Isolation and Characterization of RNA-Containing Exosomes
Summary
This paper demonstrates methods for the isolation, purification and detection of exosomes, as well as techniques for analysis of their molecular content. These methods are adaptable for exosome isolation from both cell culture media and biological fluids, and can beyond analysis of molecular content also be useful in functional studies.
Abstract
The field of exosome research is rapidly expanding, with a dramatic increase in publications in recent years. These small vesicles (30-100 nm) of endocytic origin were first proposed to function as a way for reticulocytes to eradicate the transferrin receptor while maturing into erythrocytes1, and were later named exosomes. Exosomes are formed by inward budding of late endosomes, producing multivesicular bodies (MVBs), and are released into the environment by fusion of the MVBs with the plasma membrane2. Since the first discovery of exosomes, a wide range of cells have been shown to release these vesicles. Exosomes have also been detected in several biological fluids, including plasma, nasal lavage fluid, saliva and breast milk3-6. Furthermore, it has been demonstrated that the content and function of exosomes depends on the originating cell and the conditions under which they are produced. A variety of functions have been demonstrated for exosomes, such as induction of tolerance against allergen7,8, eradication of established tumors in mice9, inhibition and activation of natural killer cells10-12, promotion of differentiation into T regulatory cells13, stimulation of T cell proliferation14 and induction of T cell apoptosis15. Year 2007 we demonstrated that exosomes released from mast cells contain messenger RNA (mRNA) and microRNA (miRNA), and that the RNA can be shuttled from one cell to another via exosomes. In the recipient cells, the mRNA shuttled by exosomes was shown to be translated into protein, suggesting a regulatory function of the transferred RNA16. Further, we have also shown that exosomes derived from cells grown under oxidative stress can induce tolerance against further stress in recipient cells and thus suggest a biological function of the exosomal shuttle RNA17. Cell culture media and biological fluids contain a mixture of vesicles and shed fragments. A high quality isolation method for exosomes, followed by characterization and identification of the exosomes and their content, is therefore crucial to distinguish exosomes from other vesicles and particles. Here, we present a method for the isolation of exosomes from both cell culture medium and body fluids. This isolation method is based on repeated centrifugation and filtration steps, followed by a final ultracentrifugation step in which the exosomes are pelleted. Important methods to identify the exosomes and characterize the exosomal morphology and protein content are highlighted, including electron microscopy, flow cytometry and Western blot. The purification of the total exosomal RNA is based on spin column chromatography and the exosomal RNA yield and size distribution is analyzed using a Bioanalyzer.
Protocol
1. Exosome isolation
- Grow cells in medium with exosome-free serum. Thus, any serum added to the cell culture medium should be depleted of exosomes by ultracentrifugation at 120 000 x g over night at 4 °C prior to use.
- Transfer the cell suspension to conical tubes.
- Centrifuge at 300 x g for 10 minutes at 4 °C to pellet the cells.
- Transfer the supernatant to ultracentrifuge tubes and if not completely full add PBS.
- Centrifuge the sample at 16 500 x g for 20 minutes at 4 °C to further remove cells and cell debris.
- Filter the supernatant through a 0.2 μm filter to remove particles larger than 200 nm.
- Transfer the filtered supernatant to new ultracentrifuge tubes and seal the tubes before ultracentrifuge at 120 000 x g for 70 minutes at 4 °C to pellet the exosomes.
- Discard the supernatant.
- For maximal exosome retrieval, resuspend the exosome enriched pellet repeatedly in a small volume (~3 x 50 μl) of an appropriate buffer. This buffer depends on the downstream experiments planned following the exosome isolation. For example, lysis buffer is used for protein and RNA isolation, PBS is used for electron microscopy and flow cytometry and for functional studies medium may be preferred.
Note; exosomes can also be isolated from different body fluids, such as plasma, using the same procedure as for cell culture media. For viscous fluids it may be necessary to dilute the sample with PBS prior to the centrifugation and filtration step. In relation to the centrifugation speed for point 5 above, it can be increased to 29 500 x g, and the ultracentrifugation in point 7 can be extended to 90 minutes18.
If the sample needs to be further purified the exosome pellet can be floated on a sucrose gradient and the exosomes will primarily be found in the fraction representing a density of 1.13-1.19 g/ml2.
