Tree Core Analysis with X-ray Computed Tomography
Summary
Here we show how to process tree cores with an X-ray computed tomography toolchain. Except for chemical extraction for some purposes, no further physical lab treatment is needed. The toolchain can be used for biomass estimations, for obtaining MXD/tree-ring width data as well as for obtaining quantitative wood anatomy data.
Abstract
An X-ray computed tomography (CT) toolchain is presented to obtain tree-ring width (TRW), maximum latewood density (MXD), other density parameters, and quantitative wood anatomy (QWA) data without the need for labor-intensive surface treatment or any physical sample preparation. The focus here is on increment cores and scanning procedures at resolutions ranging from 60 µm down to 4 µm. Three scales are defined at which wood should be looked at: (i) inter-ring scale, (ii) ring scale, i.e., tree-ring analysis and densitometry scale, as well as (iii) anatomical scale, the latter approaching the conventional thin-section quality. Custom-designed sample holders for each of these scales enable high-throughput scanning of multiple increment cores. A series of software routines were specifically developed to efficiently treat three-dimensional X-ray CT images of the tree cores for TRW and densitometry. This work briefly explains the basic principles of CT, which are needed for a proper understanding of the protocol. The protocol is presented for some known species that are commonly used in dendrochronology. The combination of rough density estimates, TRW and MXD data, as well as quantitative anatomy data, allows us to broaden and deepen current analyses for climate reconstructions or tree response, as well as further develop the field of dendroecology/climatology and archeology.
Introduction
Wood density is an easy-to-measure variable1 that reflects both the anatomical and chemical properties of the wood2. In biomass estimations of aboveground biomass, wood density is an important weighing variable 3,4,5, that is multiplied with the dimensions of the tree and a factor representing the carbon content of the wood. Wood density is tightly linked to the mechanical properties of timber6 and reflects the life history of a tree7.
Cell wall density is measured as being approximately 1500 kg/m³ and is considered fairly constant8, however intra-ring cell wall density variations should be considered as well8,9.Woody cells (in general tracheids in conifers, vessels, parenchyma and fibers in hardwoods) are oriented/shaped in different ways and cell wall thickness and lumen size of these cells varies10. Therefore, wood density varies between trees, within a tree (axial and transversal) and within short intervals within a tree ring11,12. In many cases the wood density variation at the ring scale also delimits the tree ring boundary13. Wood density and ultimately tissue fractions are generated and in this paper are broadly put into three categories (i.e., three different resolution scales), depending on the study goal (Figure 1) as described below.
Inter-ring scale: By measuring pieces of wood, a single value is obtained for that sample. This can be done through water immersion or geometrically14. This way, general biomass or wood technological variables can be obtained. To include pith-to-bark variation, these pieces of wood can be further divided into blocks that are measured manually to obtain information on the life history strategy15. When switching to low-resolution X-ray CT such as in medical scanners17,18, TRW data on medium-to-wide rings can be made in an efficient way on many samples18,19,20. This is also the scale that can be used to assess biomass from pith-to-bark from both temperate and tropical trees4,22, typically ranging in resolutions from 50 µm to 200 µm.
Ring scale: Wood is a recorder of past environmental conditions. The best known parameter is tree-ring width (TRW), but for global temperature reconstructions, maximum latewood density (MXD) records are proven to be a better proxy for temperature22. MXD is an easy-to-measure variable23, and a proxy for cell wall thickness and cell size on the last cells of a tree ring, and are at tree line and boreal sites positively linked to seasonal air temperature24: the warmer and longer the summers, more cell wall lignification occurs which thus increases the density of these last cells. Traditional measurements such as immersion and geometry are less accurate to determine this ring-level density. A previous work developed a toolchain for using X-ray film on thin-cut samples25. This sparked a revolution in both forestry and later paleoclimatology15,18, defining maximum latewood density (MXD), i.e., the peak density value often at the end of a ring, as a proxy for summer temperature. The basic principle is that the samples are sawn (approximately 1.2 mm to 7 mm13) to be perfectly parallel to the axial direction, and the sample is put on a sensitive film exposed to an X-ray source. Then these radiography films are read out through a light source that detects the intensity and saves the profiles and the annual tree ring parameters. These tools, however, require a significant amount of sample preparation and manual work. Recently this has been developed for X-ray CT in a more standardized way or based on mounted cores26. Resolution here ranges between 10 µm and 20 µm. TRW is measured on this scale as well, especially when dealing with smaller rings.
Anatomical scale: At this scale (resolution < 4 µm), the average density levels become less relevant as the main anatomical features are visualized and their width and proportions can be measured. Typically, this is done through making microsections or high-resolution optical scans or µ-CT scans. When the ultrastructure of the cell walls needs to be visualized, scanning electron microscopy is the most commonly used method27. At the anatomical scale, the individual tissue fractions become visible so that physiological parameters can be derived from the images. Based on the individual anatomical parameters and the cell wall density of wood, anatomical density can be derived for comparison with conventional estimators of wood density24.
Due to improved sectioning techniques and image software29,30, dendro-anatomy30 has been developed to have a more accurate record of the wood, both to have a closer estimate of the MXD in conifers and to measure several anatomical variables from broadleaf trees. On this scale, actual anatomical parameters are measured and related to environmental parameters31 . With µCT this level can be obtained as well32,33.
As wood is inherently hygroscopic and anisotropic, wood density needs to be carefully defined and the measurement conditions need to be specified, either as oven-dry, conditioned (typically at 12% moisture content) or green (as felled in the forest)34. For large samples and technical purposes, wood density is defined as the weight divided by its volume at given conditions. However, the value of wood density is strongly dependent on the scale at which it is measured, for instance from pith-to-bark wood density can double, and on a ring scale (in conifers) the transition of earlywood to latewood results in a significant rise in wood density as well, with a peak at the ring boundary.
Here, an X-ray CT scanning protocol of increment cores is presented in order to measure features at the aforenoted 3 scales (Figure 1). Recent developments in X-ray CT can cover most of these scales, due to a flexible set-up. The research goals will determine the eventual protocol for scanning.
A crucial limiting factor (which is inherently connected to the scaled nature of wood density and wood in general) is the resolution and time necessary for scanning. Examples demonstrate how to: (i) obtain inter-ring tree scale wood density profiles for biomass estimations in Terminalia superba from the Congo Basin, (ii) obtain density records from Clanwilliam cedar (Widdringtonia cedarbergensis) based on helical scanning on a HECTOR system35, and (iii) measure vessel parameters on sessile oak, on the Nanowood system. Both scanners are part of the suite of scanners at the UGent Center for X-ray Tomography (UGCT,
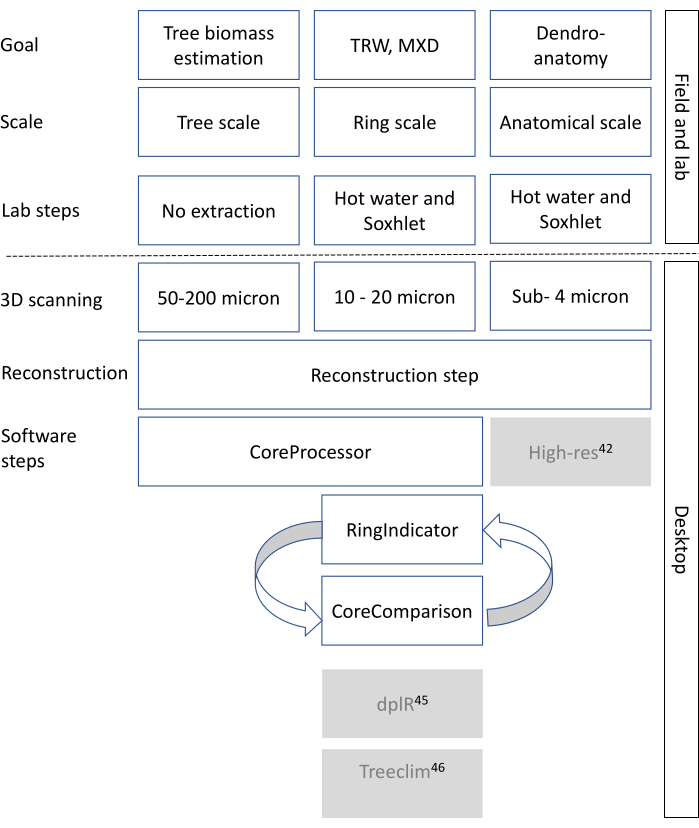
Figure 1: General methodological decision tree for X-ray CT scanning. The rows indicate the steps to take, starting from the research goal all the way to the final data format. White boxes are the steps that are relevant for this toolchain. Greyed-out boxes are steps that can be performed with other software or R packages, such as dplr47 and Treeclim48 for tree-ring analysis, and ROXAS44 as well as ImageJ42 or other (commercial) applications for deriving wood anatomical parameters based on the CT images. Please click here to view a larger version of this figure.
