CTAB (2% w/v final), 100 mL of 1M Tris-HCl, pH 8.0 (100 mM final), 40 mL of 0.5 M EDTA, pH 8.0 (20 mM final), 81.8 g of NaCl (1.4 M final) and 5 g of PVP40 (0.5% w/v final). Adjust to 1 L with double distilled water. Autoclave the solution. Store the solution for up to 1 year at room temperature.
2. Transformation of Agrobacterium with a binary vector
- Prepare the DNA of a verified binary plasmid containing the T-DNA cassette to be integrated into the plant genome. Ensure that the plasmid DNA contains low salts to avoid electric sparks as electroporation is used. Using a plasmid DNA extraction kit of choice is recommended.
- Prepare electrocompetent Agrobacterium tumefaciens C58C1 cells containing the hairy-root-inducing plasmid pRiA4b by inoculating 200 mL of liquid pre-heated (28 °C) YEB with 8 mL of a fresh overnight culture. Incubate culture at 28 °C while shaking until the optical density at 600 nm (OD600) is about 0.5, corresponding to the mid-log phase. It takes about 4 – 5 h.
- Divide the Agrobacterium culture into four pre-chilled sterile 50 mL centrifuge tubes. Centrifuge for 15 min at 4 °C at 3,200 x g.
NOTE: The cells should be kept cold from this step onward. - Remove the supernatant and resuspend the pellet gently in (4x) 2.5 mL of cold (4 °C) 10% glycerol. Add another 47.5 mL of cold 10% glycerol and gently mix.
- Pellet cells at 3,200 x g for 15 min at 4 °C. Discard supernatant and resuspend cells in (4x) 10 mL of cold 10% glycerol.
- Pellet the cells again and resuspend them in (4x) 0.75 mL of cold 10% glycerol. Pool the content of all four tubes into one (total volume = 3 mL).
- Divide the agrobacterial solution into 50 μL aliquots in pre-chilled sterile 1.5 mL microtubes. Freeze the aliquots on dry ice or liquid nitrogen. Store the tubes at -80 °C for future use.
- Place one tube of competent cells (50 μL) on ice, mix with 5 μL of plasmid DNA (1 μg in total, from step 2.1), transfer the mixture to an electroporation cuvette (0.2 cm gap), and incubate for 5 min on ice.
- Place the cuvette in the electroporator and electroporate the cells with the following settings: capacitance 25 µFD, resistance 400 Ω, electrical voltage 2.5 kV, pulse duration 9.7 ms.
- Add 950 µL of LB liquid medium to the cells, which are then cultured at 28 °C for 2 h at 300 rpm in a thermomixer. Plate 50 µL of the cell culture onto a solid LB medium with the appropriate selective antibiotic. Culture the plates for 2 days at 28 °C.
NOTE: It is recommended to plate a range of cell cultures (10 µL – 100 µL) per plate to determine a proper culture volume and avoid overgrowth of bacteria. - Prepare liquid cultures from a few selected colonies in 5 mL of LB liquid medium with antibiotics. Grow the bacteria overnight at 28 °C with shaking. Use these cultures for glycerol stocks by mixing 0.5 mL of liquid bacteria culture and 0.5 mL of 40% glycerol.
NOTE: These colonies are verified for the presence of the transgene by colony PCR. For this purpose, add a small amount of a colony from a plate to the PCR master mix containing buffer, dNTP, and Taq polymerase of choice, together with primers amplifying a part of the T-DNA from a binary vector. Inoculum from the glycerol stocks is used for the transformation of the plants.
3. Hairy root transformation of Brassica napus DH12075
- Place seeds of Brassica napus DH12075 in microtubes and sterilize them in a flow box. First, degrease the seeds using water and 0.1% detergent while shaking for 60 s. Then, rinse the seeds with water followed by 70% ethanol, both for 60 s.
- Sterilize the seeds using a 10% solution of commercial bleach containing sodium hypochlorite. Shake the seeds in this solution for 20 min.
