An Assay to Quantify Cytosolic and Vacuolar Salmonella in Infected Primary Macrophages
Abstract
Source: Meunier, E., et al. Quantification of Cytosolic vs. Vacuolar Salmonella in Primary Macrophages by Differential Permeabilization. J. Vis. Exp. (2015).
This video demonstrates an assay to quantify cytosolic and vacuolar Salmonella typhimurium inside bone marrow-derived macrophages using differential permeabilization. The macrophages are infected with a fluorescent protein-expressing Salmonella, which is internalized inside a specialized vacuolar structure called Salmonella-containing vacuole or SCV. A subset of bacteria escape the SCV and enter the cytosol. The cholesterol-rich plasma membrane of macrophages is permeabilized using digitonin, leaving the SCV intact. Upon labeling the cytosolic bacteria with a fluorescently tagged antibody, the percentage of vacuolar and cytosolic bacteria is quantified using fluorescence-activated cell sorting (FACS).
Protocol
1. Digitonin Assay with mCherry-expressing Salmonella (FACS-based Analysis)
- Preparing bacteria
Note: Salmonella wild-type SL1344 (Streptomycin resistant) expressing mCherry from a plasmid (pFPV encoding mCherry under the rpsM promoter) is used in this assay. Bacterial phagocytosis and proliferation were not altered by the constitutive expression of mCherry (data not shown).- Inoculate the strain 1 day before the infection in 3 ml of LB medium supplemented with Streptomycin (90 µg/ml) and Ampicillin (100 µg/ml, mCherry expressing vector) in a shaker at 37 °C O/N.
- Plating cells for infection.
- Seed wild-type BMDMs in a 24 well plate at a density of 1.25 x 105 cells/well in 1 ml of primary macrophage medium (DMEM supplemented with 10% Fetal Calf Serum (FCS, heat-inactivated, 56 °C, 30 min), 1% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1% Non-essential amino acids (NEAA), 1% L-Glutamine and 10% L929 supernatant (conditioned medium from macrophage colony-stimulating factor (M-CSF) producing L929 cells). Seed wells according to Table 1 account for the individual conditions. Add coverslips to the wells used as controls (Table 1).
- Incubate the cells overnight in an incubator at 37 °C and 5% CO2.
- Preparing the bacteria for infection.
- The next day, determine the concentration of Salmonella in the overnight culture by diluting the culture 1:10 in phosphate-buffered saline (PBS) and measuring the OD600 against a PBS-only control. This ensures that the OD600 is measured in the linear range of the spectrophotometer.
- Calculate the concentration of Salmonella per ml. An OD600 of 1 corresponds to 1 x 109 bacteria.
- Dilute the bacteria in a macrophage medium to reach a Salmonella concentration of 1.25 x 106 cells/ml.
- Infection of cells with Salmonella.
- Remove medium from BMDMs. Add 1 ml of macrophage medium to uninfected control wells (B1-B3, C1-C3). Add 1 ml of Salmonella suspension to wells A1 – A5 to reach a multiplicity of infection (m.o.i.) of 10 bacteria per cell.
- Centrifuge the plate for 15 min at 300 x g at 37 °C to synchronize the infection. Transfer the plate to an incubator at 37 °C and 5% CO2.
- Killing extracellular Salmonella with Gentamicin.
- At 1 hr post-infection, remove the plate from the incubator and add 0.1 ml of macrophage medium containing 0.1 mg/ml Gentamicin to kill extracellular bacteria. Add Gentamicin to all wells, even to uninfected controls, to ensure that all cells are treated the same way. Transfer the plate to an incubator at 37 °C and 5% CO2.
- Washing the cells.
- At 2 hr post-infection, remove the plate from the incubator and wash all wells 2x with 1 ml of plain DMEM (pre-warmed to 37 °C) and replace it with macrophage medium (pre-warmed to 37 °C) containing 10 µg/ml Gentamycin to prevent the growth of any remaining extracellular bacteria. Transfer the plate to an incubator at 37 °C and 5% CO2.
- Preparing fresh buffers with detergents and antibodies.