X-CT research on wood
Set-up of a scanner: A standard X-ray CT scanner consists of an X-ray tube, an X-ray detector, a rotation stage, and a set of motors to move the rotation stage, and in most cases also the detector, back and forth (Figure 2).
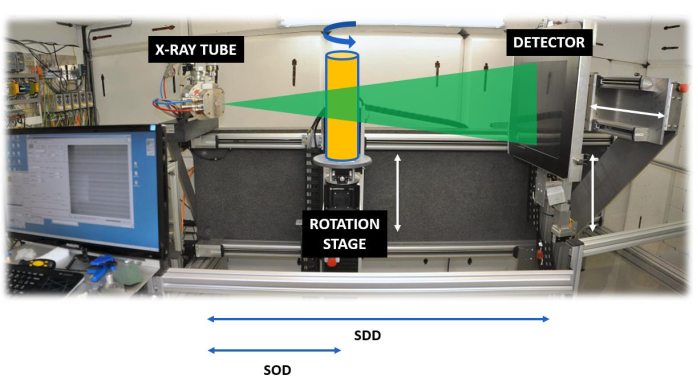
Figure 2. The HECTOR scanning system. The system35, showing the source detector distance (SDD) and the source object distance (SOD). Please click here to view a larger version of this figure.
Most lab-based systems have a cone-beam geometry, which means that the produced X-rays are distributed from the tube's exit window in a cone-beam shape, implying that by changing the distance between the object and the tube (SOD = Source-Object-Distance) and the detector and the tube (SDD = Source-Detector-Distance), the magnification is controlled (see the discussion on resolution). Due to the penetrating power of X-rays, they pass through the object, and the intensity of the attenuation beam is a function of the energy of the X-ray beam, the chemical composition of the object (the atomic number of the elements present) and the density of the material. Given a constant energy spectrum and a constant material composition of wood, the attenuation of the X-ray beam is highly dependent on the density of the material, which explains its use for densitometry. The attenuation (or transmission) can be expressed by the Beer-Lambert law:
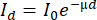
with I0 the incoming X-ray beam exponentially which decays to a transmitted X-ray beam Id when propagating through the material over a distance d. The linear attenuation coefficient µ depends on a series of interactions with the material of the object. The projections are thus recordings of the transmitted beam.
Practically, the object is mounted on the rotation stage, a proper SOD and SDD are selected, a certain power is selected as well (related to object size, density, and composition) and the object is rotated 360° and during that rotation multiple projections are taken. These projections are then used to reconstruct the interior structure of the object. There are several reconstruction algorithms available, of which the most used ones are still based on the analytical framework developed decades ago, relying on the Radon transformation and the Fourier slice theorem. For more details, the reader is referred to specialized literature36.
Conundrum of resolution, data volume and sample size: Resolution is key in X-ray CT scanning. In systems with inverse geometry, or parallel beam geometry such as synchrotron beamlines, other considerations play a role. This protocol only discusses standard lab-based X-ray CT scanning with cone-beam geometry. Here, the concept of magnification, detector pixel size and spot size are essential. Magnification is defined as the ratio of SDD/SOD. Next, the pixel size of the detector obviously also impacts the resolution: the smaller the pixel size, the higher the resolution, but in most cases the field of view (FoV) is also directly related to pixel size and size of the detector (smaller pixel size, smaller FoV for the same number of pixels). Further, the spot size of the X-ray beam is also important: the larger the spot size, the lower the resolution, which means that less details can be seen.
It is important to address that one could obtain a resolution that is higher than what is possible according to the abovementioned limits, therefore it is better to use the term voxel size (a voxel is a volume pixel) instead of resolution. Moreover, there are other factors at play, such as detector sharpness, which further limit the true resolution at which an object is scanned. Only a true calibration of the system, using established targets, supplies the truthful answer.
In most cases, however, the voxel size at which an object can be scanned is mostly limited by the size of the object. This means that the larger the object, the lower the voxel size will be. If the object does not fit within the FoV of the detector for a certain voxel size, then the voxel size can be reduced, e.g., by limiting the magnification.
Scan time and data volume are important to consider when deciding the desired voxel size. In general, the smaller the voxel size means the higher the detail one wants to see, the smaller the sample or the fewer samples that can be scanned at once, more time is required and larger the data volumes will be collected. Imagine the following theoretical example: one can scan a sample measuring 10 cm x 10 cm x 10 cm at 50 µm at once with a certain X-ray CT system and would like to scan that same volume at 10 µm, the volume that fits within the FoV would only be 2 cm x 2 cm x 2 cm, assuming that this is physically possible. This means that 125 scans are needed (5³ = 5 times higher resolution, scales to the power of 3 due to the volumetric nature of the imaging technique) to cover the entire volume, and that the data volume would increase likewise. Of course, this is only a thought experiment, and one needs to consider much more than only resolution. For more information, the reader is referred to an overview of the scanning possibilities37.
Flexibility of instruments for scanning of wooden objects: In the last decade, many companies have delivered X-ray CT systems with a similar assembly as HECTOR35. An overview of several CT systems, especially assessed for their temporal resolution, is given in38.
In all, the flexibility and user-friendliness of X-ray CT systems has improved considerably. Many systems allow scanning of a diverse range of objects, which is also the case with the systems at UGCT. The protocol below is demonstrated for the HECTOR system, which is suitable for tree-ring analysis. The protocol however is valid for any other available system if resolution and data format allows for it.
These systems allow the scanning of a variety of objects. A few pictures of different wooden objects scanned with the HECTOR system are given in Figure 3. It is this flexibility that comprises the three scales we present in Figure 1, ranging from a coarse resolution to a very fine resolution.
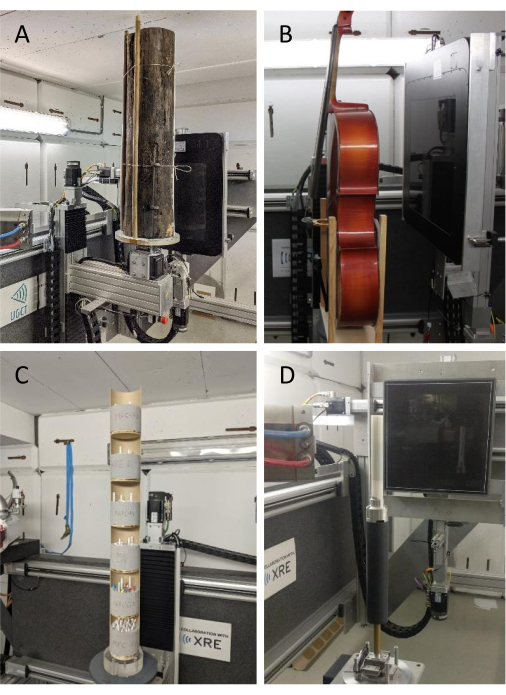
Figure 3. Scanning set-up examples. (A) A log, (B) a cello49, (C) sample holders (type 1) with tree cores for batch scanning and (D) sample holder type 2 with increment cores for helical scanning mounted on the rotation stage of HECTOR. Please click here to view a larger version of this figure.
Protocol
1. Core sampling
- Sample the tree with a Pressler borer. Consult references on how to core a tree manually39. This toolchain is presented for 5.15 mm cores.