2. Exosome identification by electron microscopy
- To further eliminate contaminating proteins, resuspend the exosome enriched pellet in PBS and ultracentrifuge at 120 000 x g for 70 minutes at 4 °C to re-pellet the exosomes.
- Take a small aliquot of the sample for protein isolation and total protein measurement. Make sure that only a small part of the sample is lysed and used for the protein measurement and keep the intact exosomes, solved in PBS, separate on ice or at -80 °C for further experiments.
- Place a drop, approximately 10 μg of exosomal protein of the intact exosomes resuspended in PBS, on a Parafilm. Then, with forceps, gently position a formvar carbon coated nickel grid on top of each drop for 30-60 minutes. Assure that the grid is positioned with the coating side facing the drop containing exosomes.
- Place three drops, each 30 μl, of PBS on the Parafilm and wash the grid by sequentially positioning the grid on top of the droplets of PBS, and use an absorbing paper in between. Use the absorbing paper gently just by holding it closely to the side of the grid, without making contact with the coated area.
- Fix the sample by deposit a drop of 2% paraformaldehyde on the Parafilm and place the grid on top of the drop for 10 minutes.
- Repeat the washing step in point 4, before immunostaining with an appropriate antibody. Commonly used antibodies are anti-CD63 and anti-MHC class II. Electron microscopy can also be performed without immunostaining as exosomes can be identified solely based on size and morphology. However, we recommend to evaluate the size, morphology and the presence of a membrane protein for a more conclusive validation.
- Transfer the grid to a 30 μl drop of the primary antibody of choice and incubate for 40 minutes. Wash by repeating point 4, but use 0.1% bovine serum albumin in PBS instead of PBS alone.
- Repeat point 7, but with the 10 nm-gold labeled secondary antibody and wash with PBS alone.
- Post-fix the sample by adding a drop of 2.5% glutaraldehyde to the Parafilm and incubate the grid on top of the drop for 10 minutes. Repeat the wash in point 4, but use five droplets of deionized water instead of three droplets of PBS.
- Contrast the sample by adding a drop of 2% uranyl acetate to the Parafilm and incubate the grid on top of the drop for 15 minutes.
- Embed the sample by adding a drop of 0.13% methyl cellulose and 0.4% uranyl acetate to the Parafilm and incubate the grid on top of the drop for 10 minutes.
- Remove excess liquid by gently using an absorbing paper, before positioning the grid on a paper with the coated side up and let it air dry for 5 minutes.
- Examine the preparations with an electron microscope or store the grids in a grid box for future work.
3. Exosome characterization by flow cytometry
As exosomes are too small to be detected by current flow cytometry equipment, it is necessary to first bind the exosomes to antibody coated beads (Figure 1). These beads can either be purchased as a ready made product or made in the laboratory with the antibody of choice. Depending on the exosomal cellular origin, different antibodies can be coupled to beads which can be either of magnetic or latex character. We currently use anti-MHC class II or anti-CD63 coated beads for this analysis.
- For the use of “homemade” antibody coated beads, we use 4 μm latex beads coated with anti-CD63 antibody. Wash 25 μl 4 μm latex beads (30 x 106 beads) twice in 100 μl MES buffer, 3 000 x g for 15-20 minutes and redissolve the pellet in 100 μl MES buffer. Prepare the antibody mixture containing a volume equal of 12.5 μg antibody with the same volume of MES buffer. Add the beads to the antibody mixture and incubate under agitation over night at room temperature. Wash the antibody coated beads three times with PBS (3 000 x g for 20 minutes) and dissolve the pellet in 100 μl of storage buffer (with a final concentration of 300 000 beads/μl).
- For each sample (each antibody), continue with a volume equal to a minimum of 30 μg of exosomal protein (of the intact exosomes solved in PBS) per ~100 000 antibody coated beads.
- Incubate exosomes and beads, in a total volume of 300 μl PBS, over night at 4 °C under gentle movement.
- Block by adding 300 μl of 200 mM glycine and incubate for 30 minutes.
- Wash the exosome-bead complexes twice in wash buffer (1-3% serum in PBS), 600 x g for 10 minutes.
