Pharmacological and Functional Genetic Assays to Manipulate Regeneration of the Planarian Dugesia japonica
Summary
An attractive model for studying stem cell differentiation within a live animal is the planarian flatworm. Regeneration is studied by simple amputation experiments that are easily performed in a basic laboratory and are amenable to pharmacological and genetic (in vivo RNAi) manipulation as detailed by protocols in this article.
Abstract
Free-living planarian flatworms have a long history of experimental usage owing to their remarkable regenerative abilities1. Small fragments excised from these animals reform the original body plan following regeneration of missing body structures. For example if a ‘trunk’ fragment is cut from an intact worm, a new ‘head’ will regenerate anteriorly and a ‘tail’ will regenerate posteriorly restoring the original ‘head-to-tail’ polarity of body structures prior to amputation (Figure 1A).
Regeneration is driven by planarian stem cells, known as ‘neoblasts’ which differentiate into ~30 different cell types during normal body homeostasis and enforced tissue regeneration. This regenerative process is robust and easy to demonstrate. Owing to the dedication of several pioneering labs, many tools and functional genetic methods have now been optimized for this model system. Consequently, considerable recent progress has been made in understanding and manipulating the molecular events underpinning planarian developmental plasticity2-9.
The planarian model system will be of interest to a broad range of scientists. For neuroscientists, the model affords the opportunity to study the regeneration of an entire nervous system, rather than simply the regrowth/repair of single nerve cell process that typically are the focus of study in many established models. Planarians express a plethora of neurotransmitters10, represent an important system for studying evolution of the central nervous system11, 12 and have behavioral screening potential13, 14.
Regenerative outcomes are amenable to manipulation by pharmacological and genetic apparoaches. For example, drugs can be screened for effects on regeneration simply by placing body fragments in drug-containing solutions at different time points after amputation. The role of individual genes can be studied using knockdown methods (in vivo RNAi), which can be achieved either through cycles of microinjection or by feeding bacterially-expressed dsRNA constructs8, 9, 15. Both approaches can produce visually striking phenotypes at high penetrance- for example, regeneration of bipolar animals16-21. To facilitate adoption of this model and implementation of such methods, we showcase in this video article protocols for pharmacological and genetic assays (in vivo RNAi by feeding) using the planarian Dugesia japonica.
Protocol
1. Trunk fragment regeneration assay
- Stop feeding a cohort of worms for at least 5 days before the regenerative assay to ensure animals are free of waste and ingested food (˜30 intact worms, each worm ˜8-10 mm long post-starvation).
- On the day of the assay, rinse a pre-frozen leveled ice dish with water and cover the flat iced surface with plastic wrap. Using forceps, place one filter paper on the plastic wrap.
- Moisten the filter paper with a few drops of spring water. Transfer planarians onto filter (<20 worms per filter) using a transfer pipette. Worms can be repositioned on filter, if necessary, by repipetting with more spring water. Remove excess fluid.
- Using a scalpel, amputate the head by a single cut made approximately halfway between the anterior apex of the animal and the anterior end of the pharynx (Figure 1A).
- Using a scalpel, amputate the tail region by a single cut made approximately halfway between the tail end of the animal and posterior end of the pharynx (Figure 1A). Remove excess mucus on the scalpel using a paper towel moistened with 70% ethanol after cutting. Wipe excess ethanol from scalpel.
- Transfer filter via forceps into a Petri dish (100 x 25mm depth) containing spring water. Wait ˜3 minutes (for wound closure, observable by a pinching and darkening of the wound).
- Rinse trunk fragments from filter paper into Petri dish containing desired medium for regenerative assay and transfer to thermostatted incubator (24 °C).
- Score regenerative phenotype after ˜1 week, when regenerated structures are identifiable (Figure 1A).
2. Pharmacological manipulation of regeneration: bipolarity induced by praziquantel
- Prepare praziquantel (PZQ) stock by solubilizing drug in DMSO (62.4mg PZQ in 1mL for 200mM stock). Use on same day.
- Make 50mL of drug-containing solution (90 μM PZQ) in buffered Montjuch salts. Vortex for ˜1 minute to ensure dispersion.
- Perform trunk fragment regenerative assay as detailed above [protocol #1], exposing a cohort of trunk fragments to PZQ at the final step [1.7].
- After 24-48 hours, exchange PZQ-containing media for spring water, washing worms several (>3) times.