- Wash the seeds 4x with sterile water for 60 s each. Place them on Petri dishes containing the seed germination medium. Cold-stratify the seeds at 4 °C overnight and move the plates to a cultivation room (21 °C, 16 h light /8 h dark).
- Transfer 5-day-old seedlings to plant culture boxes containing a plant growth medium .
NOTE: The best transformation efficiency in DH12075 was identified for 18-day-old seedlings. The seedling age may be optimized for the local growth conditions or other cultivars. - Inoculate a liquid culture of Agrobacterium tumefaciens C58C1 carrying a hairy root-inducing plasmid pRiA4b and the binary vector for transformation (from step 2.11) with an inoculation loop. Use 5 mL of LB medium. Grow this culture overnight at 28 °C until it reaches OD600 = 0.9 – 1.
NOTE: Agrobacterium containing only the Ri plasmid is used if wild-type hairy roots are produced. - Inject a small amount of culture (approximately 50 µL) with an insulin syringe into the hypocotyl of an 18-day-old seedling (from step 3.4.). Puncture the hypocotyl through with a 26G needle mounted on the syringe at about 1 cm above the surface of the medium. Inject the liquid into the wound. The tissue on the surface of the hypocotyl can also be scratched by the syringe.
NOTE: The number of inoculated seedlings must be adapted to the experimenter's needs. - Return the plants to the cultivation room at 21 °C for 2-to-4 weeks until a callus and hairy roots are formed at the wounding site.
- Cut off the callus with the emerged hairy roots from the hypocotyl and place it on a Petri dish with the hairy root growth medium containing selective antibiotics (carried by the T-DNA) and cefotaxime (200 mg/L) and ticarcillin (500 mg/L) to suppress agrobacterial growth. Seal the Petri dishes with gas-permeable tape. Culture the hairy roots at 24 °C in the dark.
NOTE: Proper concentration of specific antibiotics used for functional selection must be tested with wild-type hairy roots. For B. napus DH12075 hairy roots carrying resistance to kanamycin, a concentration of 25 mg/L kanamycin is used.
NOTE: In this step, it is possible to generate a composite plant consisting of the wild-type shoot and transgenic hairy roots supporting the growth of such plant. Instead of cutting off the hairy roots from a stem, the native roots of the plant are removed. The plant with emerging hairy roots is transferred to a plant culture box with a composite plant growth medium containing cefotaxime (200 mg/L) and ticarcillin (500 mg/L) to suppress agrobacterial growth, and the selective antibiotic (carried by the T-DNA). - After 1 – 2 weeks, isolate the hairy roots on the Petri dish from the callus and individualize them on plates with the same culture medium. Transfer the culture to a fresh plate every 4 to 5 weeks. Add 0.25 mg/L IBA to increase root branching.
- Reduce the concentrations of cefotaxime and ticarcillin gradually at each transfer by 100 mg/L (i.e., the medium for the first transfer contains 100 mg/L cefotaxime and 400 mg/L ticarcillin; for the second transfer, 300 mg/L ticarcillin, etc.). After 3 – 4 months, culture the hairy roots on a hairy root growth medium with 100 mg/L ticarcillin and the selective antibiotic.
4. Regeneration of Brassica napus DH12075 hairy roots
- Transfer independent hairy root lines to plates with regeneration medium in sterile conditions using tweezers, flamed before use. Transfer 5 – 10 roots per Petri dish and culture them at 21 °C in a long-day photoperiod (16 h light / 8 h dark). Seal the Petri dishes with gas-permeable tape.
- Every 3 to 4 weeks, transfer the hairy roots to plates with fresh regeneration medium. Note that calli are formed after approximately 2 weeks. The callus starts shooting after 2 more weeks to till 8 – 9 weeks after callus formation.
- Individualize the shoots and transfer them to plant culture boxes with shoot elongation medium for 2 to 3 weeks to promote the elongation of the shoots.