- Incubate the exosome-bead complexes with 50 μl IgG antibody at 4 °C. Wash the exosome-bead complexes twice in wash buffer as described in step 5.
- Add 90 μl wash buffer and 10 μl antibody of choice (ideally anti-CD9, anti-CD63 or anti-CD81) to the exosome-bead complexes and incubate for 40 minutes under gentle movement. Wash the exosome-bead complexes two times in wash buffer as described in step 5.
- Add 300 μl wash buffer and acquire data using flow cytometry.
As discussed above, different beads can be used, such as latex beads as described in the protocol or magnetic beads. If magnetic beads are used instead, please note, that the protocol is slightly different. The blocking step (point 4) is not required and the wash is performed by placing the tube in a magnetic stand instead of a centrifugation.
The storage buffer used for the “homemade” antibody coated beads, contain 0.1% glycine and 0.1% sodium azid in PBS, with a pH of 7.2.
Importantly, make sure to use the exosome depleted serum for the flow cytometry wash buffer (1-3% serum in PBS), to avoid any serum exosomes contaminating the analysis.
Note; when performing exosome characterization by using antibody coupled beads it is important to acknowledge that it is only a specific subpopulation, specific for the antibody used, that is isolated and characterized.
4. Exosome detection by Western blot
Western blot is a well established method and we will not go into detail regarding the method itself, but focus on the importance of a suitable protein isolation before the Western blot and the different antibodies used.
- Dissolve the exosome pellet in the protein lysis buffer of choice and pipetting thoroughly, followed by vortex-mixing. To further lyse the exosomes, sonicate the sample in a water bath 3 x 5 minutes with vortex-mixing in between. Finally centrifuge the sample, 13 000 x g for 5 minutes at room temperature, and transfer the supernatant to a new eppendorf tube.
- Measure the total protein by a method of choice and load 20-50 μg of protein per well.
- Separate and transfer the proteins by gel electrophoresis and electroblotting.
- Block and wash the membrane before performing immunostaining against proteins enriched in exosomes.
- Detect the specific protein by chemiluminescence, digital imaging system and analysis software.
As there are no exosome specific markers, proteins that are enriched in exosomes, from all different cellular origins, are commonly used for exosome detection. These are proteins such as tetraspanins (e.g. CD9, CD63 and CD81), cytoskeleton associated protein (e.g. ezrin) and proteins involved in multivesicular biogenesis (e.g. Tsg101 and alix). Other proteins that are also commonly detected in exosomes are Flotillin, Hsc70 and different rab proteins etc. However, there are also cell specific proteins found in exosomes such as A33 (intestinal epithelial cells), CD3 (T cells) and MHC class II (antigen presenting cells). Thus, depending on from what cell type the exosomes are released from the markers for detection may vary.
As other compartments of the cell also can produce vesicles, it is further recommended to determine the presence of proteins from these compartments such as the endoplasmic reticulum (e.g. calnexin and Grp78) and the Golgi apparatus (e.g. GM130). Thus, lack of these proteins indicates no or little contamination of vesicles of other compartments in the sample studied.
5. Isolation of exosomal RNA and RNA analysis
- Dissolve the exosome pellet in the RNA lysis buffer of choice and perform RNA extraction. Different RNA isolation kits can be used depending on downstream experiments, but column based methods provides samples with a wider spectrum of RNA. We are currently using miRCURY RNA Isolation Kit by Exiqon, but other kits may be suitable depending on the scientific question at hand.
- When performing RNA isolation it is important to work in a RNase-free environment. Therefore use nucleic acid and nuclease free pipette tips and wipe both bench and equipment free from contaminants and RNases.
- For RNA isolation, by using miRCURY RNA Isolation Kit, dissolve the exosome pellet in 350 μl of lysis solution and perform vortex-mixing for 15 seconds.
- Add 200 μl of 95% ethanol and vortex-mixing for 10 seconds.
- Place a column in a collection tube and transfer the lysed exosomes to the column and centrifuge 1 minute at 14 000 x g.
- Wash three times by adding 400 μl of Wash Solution to the column containing the exosomal RNA and centrifuge for 1 minute at 14 000 x g.
- Centrifuge the column for 2 minutes at 14 000 x g to make sure that the column is dry and place the dry column in an RNase-free eppendorf tube.
