Analysis of Trunk Neural Crest Cell Migration using a Modified Zigmond Chamber Assay
Summary
An approach to analyze the migration of explanted cells (trunk neural crest cells) is described. This method is inexpensive, gentle, and capable of distinguishing chemotaxis from both chemokinesis and other influences on migratory polarity such as those derived from cell-cell interactions within the primary trunk neural crest cell culture.
Abstract
Neural crest cells (NCCs) are a transient population of cells present in vertebrate development that emigrate from the dorsal neural tube (NT) after undergoing an epithelial-mesenchymal transition 1,2. Following EMT, NCCs migrate large distances along stereotypic pathways until they reach their targets. NCCs differentiate into a vast array of cell types including neurons, glia, melanocytes, and chromaffin cells 1-3. The ability of NCCs to reach and recognize their proper target locations is foundational for the appropriate formation of all structures containing trunk NCC-derived components 3. Elucidating the mechanisms of guidance for trunk NCC migration has therefore been a matter of great significance. Numerous molecules have been demonstrated to guide NCC migration 4. For instance, trunk NCCs are known to be repelled by negative guidance cues such as Semaphorin, Ephrin, and Slit ligands 5-8. However, not until recently have any chemoattractants of trunk NCCs been identified 9.
Conventional in vitro approaches to studying the chemotactic behavior of adherent cells work best with immortalized, homogenously distributed cells, but are more challenging to apply to certain primary stem cell cultures that initially lack a homogenous distribution and rapidly differentiate (such as NCCs). One approach to homogenize the distribution of trunk NCCs for chemotaxis studies is to isolate trunk NCCs from primary NT explant cultures, then lift and replate them to be almost 100% confluent. However, this plating approach requires substantial amounts of time and effort to explant enough cells, is harsh, and distributes trunk NCCs in a dissimilar manner to that found in in vivo conditions.
Here, we report an in vitro approach that is able to evaluate chemotaxis and other migratory responses of trunk NCCs without requiring a homogenous cell distribution. This technique utilizes time-lapse imaging of primary, unperturbed trunk NCCs inside a modified Zigmond chamber (a standard Zigmond chamber is described elsewhere10). By exposing trunk NCCs at the periphery of the culture to a chemotactant gradient that is perpendicular to their predicted natural directionality, alterations in migratory polarity induced by the applied chemotactant gradient can be detected. This technique is inexpensive, requires the culturing of only two NT explants per replicate treatment, avoids harsh cell lifting (such as trypsinization), leaves trunk NCCs in a more similar distribution to in vivo conditions, cuts down the amount of time between explantation and experimentation (which likely reduces the risk of differentiation), and allows time-lapse evaluation of numerous migratory characteristics.
Protocol
1. Day 1: Isolation of trunk neural tubes for overnight culture on coverslips
- Incubate chick eggs for 56 h at 38° C. Remove the eggs from incubation, mildly spray them with 70% Ethanol, and then allow them to dry. Break the eggs open into a UV-sterilized glass tray.
- Extract each embryo from its yolk and place it in chick Ringer’s. Do this by first cutting around its blood islands with curved scissors; then, with blunt forceps, pick the embryo up by its extraembryonic membrane and place it in a sterile plastic Petri dish containing chick Ringer′s solution.
- Isolate the trunk of each embryo by trimming off excess extraembryonic membranes as well as cranial, vagal, and sacral axial levels using a tungsten needle (Fig. 1). First, select about 9 embryos that are between stages HH15-17 11. For stages HH15 and up, the forebrain and hindbrain axes form an acute angle and therefore the head appears to tilt caudally. At stage HH17, the tail bud is present and tilts ventrally but does not yet contain somites. With a tungsten needle, trim off extraembryonic membranes to about 2 mm from the embryo and cut off any embryonic tissues anterior to somite 10. Also remove all caudal embryonic tissues starting from around the fifth most newly formed somite.
- Place the isolated embryonic trunks in Dispase (0.24 U/ml DMEM) and incubate them for 1 h 15 min at 37° C and 5% CO2. Once the trunks begin incubation, start preparing 6 coverslips (CS) for culturing NT explants (steps 1.5-1.8).
- Rinse 6 CS in 70% ethanol (diluted in sterile, ultrapure water), and then allow them to dry. Using a lab marker, draw a circle in the center of each CS that is about 1 cm in diameter (this circle will later help you identify where a fibronectin coat has been applied). On the same face of each CS, write the word “be” (or some other asymmetrical word or shape) outside the drawn circle (this will help you identify whether the marked side of the CS is facing up or down).