- Just before the desired time point of analysis, prepare a fresh stock solution of digitonin at 1 mg/ml in KHM Buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, pH 7.3). Filter the stock and dilute in KHM buffer to a working concentration of 50 µg/ml of digitonin. In addition, prepare a solution of 0.1% Saponin in KHM Buffer, anti-Salmonella antibody coupled to fluorescein Isothiocyanate (CSA-1-FITC) at 1/500, anti-calnexin at 1/100 or anti-protein disulfide isomerase (anti-PDI) at 1/100 in KHM Buffer supplemented with 3% bovine serum albumin (BSA).
- Cell permeabilization.
- Remove the plate from the incubator and wash wells 3x with 0.5 ml of KHM Buffer (pre-warmed to 37 °C). Remove KHM Buffer and add 0.25 ml plain KHM Buffer to wells A4 and B1/C1, KHM Buffer with digitonin to wells A1 – A3 and B2/C2, or KHM Buffer with saponin to wells A5 and B3/C3 (according to Table 1). Incubate for exactly 1 min at RT and immediately wash all wells 3x with 0.5 ml of KHM Buffer (pre-warmed to 37 °C).
- Primary antibody staining.
- Remove the KHM Buffer and add the primary antibody solutions (Goat anti-Salmonella-FITC, 250 µl/well, 0.1 µg/ml) to the appropriate wells (Figure 1, Table 1). Incubate the plate for 15 min in an incubator at 37 °C and 5% CO2.
- Wash the cells 1x with 1 mL PBS.
- Infected Cells (wells A1 – A5): FACS analysis
- Wash cells 3x with PBS and add 0.5 mL of ice-cold PBS containing 0.1% Triton. Transfer to a 5 mL FACS tube with a cell strainer cap to break up cell aggregates. Keep samples on ice.
- Analyze the samples of FACS. Using the fully permeabilized controls, set the gate for total bacteria based on forward and side scatter (FSC/SSC) first, and the mCherry signal (610 nm ± 20 nm filter).
- Next, analyze the FITC signal in the total bacterial population using the fully permeabilized and not permeabilized controls to set the gates for cytosolic bacteria (FITC+, mCherry+) and vacuolar bacteria (FITC-, mCherry+) (Figure 2).
- With the gates set, analyze the samples and determine the percentage of cytosolic vs. vacuolar bacteria using FlowJo software according to the manufacturer's protocol.
Table 1: Plating scheme for bone-marrow-derived macrophages (BMDMs) in a 24-well format for subsequent infection with Salmonella, permeabilization, and staining.
| Permeabilization | Staining | ||||||
| Glass coverslip | Salmonella | Digitonin | Saponin | anti-Salmonella | anti-Calnexin | anti-PDI | |
| A1 (sample) | -/+* | + | + | + | |||
| A2 (sample) | -/+* | + | + | + | |||
| A3 (sample) | -/+* | + | + | + | |||
| A4 (no permeabilization control) | -/+* | + | + | ||||
| A5 (complete permeabilization control) | -/+* | + | + | + | |||
| B1 (stained for calnexin) | + | + | |||||
| B2 (stained for calnexin) | + | + | + | ||||
| B3 (stained for calnexin) | + | + | + | ||||
| C1 (stained for PDI) | + | + | |||||
| C2 (stained for PDI) | + | + | + | ||||
| C3 (stained for PDI) | + | + | + | ||||
Representative Results
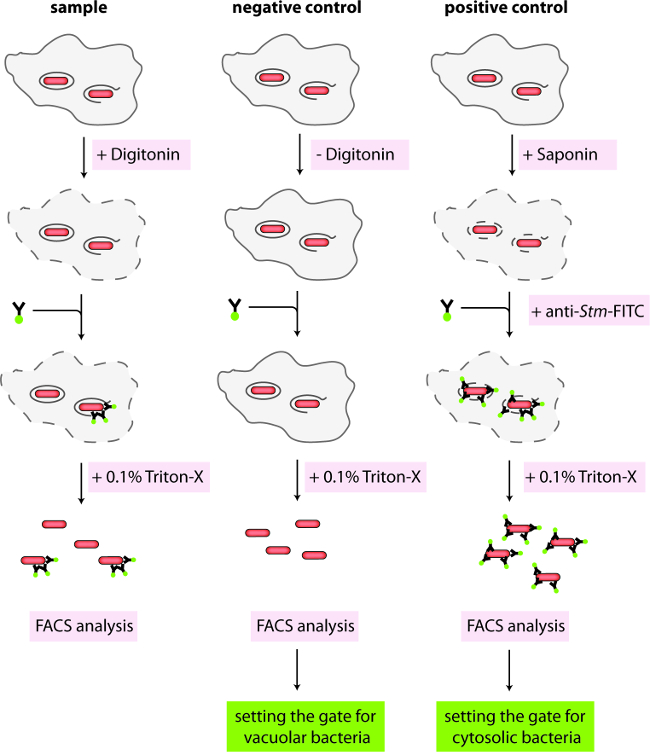
Figure 1. Schematic representation of protocol 1. Infected cells containing mCherry-positive Salmonella either in intact vacuoles or the cytosol are differentially permeabilized with digitonin. Cytosolic Salmonella are stained with anti-Salmonella coupled to FITC. Cells are washed and lysed with Triton X-100 for FACS analysis. Negative control cells are not permeabilized, while positive control cells are completely permeabilized before antibody staining.