- Add 50 μl Elution Buffer to the column and centrifuge for 2 minutes at 200 x g, followed by 1 minute at 14 000 x g.
- Continue with the isolated RNA or store it in -80 °C.
For detection and analysis of the extracted total exosomal RNA, use an Agilent 2100 Bioanalyzer with the RNA 6000 Nano or RNA 6000 Pico Kit. The analysis shows the exosomal RNA yield and size distribution.
6. Representative Results
Exosomes are too small to be detected by available flow cytometry methods, and are therefore attached to antibody-coated beads before being analyzed (Figure 1). As exosome-bead complexes can aggregate the flow cytometry scatter plot can contain different populations of single, double and triple beads (Figure 2A). The arrow indicates the single beads that are further analyzed. A CD63 positive single bead-exosome population compared to its isotype control is shown in Figure 2B.
Due to the small size of exosomes, they can only be directly visualized with electron microscopy, and not by light microscopy. Typical morphological characteristics of exosomes are round shaped and 30-100 nm sized membrane vesicles. In Figure 3, electron microscopy photographs show exosomes immunostained (A) with gold-labeled antibody for CD63 (indicated by the arrow) and non-immunostained exosomes (B).
To determine that the isolated vesicles are indeed exosomes and to further characterize the exosomal proteins, Western blot is commonly used. Figure 4 shows the absence of calnexin, an endoplasmic reticulum marker. This indicates little contamination of vesicles from the endoplasmic reticulum. Furthermore, Figure 4 shows the presences of CD81 in exosomes, which is a tetraspanin commonly enriched in exosomes.
Cellular RNA mainly contains ribosomal RNA (rRNA), seen as the two prominent peaks for the 18S and 28S rRNA subunits in a Bioanalyzer analysis (Figure 5A). However, exosomal RNA differ in their profile as exosomes lack the two rRNA peaks (18S and 28S) and are enriched in short RNA such as mRNA and miRNA (Figure 5B).
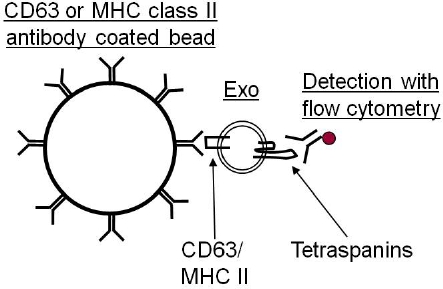
Figure 1. Due to the small size of exosomes (30-100 nm), they cannot be distinguish from noise and background when analyzed with flow cytometry. Therefore, it is necessary to attach them to antibody coated beads before the exosomes are analyzed, which then can be visualized by the laser.
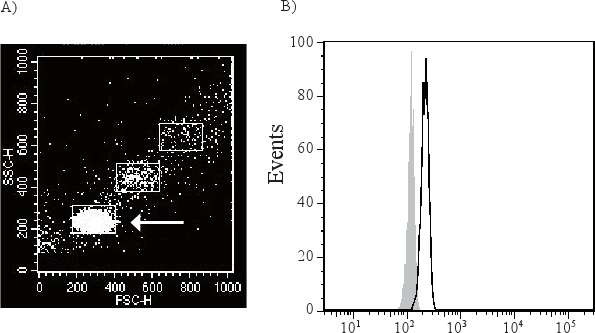
Figure 2. The exosome-beads complex can aggregate with each other and form double and triple beads (A). The arrow is demonstrating the single bead-exosome population which is gated and used for further analysis. This population will be directly compared to an isotype control (filled grey peak) to determine the presence of membrane molecules of the exosomes. CD63 positive exosomes (black open peak) are shown in figure B.
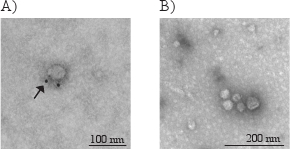
Figure 3. Electron micrographs of exosomes with the typical morphology and size (40-80 nm) (A and B). The arrow in figure A indicates the golden particle of the secondary antibody, verifying that this exosome have CD63 on its membrane surface.