- Transfer the elongated shoots to plant culture boxes with a root induction medium. Refresh the culture every 3 – 4 weeks. The rooting efficiency in DH12075 is 87% at 30 days and up to 100% after 60 days.
- Transfer the rooted plants to the soil after removing any traces of gelling agent to prevent fungal infection. Ensure that the plants are first acclimated in the phytotrons (21 °C, long-day photoperiod, 150 µE) and then transfer them to a greenhouse for flowering (21 °C / 18 °C, long-day photoperiod, 150 µE).
5. Regenerant and T1 plant selection
NOTE: The hairy root lines can be selected before going into the regeneration process. The type of selection depends on the content carried by the transgene. The hairy roots can be sampled for DNA extraction and genotyping or mutation detection, RNA extraction with subsequent cDNA synthesis and RT-qPCR for expression level analysis of the gene of choice, microscopy for fluorescence detection, or treated for GUS staining.
- After transfer to soil, genotype the T0 regenerant plants (and T1 seedlings) again to avoid escapes from the selection procedures. T0 plants exhibit an altered phenotype called Ri phenotype: extensive root growth, curly leaves, and dwarf shoots caused by the presence of the TL and/or TR-DNA of the Ri plasmid inserted into the plant genome.
- To speed up the T1 selection, use the seeds containing green mature embryos (approx. 21 – 28 days after pollination for DH12075 for a torpedo embryo or older) from T0 regenerant plants for embryo rescue.
- Work in the sterile flow box. Collect the siliques and surface-sterilize them with 70% ethanol.
- Place them on a double-sided tape taped on a plate lid or a slide.
- Using a stereo-binocular, slit the silique along the valve margins using a 26G needle. Ensure that the cut does not damage the seeds. For easy grip, mount the needle on a 1 mL syringe.
- Open the carpels and stick them to the tape. Collect the immature seeds and transfer them to plates with medium for seed germination. Seal the plates with gas-permeable tape.
- Place the plates in a cultivation room (21 °C, 16 h light /8 h dark) until germination.
- Genotype the T1 seedlings for the presence of the transgene of interest and the absence of the Ri TR/TL (Figure 1). Collect leaf material and extract their DNA using the method of choice. The CTAB method is described here:
- Collect the leaf material into a 2 mL microtube containing two ceramic beads. Freeze the tube in liquid nitrogen.
- Grind the material using a ball mill. Alternatively, pestle and mortar may be used.
- After a quick spin down, add 400 µL of CTAB buffer to the powder. Vortex briefly, spin down, and incubate at 60 °C for at least 50 min.
- Cool the solution to room temperature for 15 min. Add one volume of chloroform and mix gently.
CAUTION: When using chloroform, work under the chemical flow and use gloves for protection. Any solution containing chloroform should be discarded in the appropriate waste bin. - Centrifuge for 5 min at 18,400 x g using a tabletop centrifuge. Transfer 250 – 350 µL of the upper aqueous phase to a new microtube. Omit the interphase.
- Add one volume of isopropanol, mix well, and incubate for 5 min at room temperature. Centrifuge for 40 min at 18,400 x g.
- Discard the liquid and add 200 µL of 70% ethanol. Wash the pellet and centrifuge for 15 min at 18,400 x g.
- Discard the liquid. Air dry the pellet and add 50 – 100 µL of ultrapure water. Let the DNA dissolve for 1 h to overnight in the fridge.
- Perform a genotyping by PCR for the rolA gene (TL), the aux1 gene (TR), and the virC locus (Agrobacterium). Prepare the PCR reaction using the prepared DNA (from step 5.3.), primers, buffer, dNTP, and Taq polymerase according to the manufacturer's protocol. The amplified fragments are 200 – 500 bp long.