- Put the tree cores unglued into 6 mm paper straws or in any other receptor that does not require glue. Do not glue the tree cores on a wooden support. If the cores were already mounted, unmount them with scalpel/saw or solvent depending on the glue type.
- Avoid wrapping them in plastic tubes, try to dry them first to avoid mold growth and fungal decay.
- Use pencil to write on the paper straws, since the extraction could fade marker writings.
NOTE: This toolchain is also suited for 4 mm cores. Already mounted and sanded cores can be used, but these need to be unmounted first. For archeological or remnant wood, a core-sized sample needs to be made (by sawing/reducing the size) for scanning.
2. Core preparation before scanning
- Perform an ethanol/toluene or ethanol or acetone40 Soxhlet extraction for at least 24 h (Figure 4A,B). This is necessary to ensure a resin-free wood volume to have an accurate density calculation (resin attenuates the signal, Figure 4C13, 41).
- Make a solution of 1000 mL of ethanol with 427 mL of toluene (according to e.g., ASTM D 1107 – 96) or pure ethanol or acetone. Many universities have a lab glass workshop that can make an adapted version of the Soxhlet apparatus for tree cores (Figure 4B).
- Fill the round-bottomed flask with the solvent, and put the samples within the straws in a stainless-steel support in the Soxhlet apparatus with cooler on a laboratory heater. Allow for at least 6 siphonings per h.
NOTE: A closed water circuit is preferably used for cooling (or any other modern cooling system). In this case, a water pump circulates the water, stored in a 100 L basin with chlorine tablets (to avoid algae development), at 120 L/h (but this will depend on the size of the Soxhlet apparatus as well as the applied heat by the heat element). An air cooler is used to cool the water in the 100 L basin. To save on solvent, glass beads can be used to fill-up the void volume of the Soxhlet apparatus. - Check the temperature and number of siphonings regularly. Dry the cores under a chemical hood (check lab policy for this) to remove the solvent residue or put a drying oven under the chemical hood.
- Dry the cores for 24 h in a drying oven (103.5 °C) under exhaust ventilation and perform a hot water bath (90 °C) or hot water Soxhlet extraction for 24 h as described above.
- Keep the samples in the paper straws in the steel sample holders. Dry again in a dry oven for 24 h at 103.5 °C, then condition to 60% relative humidity (RH) before scanning.
NOTE: The 60% value is chosen in this case because these are approximately the ambient average relative humidity conditions in the X-ray CT scanning room. Oven dry samples can also be used. The most important factor is that all samples are scanned under the same conditions. - In case the water extraction degenerates the paper straws, put the tree cores back into new 6 mm paper straws21. Both 5.15 mm and 4 mm cores can be put into 6 mm paper straws.
- Make sure the entire paper straw is filled with wood. Wrap both straw-ends up and cut the ends. This enables easy insertion of the straws in the cylindrical holder.
- Make sure the cambium (bark) side is clearly indicated on the core because the CoreProcessor and RingIndicator assume that the bark side is down. Remove bits and pieces as well as cores with cracks as these are difficult to work with.
- Keep the samples in the paper straws in the steel sample holders. Dry again in a dry oven for 24 h at 103.5 °C, then condition to 60% relative humidity (RH) before scanning.
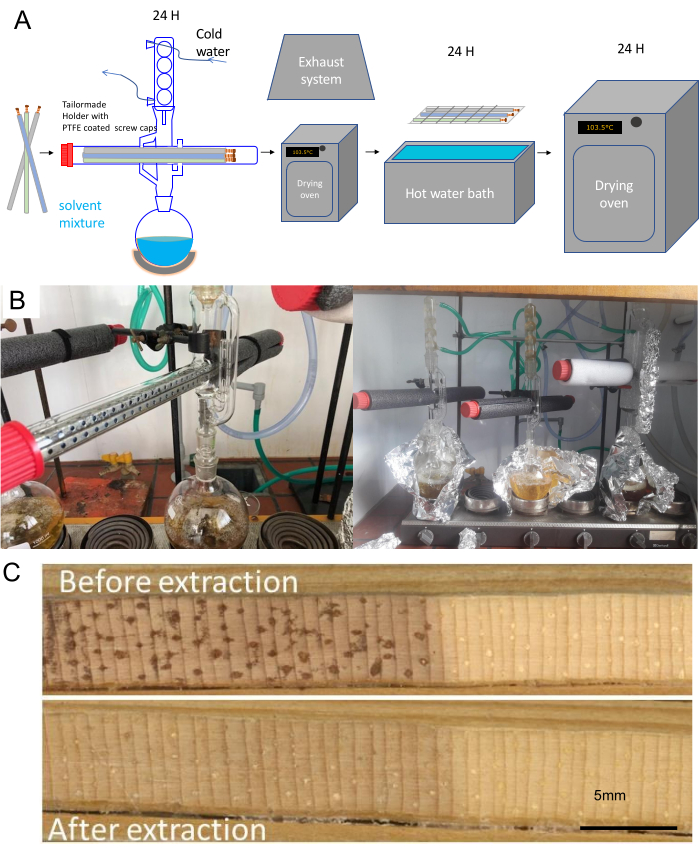
Figure 4. Workflow for extracting and drying cores prior to scanning. (A) Increment cores in paper straws are first put in a hot water bath and subsequently in a hot ethanol-toluene mixture with a Soxhlet apparatus for 24 h then dried, put in a hot water bath for 24 h, then dried again at 103.5 °C, then conditioned before scanning. (B) Images of the adapted Soxhlet set-up at Ghent University. Multiple adapted Soxhlet apparatuses are serially attached. Note the aluminum foil and insulation tubes to keep the solvent or solvent mixture warm enough for extraction. (C) Example of the surface of an increment core from Pinus longaeva before and after extraction. Resins and other extractives mask the true density signal, due to attenuation of the X-rays. Please click here to view a larger version of this figure.
3. Core scanning
- Select the proper sample holder type (Figure 5) as described below, depending on the research purpose as described in Figure 1.
- For inter-ring scale purposes, use holder type1 – cardboard and perforated floral foam.
- For the MXD data use holder type 2 – a plastic sample holder that holds 6 cores of 1-15 cm length and has a 3 cm diameter.
- Use holder type 3 – for dendro-anatomy purposes that consists of simply tying paper straws together.
- Load the tree cores in paper straws into the sample holder and fill in the spreadsheet template that can be downloaded on https://dendrochronomics.ugent.be/#software. Each number corresponds to a given position in the sample holder.
- Scanning in an X-ray µCT facility: consult an X-ray CT expert for the proper settings and scanning protocol. For the scanner used in this protocol, see37.
- Reconstruction of the scanned images: consult an X-ray CT expert for the proper settings of the reconstruction, or the settings that come with the CT scanner (software package of the X-ray CT scanner). For reconstruction parameters for this protocol, see37.
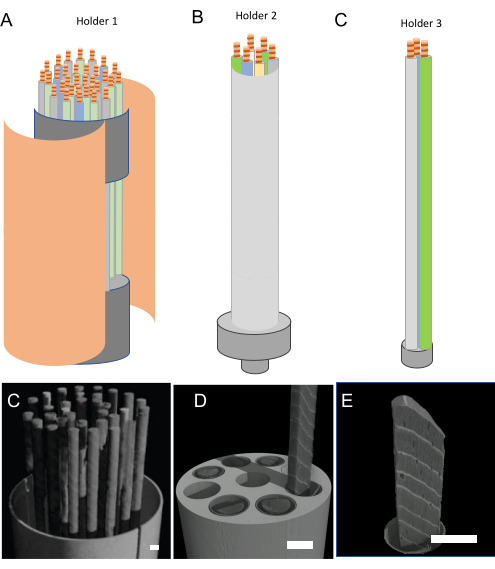
Figure 5. Sample holder types and resolution. Design of the three main holder types, that correspond to (A) inter-ring scale, (B) tree ring scale, and (C) anatomical scale. Due to the resulting size, the number of samples decreases with increasing resolution requirement. The corresponding 3D renderings for (D) inter-ring scale (holder 1), (E) ring width and MXD (holder type 2), and (F) anatomical parameters (holder type 3). Scale bar = 5 mm. Please click here to view a larger version of this figure.