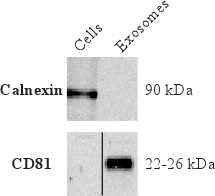
Figure 4. Western blot results showing the absence of the endoplasmic reticulum protein calnexin but the presence of the protein commonly enriched in exosomes, tetraspanin CD81.
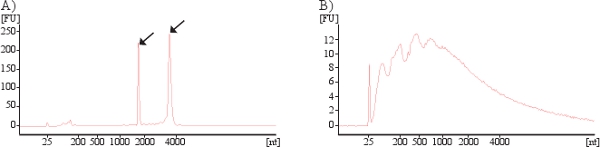
Figure 5. Bioanalyzer results showing presence RNA in cells (A) and exosomes (B). Arrows indicates the 18S and 28S rRNA subunits present in cells. As exosomal RNA manly contain small RNA no or little rRNA is identified in exosomes.
Discussion
When studying the molecular content and/or function of exosomes, it is crucial to use a high quality isolation method that provides an appropriate yield and the lowest possible degree of contamination. The methodology described in this paper is a well established approach for isolating exosomes and to study their RNA content and biological function. Furthermore, the importance of characterization of the isolated exosomes, by a combination of different methods, including electron microscopy, flow cytometry, and Western blot, is highlighted.
When isolating exosomes, the first centrifugation step (300 x g) is used to pellet cells that might be present in the cell suspension or the studied biological fluid. The second centrifugation at 16 500 x g needs to be sufficiently strong to pellet dead cells, larger debris, apoptotic bodies and other organelles. After this centrifugation, some protocols include a nano-filtration step, but some investigators have excluded this step. However, we emphasize the importance of eradicating fragments and vesicles larger than 200 nm and therefore strongly recommend including a filtration step. If a more stringent isolation is needed, 100 nm filters can be used, but note that if viscous fluids are used, these filters can easily become blocked and exosomal material is lost. Also, a 100 nm filtration may affect the morphology of the exosomes and furthermore if exosomes are on the larger scale they might be lost. Thus, the 200 nm filters seem appropriate for exosome isolation. After the filtration, the mandatory ultracentrifugation at 120,000 x g is used to pellet the exosomes. The exosomes can be further washed with PBS, bound to antibody coated beads and washed, or placed on a sucrose gradient to remove contamination proteins. However, this can be a very resource consuming process that we argue not to be necessary, especially if the starting sample volume is small and/or the RNA content of the exosomes is low, as extra steps will substantially decrease the material further. However, again, the nature of the isolated vesicles should be proven by the presence and absence of certain proteins.
To date, no absolutely unique protein marker for exosomes has been identified to verify that the sample contains exosomes and nothing else. As a result, a combination of methods are required to characterize the exosomes, including determination of size and morphology of the exosomes by electron microscopy. Also, the purity of the sample can be determined by electron microscopy, as this method can provide an overview of the level of contamination of the sample, for example with larger vesicles, such as microparticles, apoptotic bodies or cell debris. Furthermore, the protein content of the exosomes can be determined by flow cytometry and Western blot, and the combination of these two methods results in an analysis of both the membrane bound (e.g. CD63 and CD81) and internalized proteins (e.g. Tsg101 and alix) of the exosomes. Detection of proteins enriched in exosomes, such as CD63, Tsg101 and alix, and the absence of proteins such as the endoplasmic reticulum protein calnexin, is an indication that the exosome enriched pellet is indeed exosomes and not contaminating vesicles from other compartments of the cell.
The profile of the RNA that is detected in exosomes is fundamentally different to that of the RNA found in intact cells. Thus, exosomal RNA analyzed by Bioanalyzer or in gel-based RNA analysis approaches lack the two peaks for the 18S and 28S rRNA subunits, which are prominent in analysis of cellular RNA. We therefore argue that the detection of any significant amounts of rRNA in a sample indicates that the total RNA extracted is not of exosomal origin alone, and thus that the isolation method needs to be improved and quality assured.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was financed by the Swedish Research Council (K2008-57X-20 676-01-3).