Primers specific for rolA:
Forward: GTTAGGCGTGCAAAGGCCAAG
Reverse: TGCGTATTAATCCCGTAGGTC
Primers specific for aux1:
Forward: CATAGGATCGCCTCACAGGT
Reverse: CGTTGCTTGATGTCAGGAGA
Primers specific for virC:
Forward: AATGCGTCTCTCTCGTGCAT
Reverse: AAACCGACCACTAACGCGAT
NOTE: T-DNA transfer and integration might be partial, and only parts of TL and/or TR may integrate into the genome. Thus, it is recommended to analyze the presence of other ORFs of TL (e.g., rolB and rolC) and TR (aux2, mas1, ags1) in T1 seedlings. Primer sequences specific to these loci are listed in Jedličková et al.9. - Evaluate PCR reactions by gel electrophoresis.
NOTE: Including a positive control in this analysis is recommended to ensure having PCR-grade DNA. - Select for the presence (or absence) of the transgene according to the protocols based on the content of this transgene.
- Transfer selected seedlings to the soil.
6. Hairy root transformation and regeneration in Arabidopsis thaliana Col-0
- Surface-sterilize A. thaliana seeds with a method of choice (bleach, ethanol, or chlorine gas).
- Plate sterile seeds on a medium for seed germination. After 2 days of cold stratification, move the plates to a cultivation room (21 °C with long-day photoperiod and 50% humidity).
- Transfer 1-week-old seedlings to a plant culture box with plant growth medium.
- Prepare Agrobacterium cultures as indicated in step 3.5.
- Inject a small amount of culture (approximately 50 µL) with a needle mounted on an insulin syringe at the base of the primary inflorescence stem (approximately 1 – 2 cm above the rosette) of 1-month-old Arabidopsis plantlets. The tissue on the surface of the primary stem can also be scratched by the syringe.
- At 2-4 weeks after the injection, excise the emerging hairy roots and cultivate them on Petri dishes with the hairy root growth medium supplemented with the selective antibiotic (carried by the T-DNA) and cefotaxime (200 mg/L) and ticarcillin (500 mg/L) to suppress agrobacterial growth. Incubate the plates at 24 °C in the dark.
- After 1 – 2 weeks, individualize the hairy roots on plates with the same culture medium. Transfer the selected hairy root lines to a fresh medium every 4 – 5 weeks.
NOTE: A. thaliana hairy roots are thinner than those of B. napus, and caution must be taken when transferring to fresh media. - Transfer the hairy roots to plates with regeneration medium to induce callus formation. Culture the plates at 21 °C in a long-day photoperiod (16 h light / 8 h dark).
- Shoots emerge from the callus after 18 – 21 days of cultivation. Cut the shoots and transfer them to a shoot elongation medium for 2 – 3 weeks to promote growth and elongation.
- Transfer the elongated shoots to the root induction medium.
- Transfer the rooted plants to the soil. T0 plants also present a Ri phenotype. Perform the transgenic selection, as described for B. napus DH12075 (step 5).
Representative Results
We have previously optimized a protocol for injection-based hairy root induction in three cultivars of Brassica napus, namely DH12075, Topas DH4079, and Westar9. To apply this transformation protocol to the model species A. thaliana, the primary inflorescence stems of 1-month-old plantlets were injected with an agrobacterial inoculum. Hairy roots emerged at the site of injection after 2-4 weeks. Hairy roots were excised and cultivated on the solid medium. The comparison of the method in these two species is depicted in Figure 1.
Selected hairy root lines were transferred to the regeneration medium to induce shoot formation. In A. thaliana, yellow calli were induced within 14 days in all 10 tested hairy root lines. First shoot primordia visible as dark green spots emerged within 3 weeks after transfer to the regeneration medium (Figure 2). After 4 weeks of culture, shoots covered the hairy roots in 9 of 10 hairy root lines (90% regeneration efficiency). In some cases, adventitious roots were elicited from the callus (Figure 2H). One line did not regenerate even after 3 months on the Regeneration medium (every 4 weeks, the hairy roots were transferred to a fresh medium). Thus, the regeneration efficiency of A. thaliana hairy roots resembles the efficiency of B. napus DH120759.