4. Obtain TRW data, density data and anatomical data
- Getting started with the software.
- To obtain density values (holder type 1 and 2), install the following three software packages: (i) CoreProcessor, (ii) RingIndicator and (iii) CoreComparison toolboxes from
- Since these toolboxes were made in MATLAB, install the correct MATLAB compiler runtime (MCR), available on: http://nl.mathworks.com/products/compiler/mcr/. The version needed currently is MATLAB2022B. The MCR is free, just as the 3 software packages.
- Next to the folder of the reconstructed volumes, make a new empty folder for the MAT files (a file format from MATLAB that stores variables, etc.), e.g., matfiles_useful_name_sample_holder, this is necessary to store temporary parameters.
- Make sure the spreadsheet is in the folder of the reconstructed volumes, in order to fill in the samples in a later step.
- Preprocessing the core volumes with the CoreProcessor.
NOTE: For high-resolution anatomy data, following processing steps are not necessary (Figure 1), once the reconstructed volumes are available, refer to the ImageJ guidelines for 3D images42. Segmentation can be done through the Weka plugin43 for indicating vessels on individual slices for instance12, or in software plugins such as ROXAS44.- Prepare the core extraction: Prepare the automated extraction of the increment cores (depending on inter-ring scale Figure 6A or ring scale Figure 6B) as described below.
- Select the folder with reconstructed files (16-bit cross-sectional TIFF slices) and the spreadsheet file (XrayForm.xlsx).
- Select the empty folder from 4.1.3. This is the folder where CoreProcessor will create a MAT file that will be used in 4.2.2. for mass core extraction.
- Select the newly created empty MAT file folder or, if not created, select any other folder. The volume is loaded, and a longitudinal cross-section of the cylinder appears, check a few horizontal slices to ensure if all cores and references were correctly mounted into the cylinder.
- Click from bottom to top to select slices to evaluate this. These cross-sections will be used for the next step, so click across the entire length. Indicate the dark (air) and white references (Figure 6), as many times as indicated in the spreadsheet file. Double click after each selection.
- For sample holder 1, inspect the multiscreen with 9 cross-sections, generated through the reference material to check whether the density calculation was performed on the wood (and not on the paper straw or other non-wood material). This is essential for correct conversion to actual wood density values.
- In a new pop-up window, select every core separately by drawing a circle/ellipse around it. Perform labelling based on the information from the spreadsheet file, check whether the name is correct. Respect the order in which the template is filled. All info is stored in a MAT file, that will be used during the next step for extracting all cores at once.
- Click on Mass Core Extraction to carry out core extraction for all the cores in a given cylinder (that were indicated in 4.2.1.2), given that the metadata and spreadsheet files are located in the same folder.
NOTE: When using Mass core extraction, all cores will be extracted according to the labelling of the user (that are stored in the mat file folder). As a result of this operation, a new folder named Extracted will be created which contains all the separate cores. When multiple sample holders are prepared in the first step, and all the MAT files are stored in the same folder, all MAT files in this folder will be processed. - In the CoreProcessor toolbox, click on Manual Tg Correction, then select the Extracted folder to ensure proper orientation of the transversal and radial plane of every core volume, similar to mounting physical samples in a sample holder before sanding keeping the transversal plane in view (Figure 7B).
- Some cross-sections are displayed so that the user can see the grain direction. In the last plot (bottom right), inspect the slice that is presented and draw a line to indicate the grain direction. Double click to automatically rotate the core.
- Then a screen appears, here crop the volume of the core (Figure 7C), making sure that the rectangle encompasses the entire core diameter. This is necessary because the central point of the rectangle should be as close as possible to the core axis (for correct alignment in the RingIndicator module).
NOTE: If scanning at high resolution, select only material within the core at this stage and use the converter to switch to sample holder type 2. This is certainly helpful for ring-porous species for example.
- Perform the following optional steps in case of misaligned, flipped, or broken cores or incompatible image formats.
- Core tilt correction (only for sample holder 1): To automatically correct for core tilt (e.g., when the core is slightly inclined in the cylinder; Figure 7A). This is important for later steps. Select the Extracted folder created in the previous step.
NOTE: This is normally not needed if you have a sample holder type 2 or type 3 (Figure 5) or when all samples and sample holders are put perfectly upright. - Flip volumes: In case the sample was not put with the bark side down (the pith-bark direction is not left to right), change the direction by flipping cores. The function asks for a folder and will create a new folder.
- Glue cores: Stitch large parts of individual cores together virtually. The idea is not to stitch perfectly, but to make sure that, if needed, single cores are in the same multipage TIFF volume. Use the broken core indication (see step 4.3.4.9) to overcome the spaces in between the core pieces.
- Converter: If the volumes have been manipulated in another software package and are saved outside the toolbox (e.g., ImageJ), perform this step to get the header info correct. Use this step for 16-bit multipage TIFF volumes coming from another CT scanner as well and for using the RingIndicator toolbox.
NOTE: All steps from here are for structure direction corrections and finally, densitometry and tree ring analysis.
- Core tilt correction (only for sample holder 1): To automatically correct for core tilt (e.g., when the core is slightly inclined in the cylinder; Figure 7A). This is important for later steps. Select the Extracted folder created in the previous step.
- Prepare the core extraction: Prepare the automated extraction of the increment cores (depending on inter-ring scale Figure 6A or ring scale Figure 6B) as described below.
- Correcting the angles and indicating rings in RingIndicator.
NOTE: Following steps need to be taken chronologically in the RingIndicator module: (i) visualization of the transversal and radial plane, (ii) structure correction, (iii) creating the density profile, and (iv) indicating the rings manually or semi-automatically.- Perform visualization of transversal and radial plane as described below.
- To get familiar with the GUI, open the toolbox and select a multipage TIFF File (one that was created after extraction and after tg corrected, so from the folder name Tg corrected).
- Select the Half Thickness of Slice Image to display an average image of the central X slices, with X ranging from 1-10 with 1 being no averaging (i.e. only display the central slice), and 10 being an average image of the 10 central slices.
NOTE: Averaging can be beneficial to highlight ring borders better, and suppress noise, on the other hand it can also make ring borders look less distinct, especially when the fiber angle is substantial. Please tune this value according to the needs. It is important to mention that this is merely for displaying, it does not impact the underlying volume, nor does it impact the eventual calculations/results. - Inspect the graphical user interface (GUI). The GUI contains a top figure with the transversal plane of the core and a bottom figure with the radial plane of the core, and a menu (Figure 8).
- To load a new core volume, search for the Volume > Load Volume function to select a new multipage TIFF file in the directory. Now the user is ready to indicate the structure direction to subsequently indicate the actual tree rings.
- Structure correction to facilitate automatic tree ring indications.
- Manually click on the image to insert green bars for structure correction. Start with the pith (or oldest ring available) and end with the most recent ring. Along the core correct for structural deviations on both the transversal and the radial plane.
- Make sure to indicate the ring and fiber angle properly, because all subsequent indications will take an interpolated angle between the respective indications.
- Insert a green bar on one plane, it will automatically generate one on the other plane. After inserting the bar, move the nodes (white squares) at the end of the bar to change the angle. Use the middle node to change the bar position. Adjust the size of the nodes for ease of use.
- After every step, make sure to press Data > Export and Rings > Export Rings, to ensure that ring and fiber indications are written to the proper .txt files. Throughout all corrections, make sure to always perform these two actions.
- Calculating the density profile.
- Use the Densitometry > Densitometry plot to calculate the density profile. Create and plot a density profile by selecting Overlay Plotting > Plot Density Profile, that can serve as an overlay on the core planes.
- For MXD calculation, indicate a percentage (recommended is 20%, e.g., 20% means that only 20% of the highest density values are included), to exclude lower values from resin ducts, earlywood etc. For mean density values use 100%. For minimum density also use 100%.