- Place each CS in a separate 40 x 10 mm sterile dish with the marked surface facing down and allow the dish to sit open underneath a germicidal UV lamp for 10 min.
- Apply 60 μl fibronectin (FN; 10 μg/ml DMEM) to the unmarked surface of the CS while making sure the entire area within the 1 cm circle is coated. Place the dishes to incubate at 37° C for 30 min and then carefully aspirate the fibronectin from each CS.
- Add 250 μl “culture” medium [DMEM with L-Glutamine (2 mM), Penicillin (100U/ml), Streptomycin (100 mcg/ml), and 8% fetal bovine serum (FBS)] to the FN-coated area of the CS. Place the dishes containing each CS at 37° C and 5% CO2 again until the NTs have been isolated.
- Transfer all the incubated embryo trunks to one 5 cm glass Petri dish containing L15 medium and start dissecting out each NT using fine forceps and a tungsten needle (Fig. 1). Carefully slice along the border of the NT and somites with a sharp tungsten needle while being cautious not to damage the NT. It is often easier to begin isolating each NT from the caudal-most end of the trunk.
- Select 6 of the straightest and longest NTs to culture overnight (NTs between roughly 8 and 15 somites long are recommended). Using a micropipette tip primed with culture medium, transfer each of the 6 NTs to its own previously prepared CS (from steps 1.5 through 1.8). Be sure that the NT does not remain floating at the surface. If the NT is floating, drip medium onto it until it sinks using a micropipette.
- Place each dish at 37° C and 5% CO2 overnight. Be careful to ensure each NT is within the FN-coated area of its respective CS immediately prior to placing the dish in the incubator (by using the circle drawn in step 1.5 as a reference). A micropipette can be used to better adjust the position of each NT if needed.
- Place at least 2 ml of culture medium (without serum) into a sterile 15 ml centrifuge tube and incubate overnight at 37° C and 5% CO2. Leave the cap slightly unscrewed to allow the pH of the medium to adjust overnight. Pre-incubating the medium is important to help prevent bubble formation in your chamber, which may disrupt the establishment of a molecular gradient. Such “preincubated ” medium should be used in all future steps. When not being used, this medium should be incubating at 37° C.
2. Day 2: Loading the modified Zigmond chamber and time-lapse analysis of cell migration
- Out of the 6 NTs cultured, select the 3 cultures best suited for analysis. Generally, NCC cultures that have at least one long, straight edge should be selected (Fig. 2A). The 3 best cultures will be used to load and film 3 modified Zigmond chambers throughout the day, each with a different treatment. Out of the 3 cultures, choose one for loading the first chamber and return the others to the incubator for later use.
- Using a cotton swab, apply a thin, even layer of petroleum jelly surrounding the reservoirs and bridge of one modified Zigmond chamber.
- With a tungsten needle, gently remove the NT from the CS, while leaving the surrounding NCCs attached to the surface of the CS. Mark the dish with a pen to remember the orientation of the straightest edge of the NCC culture.
- Place a few drops of preincubated medium onto the bridge. Pick up the CS with fine forceps, dab the edge of the CS against a Kimwipe to remove most of the old culture medium, then immediately place the CS on the modified Zigmond chamber so that the straight edge of the culture to be filmed is centered over the length of the bridge and roughly perpendicular to the bridge-reservoir border (Fig. 2A,B).
- Using an inverted microscope, move the straight NCC border to be on the side of the bridge closest to the reservoir that will contain the suspected chemotactant (Fig. 2B; for controls this will correspond to whichever reservoir is loaded second). Also, more finely align the straight edge of the culture to be perpendicular to the bridge-reservoir border.
- Carefully, but securely press the CS into the petroleum jelly present on the Zigmond chamber, making sure it is completely sealed on to the chamber, then place additional petroleum jelly along the edge of the CS to further ensure it will be airtight. Fine-adjust the angle of the NCC border again to correct for any movement during the sealing process.
- Load the reservoir that will not contain the suspected chemotactant first (Fig. 2B). Do so by loading a 1 ml syringe (25 G x 1.5 in. attached needle) with roughly 300 μl preincubated medium and inject the medium into the reservoir until full (while being careful not to generate any bubbles in the reservoir). Plug the reservoir on both sides with a sufficient amount of petroleum jelly prior to loading the next reservoir.