- Return worms to incubator. Score bipolarity (two-heads, Figure 1B) one week after cutting.
3. Genetic manipulation of regeneration: in vivo RNAi feeding protocol
- Prior to assay, prepare chicken liver homogenate. Discard fatty, yellow colored sections from foodstock, and purée remaining liver. Filter purée through mesh strainer and aliquot suspension for storage (1.5mL tubes, -20 °C).
- Prior to assay, prepare bovine red blood cells (RBCs) by aliquoting supply into 1mL samples, stored at -20°C.
- Select worms for RNAi. Three cohorts are used: gene of interest, positive control (gene with known RNAi phenotype; Figure 1C, D & E) and negative control (gene with no RNAi phenotype, also useful for assessment of knockdown levels compared to experimental cohort). For each cohort, use ˜250 worms (˜8-10 mm long after starving for at least 5 days). Store in spring water in plastic tub.
- For each RNAi cohort, thaw stock of E. coli expressing dsRNA targeting gene of interest [8], along with a tube of chicken liver homogenate [3.1] and a tube of RBCs [3.2].
- Pellet bacteria (13,000g, 1 minute). Remove supernatant and resuspend pellet in 2xYT media (˜700μl). Recentrifuge, discard supernatant, place pellet on ice.
- Create a large bore P200 pipette tip by cutting off the end of the tip. Thoroughly resuspend the bacterial pellet in a mixture of chicken liver homogenate (150 μL) and RBCs (50 μL) to create RNAi feeding mix. Remove air bubbles by centrifugation.
- For feeding, remove the majority of the water from the plastic tub containing worms (leave ˜1 inch depth). Swirl tub to concentrate worms at the center of this container, and pipette RNAi feeding mix in a circle corralling worms. Use a transfer pipette to carefully coax escapees back onto the RNAi feeding mix without perturbing other diners!
- After feeding (˜1 hour), identify planarians that fed well. These worms show a deep red coloration from ingested RBCs. Discard others.
- Carefully replace feeding solution with fresh spring water, minimizing disturbance to prevent egestion of food. To do this, gently localize the worms to one side of the tube using a transfer pipette, pour out turbid water, and carefully replace with fresh water poured down the opposite wall of the tub. Resubmerge planarians quickly as prolonged exposure to air also results in food egestion.
- Loosely seal container lid and return to incubator (24°C)
- Repeat RNAi feeding protocol over multiple cycles (˜2-3 days apart), interspersed with regenerative cycles [protocol #1] to screen for RNAi phenotypes. A standard protocol for feeding and regeneration effective for many genes is shown in Figure 2, which takes ˜1 month in total duration.
Modify this scheme for different genes, as optimal protocol will depend on factors such as mRNA stability, tissue localization, protein perdurance, or development of a phenotype that precludes multiple feeding cycles after regeneration. Assess knockdown levels by simply screening for penetrance of phenotype (if apparent), or by qPCR approaches to compare targeted mRNA levels with negative control cohort.
4. Representative results:
The trunk fragment regeneration assay is robust such that all worms should regenerate with normal anterior-posterior (‘head to tail’) polarity. This can be scored as soon as five days after cutting, facilitated by the emergence of the anterior eyespots ( asterixed, Figure 1A). However, complete morphological restoration of lost structures occurs after one week. Pharmacological manipulation of trunk fragment regeneration by PZQ to yield two-headed worms (Figure 1B) is also robust (EC50=87 (+-) 11% bipolar fragments, 70μM PZQ for 48 hours20). Bipolarity occurs over a single regenerative cycle. Lower efficacy in these assays likely relates to issues with drug solubility and/or storage, or use of a different flatworm species where PZQ is ineffective. For drugs with lower penetrance, an anteriorization index is often used to score intermediate phenotypes aside from complete bipolarity22. Phenotypes resulting from RNAi of positive control constructs are shown in Figure 1: RNAi of Dugesia japonica six-1 (Dj-six-1) results in an eyeless phenotype23 (Figure 1C), RNAi of Dugesia japonica PC2 (Dj-PC2) results in a loss of the light aversion response24 (Figure 1D) and RNAi of Dugesia japonica βcatenin-1 (Dj-βcatenin-1) results in a two-headed phenotype16-19, 21 from trunk fragments (Figure 1E). These phenotypes can be achieved using the following simple RNAi protocols: Dj-six-1 (FFxFx), Dj-PC2 (FFFx), Dj-βcatenin-1 (FFxFx), where F=feeding cycle and x=regenerative cycle respectively.