- Perform manual and/or automatic indication of ring boundaries using one left mouse click per ring as described below.
NOTE: Insert the same green bars as in step 4.3.2.1., this time the user should do this now ring per ring (Figure 9).- Use Overlay Plotting > Plot Density Profile, to visualize the density profile on the image. It is best to indicate the ring boundary also in conjunction with this profile.
NOTE: Open as many instances from the toolboxes as needed, in combination with the CoreComparison toolbox (see further). - For automatic indications (recommended for conifers for instance), use Auto > Max/Min/Inflection detection. This function will automatically indicate tree ring boundaries based on the densitometry profile created in the previous step.For automatic indications (works best for boreal conifers or species with clear ring boundaries defined by the density profile), use Auto > Max/Min/Inflection detection. This function will automatically indicate tree ring boundaries based on the densitometry profile created in the previous step.
- Depending on the species (diffuse-porous/ring-porous or coniferous), use the maximum/minimum, or curve inflection (right after the density peak) as the tree ring boundary criterion. The first parameter is a threshold for the peak function, the second is a smoother function.
- After performing the automated ring indications, perform automatic shifting (with window size of 1 pixel and user-defined iterations) , while making sure that no ring indications are plotted on each other due to this shift and re-run the densitometry plot.
- Change the felling date to the year at which the increment cores were taken on living trees or any other date that is suitable. The default is set at 1900.
- In Overlay Plotting, select Plot Rings and the years are displayed. Also select the plane on which to plot rings and/or the density profile.
- Once there are two cores fully indicated (e.g. from the same tree), do pairwise comparisons of core ring width curves by opening the CoreComparison toolbox. Open one RingIndicator instance per core and edit the indications if necessary. Export them again and arry out this iterative and interactive approach to obtain good quality data. In the next step (step 4.4.1.), do this for all cores to export both the TRW data and the corresponding density values.
- Check double-ring positions in the Rings > Check Rings option; this will check if any indications have a position outside the images as well as rings that are overlapping, e.g., in case if a double click has happened twice on the same position.
- For broken cores that have bits and pieces, start at the pith, and demarcate anomalies (air due to broken cores, bright spots due to resin pockets, contamination, and decayed parts) as if they were rings to delete them afterwards. End with the tree ring boundary of the last formed ring and export the data. Try to avoid broken cores during sampling (see step 1.1. and39).
NOTE: Both missing as well as broken rings can be indicated, see Figure 10. Two cases are defined, one where the core breaks in the middle of a ring, and one where the core breaks on a ring boundary. For the first one (Figure 10A), indicate the gap, note the number of that gap followed by number 1 and close the brackets. Then the program will sum the two parts of the ring to obtain a correct TRW value. In the second case, the gap number is indicated followed by number 2, and the program will omit this section (Figure 10B). - By indicating rings and generating density profiles, several .txt files are created and stored in the same folder as the multipage TIFF files, and the most important ones are listed in Table 1. Keep these .txt files in the same folder as the TIFF images, in order to further treat them in the CoreComparison module (step 5.4).
NOTE: For calculating density values in a different way based on the raw density profile (for instance latewood width vs earlywood width45 or other metrics not supported by the CoreComparison and CoreProcessor toolboxes), use the zpos_corr and density_corr text files and use these for such calculations.
- Use Overlay Plotting > Plot Density Profile, to visualize the density profile on the image. It is best to indicate the ring boundary also in conjunction with this profile.
- Perform visualization of transversal and radial plane as described below.
- For comparing TRW series and exporting the data via CoreComparison , follow the steps below.
NOTE: When using this toolbox, two windows are opened simultaneously, the CoreComparison and the PatternMatching module. The CoreComparison deals with cross dating and exporting of TRW data, whereas the PatternMatching module can be used for exporting density profiles and density values (MXD, minimum density (MND), mean density, and 4 quartile values). The main goal of CoreComparison is to generate the TRW values and density values, and to have a rough estimate of cross dating quality, with a coupling to RindIndicator in case of missing rings or wrong indications.- Perform the following steps for cross dating or matching the ring width pattern with other series.
- Once the RingIndicator section is completed, select the text files to compare, and a screen will open with ring widths, together with cross dating or statistical parameters such as Gleichlaüfigkeit (GLK)46 and or Spearman correlation between individual series (Figure 11).
- To make changes in the ring indications (for instance during cross dating), re-open RingIndicator, import data, export the modified data and rings, and use the Refresh button in the CoreComparison module (see top left-hand side) to see the changes.
- Open multiple RingIndicator instances to make changes to different tree-ring series and use multiple screen monitors to do this.
- In the GLK and CORR menu (Figure 11), perform a series of actions related to GLK, such as thresholding the figures, exporting the GLK values to a spreadsheet and ranking them.
- Obtain TRW values from CoreComparison as described below.
- Use the plotting and exporting function in CoreComparison to change the offset between the curves (a larger value result in a smaller offset). Visualize the TRW data in a plot by selecting Plotting and Exporting > Ring width.
- Export the TRW data by clicking Plotting and Exporting > Export RW Data. Export ring widths in spreadsheet format or in Tucson format.
- Obtain the density profiles from the PatternMatching module as described below.
- To obtain the density profiles in spreadsheet, go to the PatternMatching module, and after loading in the profiles, in the menu Other select Plotting > Export Corrected Density Profiles.
- To batch process profiles or to assess the data using different methods, directly use the .txt files from Table 1 (see step 4.3.4.11).
- Obtain MXD, MND, and other density values from the PatternMatching module as described below.
- For obtaining the mean density, MXD, MND and quartile data per tree ring, use Other Plotting > Export Clustered Data. A spreadsheet will be generated and can be found in the same folder as the TIFF files.
- The tabs in the spreadsheet are detailed in Table 2. Export these tree ring variables to Rstudio or other software for further analysis.
- Perform the following steps for cross dating or matching the ring width pattern with other series.
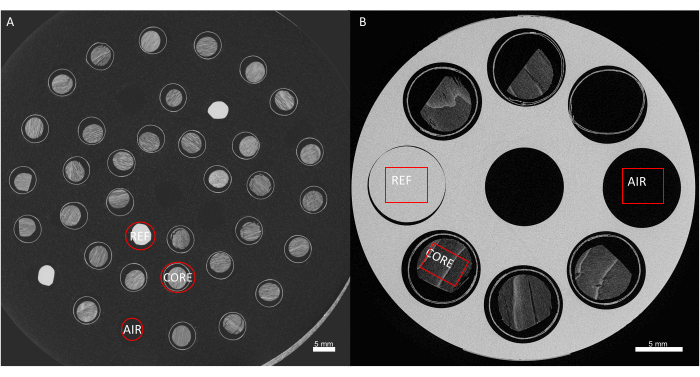
Figure 6. Obtaining wood density values from grey values. Conversion of the grey values of the image to actual wood density estimates, through calibration with a reference material. (A) Sample holder 1, at 60 µm resolution, showing the air reference, white reference, and the core. (B) Sample holder, at 20 µm, air, reference and core are shown as well. Please click here to view a larger version of this figure.
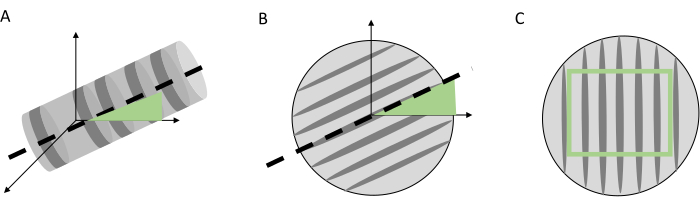
Figure 7. Pre-processing steps of the 3D core volume. Pre-processing steps of the 3D core volume before visualization and indicating the tree rings. (A) Tilt (axial) correction, (B) tangential correction, with the grey bars indicating the fiber direction, (C) the cropping of the core volume to obtain a volume with only woody voxels. Please click here to view a larger version of this figure.