Hairy root regenerants of B. napus and A. thaliana display a dwarf phenotype (Figure 3), a typical feature of the hairy root-derived plants2. We also observed dense root systems, wrinkled leaves, and changes in flowering time. This so-called hairy root (or Ri) phenotype is caused by the rol genes from the Ri plasmid inserted into the plant genome. The insertion of the Ri T-DNA and the transgene encoded on a binary vector may be independent or linked. Thus, a segregation analysis of T1 progeny created by self-pollination helps to identify rol-free plants expressing the transgene of interest. Genotyping of the T1 plants is performed by PCR primers specific for the ORFs of TL and TR and the transgene of interest. The absence of agrobacterial contamination is verified by the absence of PCR products of virC primers (Figure 1).
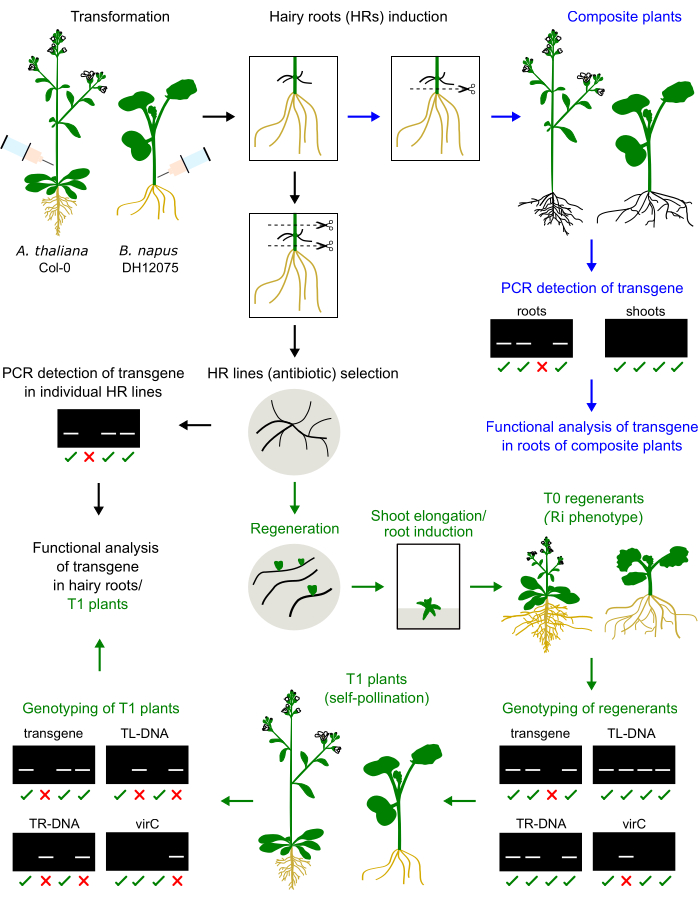
Figure 1: Summary of the procedure in A. thaliana and B. napus. The injection of the Agrobacterium inoculum in the hypocotyl or primary inflorescence stem induces the development of hairy roots. The hairy roots can replace the native roots to generate a composite plant that will be genotyped and analyzed (blue arrows). Cultured hairy roots can be regenerated into T0 plants, propagated in T1 plants, and genotyped (green arrows). The hairy roots can also be sub-cultured for functional analysis (black arrow). Examples of genotyping results are presented. Please click here to view a larger version of this figure.