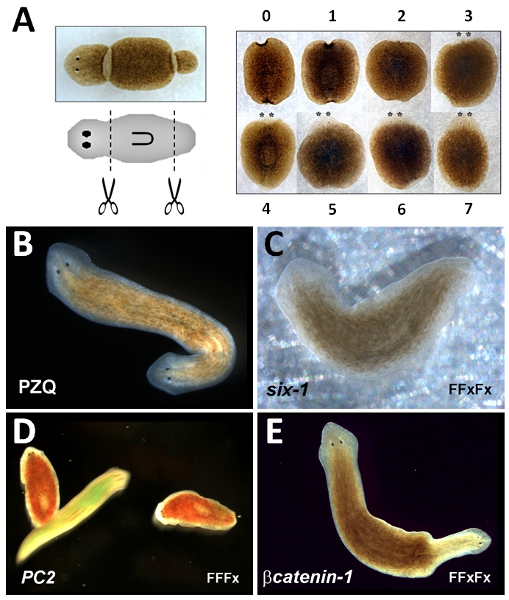
Figure 1: Trunk fragment regeneration assay. A Left, Image of planarian (top) and schematic (middle) to show location of excised trunk fragment. Right, timecourse of trunk fragment regeneration showing appearance of regenerating fragment at indicated times (days). B Image of two-headed planarian produced by PZQ exposure. C ‘Eyeless’ worm produced by Dj-six-1 RNAi. D Loss of light aversion response in immobile Dj-PC2 RNAi worms (stained red), compared to mobile control worm (stained green). E Bipolar planarian produced by Dj-βcatenin-1 RNAi. In B-E, the original anterior end of the RNAi worms is oriented to the left.
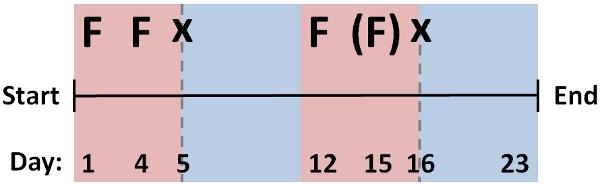
Figure 2: Sequence of feeding and cutting cycles for typical RNAi protocol comprising multiple feedings (F), and regenerative cycles (x) that is effective for knockdown of many genes in D. japonica. Modification of this protocol for individual genes may be needed to yield optimal results, for example using a fewer (parentheses) or a greater number of feeding and regenerative cycles.
Discussion
The protocols described here detail assays for studying and manipulating regeneration of the planarian Dugesia japonica. They are simple and do not require specialized equipment such that they can be easily performed in the laboratory or classroom. Assays can be performed individually, or combined (for chemical genetic screening of drug efficacies in vivo) and can be performed at the candidate gene level, or are adaptable to unbiased, higher throughput screening8. Whether for studying the intriguing biology of planarians in their own right, or for assessing in vivo function of mammalian homologues in a alternative model for studying tissue regeneration, these approaches should catalyze interest from a diverse range of researchers.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Work in the lab is supported by NSF (MCB0919933) and NIH (GM088790).