- Repeat step 2.7, except this time using preincubated medium containing the candidate chemotactant. It is critical when generating a molecular gradient across the culture to always load the reservoir containing the molecule to be tested after loading the reservoir lacking the tested molecule.
- Place the loaded Zigmond chamber at 37° C to incubate for 1 h prior to filming. Image the straightest border of the NCC culture for 3 h at 90 s intervals while incubating at roughly 37° C (Fig. 2A,B). Prior to creating any movie, be sure to align the camera so that the edge of the images to be obtained are aligned with and touching the edge of the bridge that borders the last reservoir loaded (Fig. 2B, upper panel; dashed box represents ideal position for imaging). This will facilitate later software analysis by standardizing the directionality of the molecular gradient applied and the distance from the reservoir filmed in each movie produced.
- For controls, repeat steps 2.2-2.9 for each of the two other NCC cultures selected (in step 2.1), but fill each reservoir with an appropriate medium. For one type of control fill both reservoirs with preincubated medium not containing the molecule to be tested. For a second control treatment, prime the bridge with a few drops of preincubated medium containing the suspected chemotactant prior to mounting the CS. Then, load both reservoirs with the same medium containing the suspected chemotactant.
- Use ImageJ (NIH) Manual Tracking (rsb.info.nih.gov/ij/plugins/track/track.html) and Chemotaxis and Migration Tool v1.01 (www.ibidi.de/applications/ap_chemo.html) plugins to track the migration of peripheral NCCs along the straight border of the culture just imaged and to analyze various parameters of the migratory trajectories obtained (Fig. 2B-C).
3. Representative Results:
A sample of cell trajectories from a movie where many trunk NCCs were responsive to a candidate chemoattractant using the above technique is shown (Fig. 2D). Most cells in this example of a positive response displayed a net movement up the chemoattractant gradient (as shown in red). Trajectory data can be used to analyze other properties of cell migration as well.
In order to visually evaluate an applied gradient in a modified Zigmond chamber, an Alexa Fluor 488 IgM conjugate (MW ~ 900 kDa) was loaded into the second reservoir of a modified Zigmond chamber (at roughly 40 μg/ml H2O). A gradient was established by 1 h and still somewhat present after 26 h, but greatly diminished by 50 h (Fig. 3). If the molecule to be tested is smaller, then the applied gradient will degrade faster than what is shown.
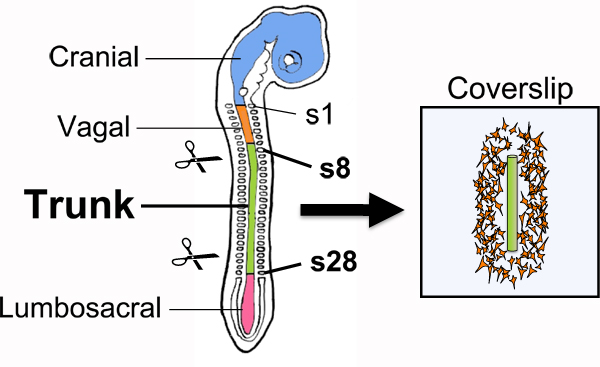
Figure 1. Explantation of trunk-level NTs for overnight culturing on fibronectin-coated coverslips. Because trunk NCCs delaminate from the dorsal NT located adjacent to somites 8-28, this segment of the NT is isolated by microdissection and cultured overnight on a fibronectin-coated CS to allow for emigration of NCCs from the NT explant. Isolated NTs that are between 8-15 somites long and relatively straight are best suited for overnight culturing as they tend to yield NCC cultures with longer straight borders. Regions of the neural tube that give rise to other neural crest axial levels are shown in a smaller font. s, somite.