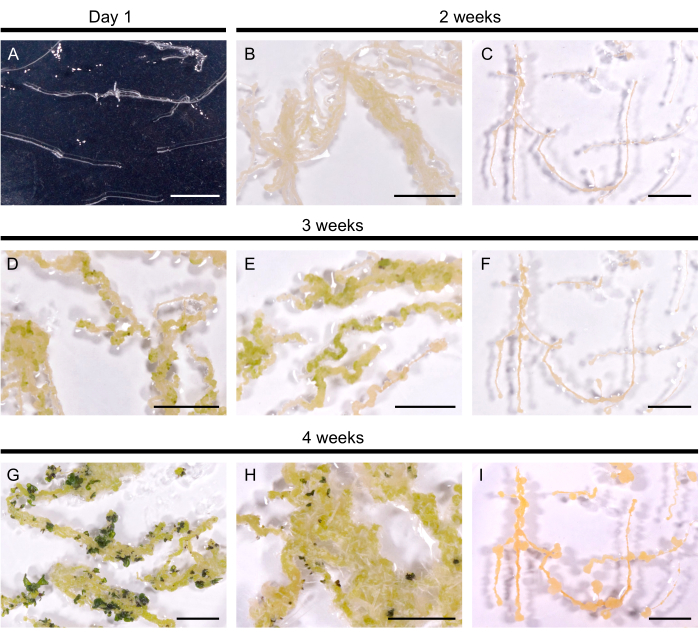
Figure 2: Regeneration of hairy roots in A. thaliana. (A) Hairy root culture 1 day after its transfer to plates. (B, C) Calli developed within 2 weeks of culture on regeneration medium. (D, E) Shoot primordia emerged after 3 weeks of culture. (G, H) Shoots are formed after 4 weeks. (H) Adventitious roots developed from the callus. (C, F, I) Non-regenerating hairy root line. Scale bars represent 1 cm. Please click here to view a larger version of this figure.

Figure 3: Representative photos of B. napus and A. thaliana wild-type plants and hairy-root-derived regenerants (T0 plants). Note the Ri phenotype of the regenerants. Scale bars represent 2 cm. Please click here to view a larger version of this figure.
Discussion
We developed a simple protocol for hairy root transformation and subsequent regeneration in B. napus and A. thaliana. This process includes injection-based hairy root induction in the hypocotyl (B. napus) or primary inflorescence stem (A. thaliana). The method of injecting the hypocotyl with agrobacterial strain C58C1 carrying a Ri plasmid was also effective in the Fabaceae family10,11, besides the members of Brassicaceae presented in this study.
An alternative to the injection-based method is the immersion-based transformation consisting of explant immersion in a bacterial suspension, followed by co-cultivation of the explant with agrobacteria. The advantage of the injection-based method over the immersion method is the time saved by the absence of some protocol steps: explant preparation, a test of the co-cultivation time, and culture on a medium containing hormone for induction of hairy roots. Although both approaches are effective for hairy root induction, higher transformation efficiency was observed in some species with the injection-based method compared to the explant immersion one12,13. Moreover, injection-based transformation is also useful for generating composite plants (transgenic hairy roots and wild-type shoots). After cutting off the original roots of the transformed plant, hairy roots support the plant growth, and the transgene can be studied in the context of the whole plant.
The critical step of hairy root induction is injecting the inoculum into the hypocotyl, or primary inflorescence stem. The hypocotyls of B. napus are breakable, and cutting the whole hypocotyl can easily happen. The same can be observed with A. thaliana because of the inflorescence stem thinness. If a comparison of the transformation efficiency of different species/cultivars is required, we recommend that one person perform all experiments to avoid the error caused by manipulation and skill in injecting the plants.
We developed an effective protocol for hairy root regeneration in B. napus DH12075 and A. thaliana Col-0. As regeneration is a highly variable process, some protocol modifications may be applied to a species or cultivar of choice. For instance, the hairy root-derived shoots can be elicited by a different auxin/cytokinin ratio (1:1) in B. oleracea14. Alternatively, cytokinin thidiazuron can be used instead of BAP, such as in the case of B. campestris hairy roots15.
Multiple insertions of the Ri plasmid T-DNA into the plant genome represent a potential limitation of the hairy root transformation and regeneration system. In such cases, no plants free of TL/TR from the Ri plasmid are uncovered after a segregation analysis of T1 seedlings. Thus, we recommend generating several independent hairy root lines for each transgene.