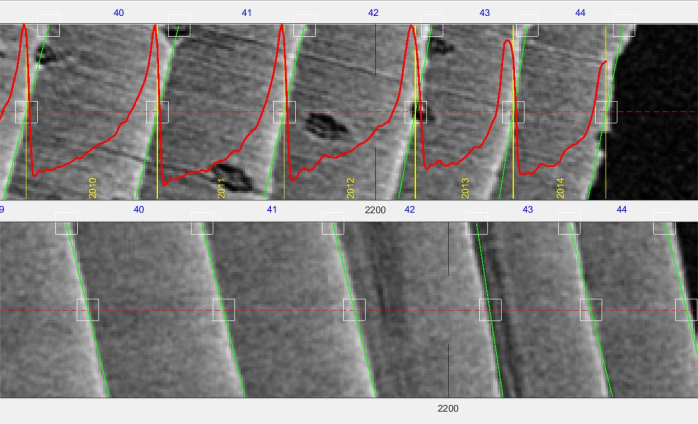
Figure 8. Screenshot of a core. Screenshot of a core (tangential width approximately 3 mm, resolution µm) opened in the RingIndicator toolbox. Top pane is the transversal view, bottom pane is the radial view. Green bars show the structure indication (in this case on the ring boundary), red line is the density profile. The yellow numbers indicate calendar year, and the blue numbers the generic ring number (counting from the first indication). Please click here to view a larger version of this figure.
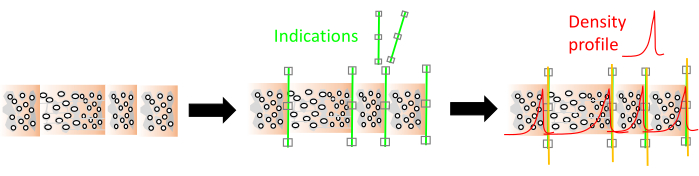
Figure 9. Ring indication and density profile visualization in the RingIndicator module. Green lines are indications, red line is the wood density profile. Please click here to view a larger version of this figure.
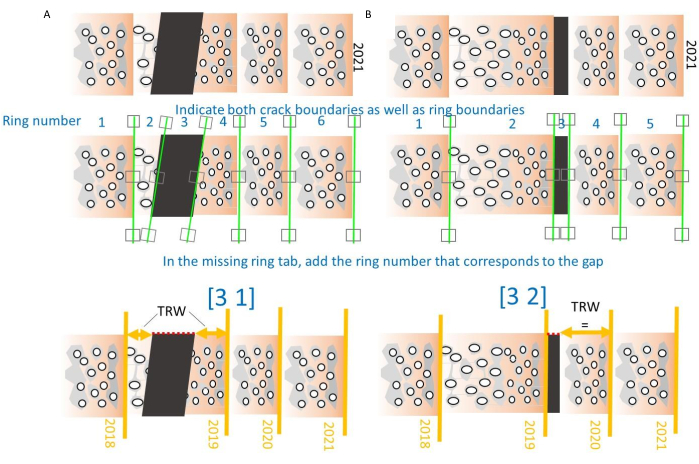
Figure 10. How to deal with cracks in the cores. (A) When a crack occurs in the middle of the ring, the generic ring number (starting from the pith) will be labeled as a gap, and TRW will be calculated by summing the widths of the two ring parts. Parameters are 3 (number of the ring) and 1 (case 1: crack in the middle of a ring)31. (B) When a crack occurs at a ring boundary, the gap will be omitted from the TRW calculation. Parameters are 3 (number of the ring) and 2 (case 2: crack at the ring boundary)31. Please click here to view a larger version of this figure.
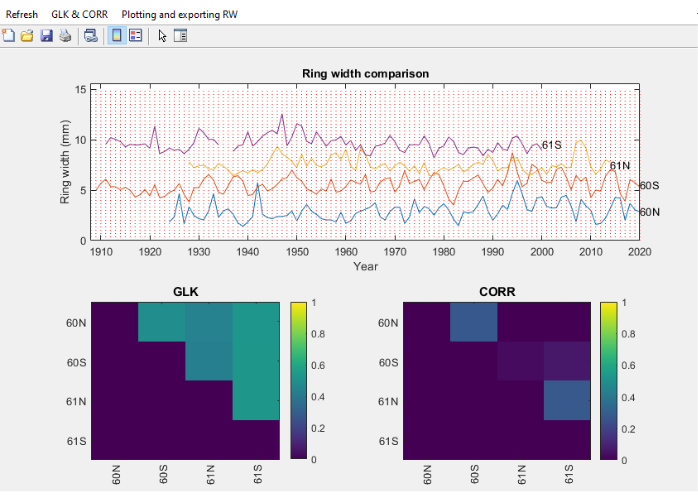
Figure 11. Screenshot of the Core Comparison module. Screenshot of the Core Comparison module, where ring width series are distributed vertically for easy visual comparison. The top pane shows the ring width of 4 cores from Fagus sylvatica, the bottom left pane shows the Gleichlaüfigkeit (GLK) and the bottom right pane the Spearman correlation coefficient. Please click here to view a larger version of this figure.
| File name | Explanation | ||
| SAMPLENAME_density.txt | Uncorrected density values for the entire length of the image, thus not corrected for ring and fibre angle | ||
| SAMPLENAME _density_corr.txt | cf. previous txt but then with corrected density values | ||
| SAMPLENAME _ring-and-fibre.txt | ring and fibre indication positions and angles | ||
| SAMPLENAME _zpos.txt | position of the tree ring boundaries on your core, without ring and fibre correction | ||
| SAMPLENAME _zpos.corr.txt | idem as zpos, but then corrected for ring and fibre angle | ||
| SAMPLENAME _ringwidth.txt | calculated ring widths, considering ring and fibre angle | ||
Table 1. Text files generated by RingIndicator. List of text files that are generated after indicating rings and exporting the density profile in RingIndicator. These should remain in the same folder as the .tiff file in order to be opened in the CoreComparison module.
| Tree-ring variable | Explanation | ||
| Mean density | mean density per tree ring | ||
| Max density half-based | MXD but only based on the second half of the tree ring to avoid that high density values of the previous year are not included | ||
| Min density | the minimum density per tree ring | ||
| Q1-Q4 | mean and maximum values for each quartile of the intra-annual ring profile | ||
Table 2. Spreadsheet details. Tree-ring variables that are exported into a spreadsheet named export clustered data.
Representative Results
If the goal is biomass estimation or tree growth increment of many samples, i.e., inter-ring scale (Figure 1), then sample holder 1 (Figure 5) is used to scan samples to obtain density profiles (see step 5.4.3) and estimates of tree growth, e.g., for fast-growing trees with large TRW, which allows for a coarser resolution. Figure 12 shows an example of both pith-to-bark and axial density trends of T. superba, a species from the Congo basin. These diagrams were based on scans of 110 µm from tree increment cores taken at various height positions in the stem.
For the ring scale (Figure 1) that is targeted by scanning cores with sample holder 2 (Figure 5B), tree ring analysis and MXD calculation is possible. An example of this is Figure 13, where a minimum density (in this case linked to wet season precipitation) and maximum density chronology is plotted for cores of W. cedarbergensis from the Cedarberg mountains in South Africa26. This was based on scanning mounted cores at 10 µm resolution.
For the anatomical scale (Figure 1), scanning cores with holder 3, i.e., multiple cores in papers straws wrapped together, allows a resolution of up to 4 µm. The result is shown in Figure 14, where an X-ray CT image slice of oak (Q. robur) is shown, with both earlywood and latewood vessels segmented.
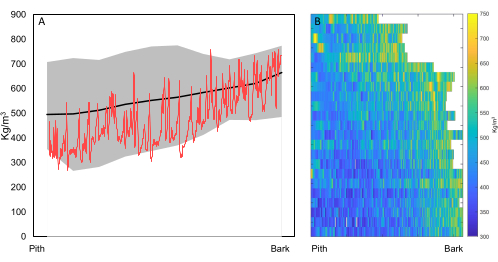
Figure 12. Radial and axial wood density trends. (A) Radial variation of ovendry wood density (kg/m3) from 46 Terminalia superba trees from the Congo Basin (DRC; data extracted from 21), with minimum (min), mean, and maximum (max) values, with a raw profile (red) superposed. (B) Radial variation and axial variation from a single Terminalia superba tree, where at every height interval of 1 m an increment core was taken and scanned (23 cores in total). There is an increasing density trend from pith to bark (highlighted according to colorbar) , and a higher wood density in the upper stem. Scanning resolution = 110 µm. Please click here to view a larger version of this figure.