Materials
| Reagent | Vendor | Catalogue Number | Comments |
|---|---|---|---|
| Spring water | Kandiyohi. Premium Waters Inc. Minneapolis, MN | n/a | Other forms of spring water work well also. Trial first in viability assays. |
| 1 x buffered Montjuïch salts: NaCl (1.6mM), CaCl2 (1mM), MgSO4 (1mM), MgCl2 (0.1mM), KCl (0.1mM), NaHCO3 (1.2mM), HEPES (1.5mM). pH 7.4 at 24°C. | Multiple vendors | n/a | Artificial water for drug treatments during regenerative assays to ensure pH buffering. 5/8 Holtfreter’s solution is an alternative. |
| 2xYT Broth | Fisher Scientific | BP2467-500 | Media = 31 g/L . Autoclaved. |
| Petri Dish (100x25mm) | Fisher Scientific | 08-757-11 | Housing worms during regeneration cycles |
| Square Dish (100x100x15mm) | Fisher Scientific | 08-757-11A | Fill with water, freeze for ice tray used as worm cutting surface |
| Plastic tub: Ziploc Twist ‘n Loc (16oz). | Various retailers | n/a | Convenient water tight containers for RNAi cohorts |
| Chicken Liver | Commercial grocery | n/a | Bias towards organic supplies, owing to avoidance of antibiotics. |
| Hand Blender | Any kitchen supplier | n/a | Use for making chicken liver puree |
| Wire 1mm Mesh strainer | Any kitchen supplier | n/a | Use for straining chicken liver puree |
| Bovine red blood cells | Lampire Biological Laboratories | 7240807 | 100% Washed and pooled RBC suspension |
| Circular filter papers | Whatman #3 | 1003 055 | |
| Transfer Pipette | Fisher Scientific | 13-711-41 | |
| Sterile, surgical blades | Multiple Vendors | n/a | |
| ±Praziquantel | Sigma Aldrich | P4668 | Store powder aliquots in the dark at 4°C. Desiccate. |
| Dj-six-1 | GenBank AJ557022.1 | RNAi positive control for “eye-less” phenotype23 | |
| Dj-βcatenin-1 | GenBank AB181913.1 | RNAi positive control for two-headed phenotype17 | |
| Dj-PC2 | RNAi positive control for loss of light aversion phenotype24 |
References
- Morgan, T. H. Experimental studies of the regeneration of Planaria maculata. Arch Entwm Org. 7, 364-397 .
- Robb, S. M., Ross, E., Sánchez Alvarado, A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36, 599-606 (2008).
- Forsthoefel, D. J., Newmark, P. A. Emerging patterns in planarian regeneration. Curr Opin Genet Dev. 19, 412-4120 (2009).
- Adell, T., Cebrià, F., Saló, E. Gradients in Planarian Regeneration and Homeostasis. Cold Spring Harb Perspect Biol. 2, (2010).
- Agata, K., Watanabe, K. Molecular and cellular aspects of planarian regeneration. Semin Cell Dev Biol. 10, 377-383 (1999).
- Newmark, P. A. &. a. m. p. ;. a. m. p., Sánchez-Alvarado, A. Not your father’s planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 3, 210-219 (2002).
- Reddien, P. W., Sánchez-Alvarado, A. Fundamentals of planarian regeneration. Ann Rev Cell Dev Biol. 20, 725-757 (2004).
- Reddien, P. W., Bermange, A. L., Murfitt, K. J., Jennings, J. R., Sánchez-Alvarado, A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 8, 635-649 (2005).
- Newmark, P. A., Reddien, P. W., Cebria, F. &. a. m. p. ;. a. m. p., Sánchez-Alvarado, A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci USA. 100, 11861-11865 (2003).
- Collins, J. J. Genome-Wide Analyses Reveal a Role for Peptide Hormones in Planarian Germline Development. PLoS Biology. 8, e10000509-e10000509 (2010).
- Cebrià, F. Regenerating the central nervous system: how easy for planarians!. Dev Genes Evol. , 733-748 (2007).
- Mineta, K. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci USA. 100, 7666-7671 (2003).
- Oviedo, N. J., Nicolas, C. L., Adams, D. S., Levin, M. Emerging Model Organisms. A Laboratory Manual Chapter. 8, (2009).
- Buttarelli, F. R., Pellicano, C., Pontieri, F. E. Neuropharmacology and behavior in planarians: translations to mammals. Comp Biochem Physiol. 147, 399-408 (2008).
- Sánchez-Alvarado, A., Newmark, P. A. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A. 96, 5049-5054 (1999).
- Gurley, K. A., Rink, J. C., Sánchez-Alvarado, A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 319, 323-327 (2008).
- Yazawa, S., Umesono, Y., Hayashi, T., Tarui, H., Agata, K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A. 106, 22329-22334 (2009).
- Petersen, C. P., Reddien, P. W. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 319, 327-330 (2008).
- Iglesias, M., Gomez-Skarmeta, J. L., Saló, E., Adell, T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 135, 1215-1221 (2008).
- Nogi, T., Zhang, D., Chan, J. D., Marchant, J. S., S, J. A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel beta subunits: subversion of flatworm regenerative polarity. PLoS Negl Trop Dis. 3, e464-e464 (2009).
- Oviedo, N. J. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol. 339, 188-199 (2010).
- Nogi, T., Levin, M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev Biol. 287, 314-335 (2005).
- Mannini, L. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol. 269, 346-359 (2004).
- Agata, K. Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zoolog Sci. 15, 433-440 (1998).