Hairy root cultures are an extremely powerful tool for gene functional studies mainly because of their rapid establishment and cheap maintenance (no hormones needed in cultivation media). This protocol covers the methods for hairy root induction and regeneration in B. napus and A. thaliana, which can be used to study the transgene of interest directly in hairy root cultures, in the context of the whole plant using composite plants, or after the regeneration of the transgenic plants.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We acknowledge Jiří Macas (Biology Centre CAS, České Budějovice, Czech Republic) for providing the agrobacterial strain. The Core Facility Plants Sciences of CEITEC MU is acknowledged for its technical support. This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic with the European Regional Development Fund-Project "SINGING PLANT" (no. CZ.02.1.01/0.0/0.0/16_026/0008446) and the INTER-COST project LTC20004.
Materials
| 1.50 mL tubes | Eppendorf | 125.215 | |
| 10% solution of commercial bleach | SAVO | ||
| 1-naphthaleneacetic acid (NAA) | Duchefa | N0903 | Callus regeneration medium |
| 2.0 mL tubes | Eppendorf | 108.132/108.078 | |
| 3M micropore tape | Micropore | ||
| 6-Benzylaminopurine (BAP) | Duchefa | B0904 | Callus regeneration medium, Shoot elongation medium |
| 70% ethanol | |||
| bacteriological agar | HiMedia | RM201 | LB medium |
| Bacteriological peptone | Oxoid | LP0037 | LB and YEB media |
| Beef extract | Roth | X975.1 | YEB medium |
| Bottles | DURAN | L300025 | |
| Cefotaxime sodium | Duchefa | C0111 | Hairy root growing medium, Callus regeneration medium, Shoot elongation medium, Root induction medium |
| chloroform | Serva | 3955301 | |
| CTAB Hexadecyltrimethylammonium bromide | Sigma | 52365 | |
| dNTP mix | Thermo Fisher Scientific | R0193 | |
| EDTA – Titriplex III, (Ethylenendinitrilo)tetraacetic Acid, Disodium Salt, Dihydrate | Sigma | ES134-250G | |
| elctroporation cuvette | |||
| electrophesis agar, peqGOLD universal | VWR | 732-2789 | |
| electrophoresis chamber | BIO-RAD | ||
| electrophoresis gel reader | BIO-RAD | ||
| electroporator GenePulser Xcell | BIO-RAD | ||
| ethidium bromide | AppliChem | ||
| Gene Pulser/MicroPulser electroporation cuvettes, 0.2 cm gap | BIO-RAD | 1652082 | |
| Gene Ruler DNA ladder mix | Thermo Fisher Scientific | SM0331 | |
| Gibberellic acid (GA3) | Duchefa | G0907 | Shoot elongation medium |
| glycerol | Sigma | G5516-1L | |
| HEPES (2-(4-(2-hydroxyethyl)-1-pirerazinyl)-ethansulfonique | Merck | 1101100250 | |
| indole-3-butyric acid (IBA) | Duchefa | I0902 | Root induction medium |
| kanamycin monosulfate | Duchefa | K0126 | |
| Magenta GA-7 Plant Culture Box w/ Lid | Plant Media | V8505-100 | |
| Measuring cylinder | |||
| MES monohydrate | Duchefa | M1503 | Hairy root growing medium, Callus regeneration medium, Shoot elongation medium, Root induction medium, Medium for germination, Plant growing medium |
| Murashige and Skoog medium (MS) | Duchefa | M0237 | Medium for germination, Plant growing medium |
| Murashige and Skoog medium (MS) + B5 vitamins | Duchefa | M0231 | Hairy root growing medium, Callus regeneration medium, Shoot elongation medium, Root induction medium |
| needle Agani 26G x 1/2 – 0.45 x 13mm | Terumo | ||
| pH meter | |||
| Phytagel | Sigma | P8169 | Callus regeneration medium, Root induction medium, Medium for germination |
| PVP 40 (polyvinylpyrolidone Mr 40000) | Sigma | 9003-39-8 | |
| Redtaq DNA Polymerase,Taq for routine PCR with inert dye, 10X buffer included | Sigma | D4309-250UN | |
| Retsh mill | Qiagen | ||
| sodium chloride | Lachner | 30093-APO | LB medium |
| square Petri Dishes | Corning | GOSSBP124-05 | |
| sucrose | Penta | 24970-31000 | Hairy root growing medium, Callus regeneration medium, Shoot elongation medium, Root induction medium, Medium for germination, Plant growing medium |
| Syringe filter | Carl Roth | P666.1 | Rotylabo syringe filters 0.22 µm pore size |
| thermomixer | Eppendorf | ||
| Ticarcillin disodium | Duchefa | T0180 | Hairy root growing medium |
| Tris(hydroxymethyl)aminomethan | Serva | 3719003 | |
| ultrapure water | Millipore Milli-Q purified water | ||
| Yeast extract | Duchefa | Y1333 | LB medium |
References
- Lee, L. Y., Gelvin, S. B. T-DNA binary vectors and systems. Plant Physiology. 146 (2), 325-332 (2008).