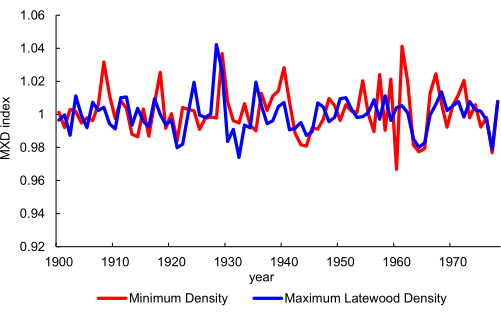
Figure 13. Chronology development. A minimum density and a maximum latewood density chronology from Widdringtonia cedarbergensis. Data extracted from26. The scanning resolution is 10 µm. Please click here to view a larger version of this figure.
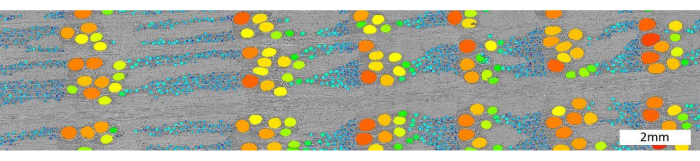
Figure 14. High-resolution scan of an oak sample. Example of a segmentation of oak vessels on Quercus robur scanned at 4 µm. Segmentation was performed using the Octopus analysis software, using bilateral filtering, thresholding, a series of binary shrink and expand operations, Euclidian distance transform and finally labelling of the vessels. Color represents vessel size, ranging from small (dark blue) to large (dark orange). Please click here to view a larger version of this figure.
Discussion
Critical steps within the protocol
Critical steps within the protocol include proper handling of the increment borer to obtain high-quality increment cores (step 1.1. and see39) to avoid bits and pieces. Next, it is essential that cores be left unmounted (but see26), both for insertion in the sample holder (Figure 5, see21) as well as for proper resin extraction50 and for possible future analysis. Right before scanning (step 2.2.2.), the conditioning of the samples in the scanner room is essential to avoid dimensional deformations due to fluctuations of moisture. Then, once the rings can be indicated in the RingIndicator module, it is essential that the ring boundaries and angles are well indicated, because the density peaks can flatten, similar to inaccurate indications on classic densitometry systems13. The indication is also important to have correct ring width measurements21. The next critical step is proper cross dating through the CoreComparison module (step 4.4.1.). If some cores have wrong indications, there could be an environmental signal loss51.
Modifications and troubleshooting of the technique
Note that the toolboxes RingIndicator and CoreComparison can be used for flatbed images as well52 after converting the data to the proper resolution in the CoreProcessor module. Therefore, the DICOM format (common format for medical purposes and is also used for wood samples53) needs to be converted to .tiff. Departments that have medical CT scanner can use this software only for the type 1 inter-ring scale analysis type with resolutions of around 200 µm.
Limitations of the technique
The drawback of the method is that there are only a few facilities available for scanning for tree ring scale and anatomical scale (which require a higher resolution). Data volume and data handling can be challenging for larger volumes54. For very high-resolution images of wood anatomy (e.g., to further elucidate pit structures etc.), refer to classic procedures of using microsections for classic transmitted light microscopy27, scanning electron microscopy or confocal microscopy55.
Significance with respect to existing methods
Using the X-ray CT toolchain for increment cores presented here has been tested as showing reliable density values due to the calibration step (Figure 6)56 and generates MXD values comparable to existing densitometry procedures13.
Future applications of the technique
Any future applications of the technique might include the application of deep learning, amongst others for the automated ring indications and wood tissue quantifications57, specifically for 3D images.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank the three anonymous reviewers for their feedback and suggestions. This research was funded by the BOF Special Research Fund for JVdB (BOF Starting Grant BOF.STG.2018.0007.01), for the UGCT as a Center of Expertise (BOF.EXP.2017.0007) and as a Core Facility (BOF.COR.2022.008), The authors also acknowledge the Research Foundation Flanders (G019521N and G009720N), and the UGent Industrial Research Fund (IOF) for the financial support to the infrastructure through grant IOF.APP.2021.0005 (project FaCT F2021/IOF-Equip/021).
Materials
| 6 mm paper straws | http://artstraws.com/education/product/artstraws-thick-asst-cols/ | Coring | |
| Aluminium core holders | |||
| ASTM standard ASTM D 1107 – 96 | |||
| Cardboard postal tubes | https://www.rajapack.co.uk/envelopes-mailing-bags/postal-tubes/1-5-2mm-brown-cardboard-postal-tubes_PDT05623.html | ||
| Column drill | |||
| Computer hardware for reconstruction and analysis | X-ray CT scanning | ||
| Cooling | |||
| Drying oven | |||
| Ethanol | |||
| Flask for under soxhlet (2000 ml) | |||
| Floral foam | https://www.oasisfloral.eu/ | Sample holder type 1 | |
| Glass beads to fill void volume of Soxhlet to save solvent | |||
| Glue | |||
| Hot water bath | https://www.memmert.com/products/water-baths/water-bath/#!filters=%7B%7D | Soxhlet extraction | |
| Increment borer | https://haglofsweden.com/project/increment-borers/ | ||
| Plastic cylinder | Moonen et al. 2022 | Sample holder type 2 | |
| Plastic cylinders | |||
| Reservoir | |||
| Tailored soxhlet apparatus | |||
| Toluene | |||
| Water pump | |||
| X-ray CT scanner |
References
- Björklund, J., et al. The utility of bulk wood density for tree-ring research. Dendrochronologia. 69 (September), 125880 (2021).
- Lachenbruch, B., Mcculloh, K. A. Traits, properties, and performance: How woody plants combine hydraulic and mechanical functions in a cell, tissue, or whole plant. New Phytologist. 204 (4), 747-764 (2014).
- Baker, T. R., et al. Variation in wood density determines spatial patterns in Amazonian forest biomass. Global Change Biology. 10 (5), 545-562 (2004).
- Bastin, J. F., et al. Wood specific gravity variations and biomass of central African tree species: The simple choice of the outer wood. PLoS ONE. 10 (11), 1-16 (2015).
- Chave, J., et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Global Change Biology. 20 (10), 3177-3190 (2014).
- Chave, J., et al. Towards a worldwide wood economics spectrum. Ecology letters. 12, 351-366 (2009).
- Plourde, B. T., Boukili, V. K., Chazdon, R. L. Radial changes in wood specific gravity of tropical trees: inter- and intraspecific variation during secondary succession. Functional Ecology. 29 (1), 111-120 (2015).
- Decoux, V., Varcin, &. #. 2. 0. 1. ;., Leban, J. -. M. Relationships between the intra-ring wood density assessed by X-ray densitometry and optical anatomical measurements in conifers. Consequences for the cell wall apparent density determination. Annals of Forest Science. 61, 251-262 (2004).
- Rathgeber, C. B. K., Decoux, V., Leban, J. M. Linking intra-tree-ring wood density variations and tracheid anatomical characteristics in Douglas fir (Pseudotsuga menziesii (Mirb.) Franco). Annals of Forest Science. 63 (7), 699-706 (2006).
- Ziemińska, K., Butler, D. W., Gleason, S. M., Wright, I. J., Westoby, M. Fibre wall and lumen fractions drive wood density variation across 24 Australian angiosperms. AoB PLANTS. 5, plt046 (2013).
- Ziemińska, K., Westoby, M., Wright, I. J. Broad anatomical variation within a narrow wood density range – A study of twig wood across 69 Australian angiosperms. PLoS ONE. 10 (4), 1-25 (2015).
- De Mil, T., et al. Wood density profiles and their corresponding tissue fractions in tropical angiosperm trees. Forests. 9 (12), 763 (2018).