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| Exosome isolation | |||
| Optima L-90K Ultracentrifuge | Beckman Coulter | 65670 | |
| Quick-seal ultracentrifuge tubes | Beckman Coulter | Depending on the rotor used different tubes are needed. Ti70 (342414), Ti45 (345776) | |
| Quick-Seal Cordless Tube Topper kit | Beckman Coulter | 358312 | |
| Filtropur Syringe Filter, 0.20μm | Sarstedt | 83.1826.001 | For viscose fluids and/or large volumes, a Vacuum filtration system, 0.2 μm, from VWR International can be used instead (514-0600). |
| Electron microscopy | |||
| Formvar/carbon coated nickel grids | Ted Pella Inc | 1GN200 | |
| Mouse-anti-human CD63 | BD Bioscience | 556019 | Final conc: 0.05μg/μl |
| Isotype control; IgG1κ (MOPC 21) | Sigma-Aldrich | M9269 | Final conc: 0.05μg/μl |
| Anti-Mouse IgG Gold Conjugate, 10 nm | Sigma-Aldrich | G7777 | Final conc: 1% |
| LEO 912AB Omega Electron microscope | Carl Zeiss NTS | Or equivalent equipment. | |
| Flow cytometry | |||
| 4μm aldehyd/sulphate latex beads | Interfacial Dynamics | 12-4000 | |
| Antibody for coating of the latex beads | For example anti-CD63 from BD Bioscience (556019) | ||
| Intelli-Mixer RM-2L | ELMI | Or equivalent equipment. | |
| IgG from human serum | Sigma-Aldrich | I4506 | |
| Antibodies for detection | BD Bioscience | For example: PE anti-CD63 (556020) PE anti-CD9 (555372) PE anti-CD81 (555676) Isotype control (555749) | |
| BD FACSAria | BD Bioscience | ||
| Western Blot | |||
| Transsonic T 490 DH | ELMA | Or equivalent equipment. | |
| Isolation of exosomal RNA and RNA analysis | |||
| miRCURY RNA Isolation Kit | Exiqon A/S | 300110 | |
| Agilent 2100 Bioanalyzer | Agilent Technologies | G2939A |
References
- Pan, B. -. T., Johnstone, R. M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell. 33, 967-978 (1983).
- Chaput, N., Thery, C. Exosomes: immune properties and potential clinical implementations. Semin. Immunopathol. , 10-1007 (2010).
- Lässer, C. RNA-containing exosomes in human nasal secretions. Am. J. Rhinol. Allergy. 25, 89-93 (2011).
- Caby, M. -. P., Lankar, D., Vincendeau-Scherrer, C., Raposo, G., Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 17, 879-887 (2005).
- Palanisamy, V. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS ONE. 5, e8577-e8577 (2010).
- Admyre, C. Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 179, 1969-1978 (2007).
- Almqvist, N., Lönnqvist, A., Hultkrantz, S., Rask, C., Telemo, E. Serum-derived exosomes from antigen-fed mice prevent allergic sensitization in a model of allergic asthma. Immunology. 125, 21-27 (2008).
- Prado, N. Exosomes from Bronchoalveolar Fluid of Tolerized Mice Prevent Allergic Reaction. J. Immunol. 181, 1519-1525 (2008).
- Zitvogel, L. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nature. Medicine. 4, 594-600 (1998).
- Ashiru, O. Natural Killer Cell Cytotoxicity Is Suppressed by Exposure to the Human NKG2D Ligand MICA*008 That Is Shed by Tumor Cells in Exosomes. Cancer. Research. 70, 481-489 (2010).
- Liu, C. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function. The Journal of Immunology. 176, 1375-1385 (2006).
- Viaud, S. Dendritic Cell-Derived Exosomes Promote Natural Killer Cell Activation and Proliferation: A Role for NKG2D Ligands and IL-15Rα. PLoS ONE. 4, e4942-e4942 (2009).
- Szajnik, M., Czystowska, M., Szczepanski, M. J., Mandapathil, M., Whiteside, T. L. Tumor-Derived Microvesicles Induce, Expand and Up-Regulate Biological Activities of Human Regulatory T Cells (Treg). PLoS ONE. 5, e11469-e11469 (2010).
- Raposo, G. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161-1172 (1996).
- Abusamra, A. J. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells, Molecules, and Diseases. 35, 169-173 (2005).
- Valadi, H. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654-659 (2007).
- Eldh, M. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One. 5, e15353-e15353 (2010).
- Lässer, C. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 9, 9-9 (2011).