- Christey, M. C. Use of Ri-mediated transformation for production of transgenic plants. In Vitro Cellular & Developmental Biology – Plant. 37 (6), 687-700 (2001).
- Gelvin, S. B. Agrobacterium-Mediated Plant Transformation: the Biology behind the “Gene-Jockeying” Tool. Microbiology and Molecular Biology Reviews. 67 (1), 16-37 (2003).
- Georgiev, M. I., Agostini, E., Ludwig-Müller, J., Xu, J. Genetically transformed roots: from plant disease to biotechnological resource. Trends in Biotechnology. 30 (10), 528-537 (2012).
- Gutierrez-Valdes, N., et al. Hairy root cultures—a versatile tool with multiple applications. Frontiers in Plant Science. 11, 33 (2020).
- Niazian, M., Belzile, F., Torkamaneh, D. CRISPR/Cas9 in planta hairy root transformation: a powerful platform for functional analysis of root traits in Soybean. Plants. 11 (8), 1044 (2022).
- Petit, A., et al. Further extension of the opine concept: plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Molecular and General Genetics MGG. 190 (2), 204-214 (1983).
- Ozyigit, I. I., Dogan, I., Tarhan, E. A. Agrobacterium rhizogenes-mediated transformation and its biotechnological applications in crops. Crop improvement. , (2013).
- Jedličková, V., et al. Hairy root transformation system as a tool for CRISPR/Cas9-directed genome editing in oilseed rape (Brassica napus). Frontiers in Plant Science. 13, 919290 (2022).
- Steinbauerová, V., Neumann, P., Macas, J. Experimental evidence for splicing of intron-containing transcripts of plant LTR retrotransposon Ogre. Molecular Genetics and Genomics. 280 (5), 427-436 (2008).
- Neumann, P., et al. Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Molecular Biology and Evolution. 32 (7), 1862-1879 (2015).
- Montazeri, M., et al. A Comparative analysis of the hairy root induction methods in Hypericum perforatum. Journal of Plant Molecular Breeding. 7 (1), 67-76 (2019).
- Zhang, X., et al. Peat-based hairy root transformation using Rhizobium rhizogenes as a rapid and efficient tool for easily exploring potential genes related to root-knot nematode parasitism and host response. Plant Methods. 19 (1), 22 (2023).
- Christey, M. C., Sinclair, B. K. Regeneration of transgenic kale (Brassica oleracea var. acephala), rape (B. napus) and turnip (B. campestris var. rapifera) plants via Agrobacterium rhizogenes mediated transformation. Plant Science. 87 (2), 161-169 (1992).
- Christey, M. C., Sinclair, B. K., Braun, R. H., Wyke, L. Regeneration of transgenic vegetable brassicas (Brassica oleracea and B. campestris) via Ri-mediated transformation. Plant Cell Reports. 16 (9), 587-593 (1997).