- Björklund, J., et al. Scientific Merits and Analytical Challenges of Tree-Ring Densitometry. Reviews of Geophysics. 57 (4), 1224-1264 (2019).
- Maniatis, D., Saint André, L., Temmerman, M., Malhi, Y., Beeckman, H. The potential of using xylarium wood samples for wood density calculations: A comparison of approaches for volume measurement. IForest. 4 (1), 150-159 (2011).
- Lehnebach, R., et al. Wood density variations of legume trees in French Guiana along the shade tolerance continuum: Heartwood effects on radial patterns and gradients. Forests. 10 (2), 1-22 (2019).
- Longuetaud, F., et al. Within-stem maps of wood density and water content for characterization of species: a case study on three hardwood and two softwood species. Annals of Forest Science. 73 (3), 601-614 (2016).
- Steffenrem, A., Kvaalen, H., Dalen, K. S., Høibø, O. A. A high-throughput X-ray-based method for measurements of relative wood density from unprepared increment cores from Picea abies. Scandinavian Journal of Forest Research. 29 (5), 506-514 (2014).
- Vannoppen, A., et al. Dendrochronologia Using X-ray CT based tree-ring width data for tree growth trend analysis. Dendrochronologia. 44, 66-75 (2017).
- Maes, S. L., et al. Evaluating the robustness of three ring-width measurement methods for growth release reconstruction. Dendrochronologia. 46 (May), 67-76 (2017).
- Van Den Berge, S., et al. Biomass increment and carbon sequestration in hedgerow-grown trees. Dendrochronologia. 70 (September), 125894 (2021).
- De Mil, T., Vannoppen, A., Beeckman, H., Van Acker, J., Van den Bulcke, J. A field-to-desktop toolchain for X-ray CT densitometry enables tree ring analysis. Annals of Botany. 117 (7), 1187-1196 (2016).
- St. George, S., Esper, J. Concord and discord among Northern Hemisphere paleotemperature reconstructions from tree rings. Quaternary Science Reviews. 203, 278-281 (2018).
- Schweingruber, F., Fritts, H., Braker, O., Drew, L., Schar, E. The X-ray technique as applied to dendroclimatology. Tree-Ring Bulletin. 38, 61-91 (1978).
- Björklund, J., et al. Cell size and wall dimensions drive distinct variability of earlywood and latewood density in Northern Hemisphere conifers. New Phytologist. 216 (3), 728-740 (2017).
- Polge, H. Applications dans les domaines Technologique et Physiologique. Annales des sciences forestières. 23 (1), 215 (1966).
- De Mil, T., et al. A lonely dot on the map: Exploring the climate signal in tree-ring density and stable isotopes of clanwilliam cedar, South Africa. Dendrochronologia. 69 (November 2020), 125879 (2021).
- Jansen, S., et al. Preparation of wood specimens for transmitted light microscopy and scanning electron microscopy. Belgian Journal of Botany. 131 (1), 41-49 (1998).
- Gärtner, H., Nievergelt, D. The core-microtome: A new tool for surface preparation on cores and time series analysis of varying cell parameters. Dendrochronologia. 28 (2), 85-92 (2010).
- von Arx, G., Crivellaro, A., Prendin, A. L., Čufar, K., Carrer, M. Quantitative Wood Anatomy-Practical Guidelines. Frontiers in Plant Science. 7 (June), 781 (2016).
- Seftigen, K., et al. Prospects for dendroanatomy in paleoclimatology — a case study on Picea engelmannii from the Canadian Rockies. Climate of the Past. 18 (5), 1151-1168 (2022).
- Castagneri, D., Regev, L., Boaretto, E., Carrer, M. Xylem anatomical traits reveal different strategies of two Mediterranean oaks to cope with drought and warming. Environmental and Experimental Botany. 133 (October), 128-138 (2017).
- Brodersen, C. R., et al. Automated analysis of three-dimensional xylem networks using high-resolution computed tomography. The New phytologist. 191 (4), 1168-1179 (2011).
- Van den Bulcke, J., et al. X-ray tomography as a tool for detailed anatomical analysis. Annals of Forest Science. 66 (5), 508 (2009).
- Williamson, G. B., Wiemann, M. C. Measuring wood specific gravity…Correctly. American journal of botany. 97 (3), 519-524 (2010).
- Masschaele, B., et al. HECTOR: A 240kV micro-CT setup optimized for research. Journal of Physics: Conference Series. 463 (1), 012012 (2013).
- Kak, A. C., Slaney, M. . Principles of Computerized Tomographic Imaging. , (2001).
- Van Den Bulcke, J., et al. Advanced X-ray CT scanning can boost tree ring research for earth system sciences. Annals of Botany. 124 (5), 837-847 (2019).
- Zwanenburg, E. A., Williams, M. A., Warnett, J. M. Review of high-speed imaging with lab-based x-ray computed tomography. Measurement Science and Technology. 33 (1), 012003 (2022).
- Gärtner, H., Cherubini, P., Schneider, L., Lucchinetti, S. Advanced Workflow for Taking High-Quality Increment Cores – New Techniques and Devices. JoVE. (193), e64747 (2023).
- Schweingruber, F. H., Fritts, H. C., Bräker, O. U. The X-ray technique as applied to dendroclimatology. Tree-Ring Bulletin. 38, (1978).
- Grabner, M., Wimmer, R., Gierlinger, N., Evans, R., Downes, G. M. Heartwood extractives in larch and effects on X-ray densitometry. Canadian Journal of Forest Research. 35 (12), 2781-2786 (2005).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- Arganda-Carreras, I., et al. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics. 33 (15), 2424-2426 (2017).
- von Arx, G., Carrer, M. ROXAS – A new tool to build centuries-long tracheid-lumen chronologies in conifers. Dendrochronologia. 32 (3), 290-293 (2014).
- Koubaa, A., Zhang, S. Y. T., Makni, S. Defining the transition from earlywood to latewood in black spruce based on intra-ring wood density profiles from X-ray densitometry. Annals of Forest Science. 59 (5-6), 511-518 (2002).
- Buras, A., Wilmking, M. Correcting the calculation of Gleichläufigkeit. Dendrochronologia. 34, 29-30 (2015).
- Bunn, A. G. Statistical and visual crossdating in R using the dplR library. Dendrochronologia. 28 (4), 251-258 (2010).
- Zang, C., Biondi, F. Treeclim: an R package for the numerical calibration of proxy-climate relationships. Ecography. (November 2014), 1-6 (2014).
- Van den Bulcke, J., et al. Nondestructive research on wooden musical instruments: From macro- to microscale imaging with lab-based X-ray CT systems. Journal of Cultural Heritage. 27, S78-S87 (2017).
- Helama, S., Vartiainen, M., Kolström, T., Meriläinen, J. Dendrochronological investigation of wood extractives. Wood Science and Technology. 44 (2), 335-351 (2010).
- Black, B. A., et al. The value of crossdating to retain high-frequency variability, climate signals, and extreme events in environmental proxies. Global Change Biology. 22 (7), 2582-2595 (2016).
- Hubau, W., et al. The persistence of carbon in the African forest understory. Nature plants. 5 (2), 133-140 (2019).
- Stoffel, M., Klinkmüller, M. 3D analysis of anatomical reactions in conifers after mechanical wounding: First qualitative insights from X-ray computed tomography. Trees – Structure and Function. 27 (6), 1805-1811 (2013).
- Van den Bulcke, J., et al. Advanced X-ray CT scanning can boost tree-ring research for earth-system sciences. Annals of Botany. , 1-11 (2019).
- Ziaco, E., Biondi, F., Heinrich, I. Wood Cellular Dendroclimatology: Testing New Proxies in Great Basin Bristlecone Pine. Frontiers in Plant Science. 7 (October), 1-13 (2016).
- De Ridder, M., et al. High-resolution proxies for wood density variations in Terminalia superba. Annals of botany. 107 (2), 293-302 (2011).
- Resente, G., et al. Repeat! Artificial Intelligence for Quantitative Wood Anatomy. Frontiers in Plant Science. 12 (November), 1-14 (2021).