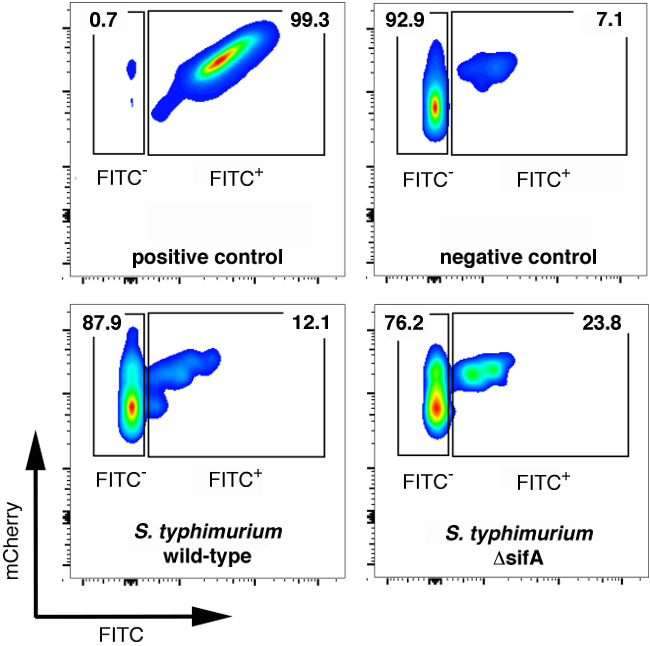
Figure 2. FACS analysis of fully permeabilized, unpermeabilized, or differentially permeabilized samples infected for 6 hr with mCherry+ wild-type or ΔsifA Salmonella for 6 hr.
Disclosures
The authors have nothing to disclose.
Materials
| Digitonin | Sigma | D5628 | 50ug/mL |
| PFA | mpbio | 219998380 | |
| HEPES | Life Technologies | 15630 | |
| Potassium Acetate | Sigma | 791733 | |
| MgCl2 | Sigma | M8266 | |
| BSA | Sigma | A2153 | |
| anti-Salmonella CSA-1-FITC | KPL | 01-91-99-MG | 1/500 |
| anti-Salmonella CSA-1 | KPL | 02-91-99-MG | 1/500 |
| anti-Calnexin | Enzo Lifesciences | SPA-860D | 1/100 |
| anti-PDI | Enzo Lifesciences | SPA-890 | 1/100 |
| Saponin | Sigma | 47036 | |
| Vectashield mounting medium | Vectorlabs | H-1200 | |
| Anti Rabbit antibody-488 | Molecular Probes | A-11070 | 1/500 |
| Gentamicin | Life Technologies | 15710-49 | 100ug/mL and 10ug/mL |
| Triton X-100 | Promega | H5141 | |
| PBS | Gibco | 20012-019 | |
| DMEM | Sigma | D6429 | |
| NEAA | Amimed | 5-13K00-H | |
| FACS Fortessa | BD technologies | detection mCherry 610 nm | |
| FACS Fortessa | BD technologies | detection FITC 530 nm |