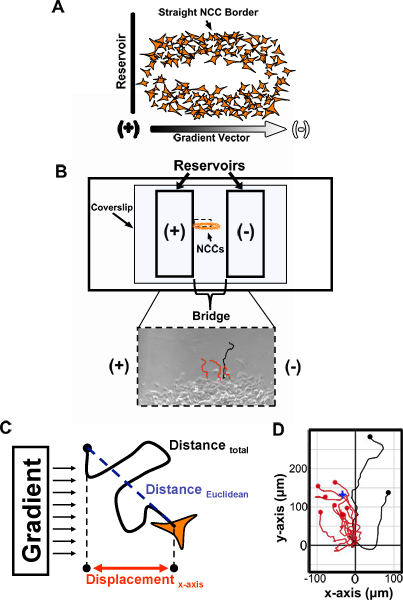
Figure 2. Method for evaluating the migration of explanted trunk NCCs using a modified Zigmond chamber. (A) Elongated trunk NCC cultures are prepared by overnight culturing of NTs and resultant NCC cultures with at least one long, straight border are selected for experimentation. The longest straight border of a selected culture is then positioned perpendicular to the bridge-reservoir border, and therefore parallel to the vector of the future applied gradient. (B) After fine-adjusting the position of the NCC culture on the Zigmond chamber and sealing the coverslip to the chamber, the chamber is loaded. When testing chemotaxis, the reservoir that will not contain the suspected chemotactant (-) is loaded first and sealed. Then, the other reservoir is loaded with the suspected chemotactant (+) and sealed. Peripheral NCCs along the previously selected border can then be imaged and tracked using the Manual Tracking plugin for ImageJ (bottom panel). (C) Numerous migratory characteristics in response to the applied gradient can be assessed based on tracking data. For instance, a chemotaxis index can be derived by dividing a cell’s displacement along the x-axis by the total distance it has migrated. (D) An example of an attractive response is shown by a cell trajectory plot initially generated by the Chemotaxis and Migration Tool plugin for ImageJ. The starting point of each trajectory is set to the origin (0,0). Note how many more cells migrate toward the chemoattractant source.The center of mass of all cells at their final position (blue cross; all cells equally weighted) is also closer to the chemoattractant source. NCCs, neural crest cells; Red tracks, cells that migrated toward the reservoir loaded with a suspected chemoattractant; Black tracks, cells that migrated away; (+), higher chemotactant concentration; (-), lower chemotactant concentration.
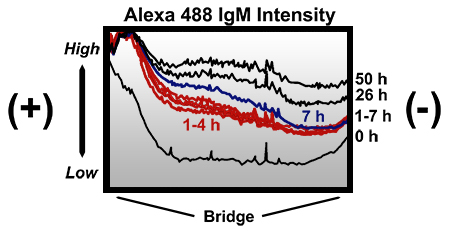
Figure 3. Intensity profiles across the bridge of a modified Zigmond chamber at different times following the addition of an Alexa Fluor 488 IgM conjugate. The chamber was loaded in a similar manner to that described in the protocol with the main exceptions that preincubated water (instead of preincubated medium) was used to dilute the antibody to 40 μg/ml, and small air pockets were present at the ends of the bridge (away from the slice where the above intensity profiles where taken). Initially, no gradient was present across most of the bridge. By 1 h a gradient was established and remained present through to 26 h. By 50 h the presence of the gradient was inconsistent at different areas of the bridge, and when present, the steepness of the gradient was greatly diminished. All profiles were generated from an identical slice across the bridge (from one bridge-reservoir border to the other) using AxioVision 4.6 software. Note that even while air pockets were present, the gradient was not disrupted. High, high intensity; Low, low intensity; x-axis, distance across entire width of bridge (2 mm); (+), reservoir loaded with the Alexa Fluor 488 IgM conjugate; (-), reservoir not loaded with the conjugate.

Figure 4. Modified Zigmond chamber specifications. Shown is a diagram of the modified Zigmond chamber used here along with its dimensional specifications (±0.2 mm). Measurements can be moderately adjusted in order to match individual preferences.
Supplemental Protocol: Fabrication of a Modified Zigmond Chamber
Please refer to Figure 4 as a reference for the protocol below:
- Purchase a sheet of 3/16″ thick polished acrylic (4.45 mm actual thickness).
- Using a table saw, cut chamber blanks oversized to the rough dimensions of 33.25 mm x 64.57 mm. This allows 3.175 mm extra material for machining.
- Set the chamber blank on a vise. With a milling machine and a 6.35 mm (1/4″) end mill bit, finish machining the sides of the chamber to their exact dimensions: 30.07 mm x 61.39 mm.
- Position the chamber blank on the milling machine and locate the center of the blank along both the x and y axes with an edge finder; then zero the center location.
- Acquire the chamber height (z-axis) by touching the end mill bit to the top surface and zero the height.
- Using a 3.91 mm (0.154″) end mill bit, offset the bit 3.03 mm along the x-axis (positive direction) for the first reservoir. Begin machining into the chamber to a depth of 2.84 mm while moving along the y-axis (positive direction) to 7.62 mm (0.300″) and then traverse to 7.62 mm (0.300″) in the opposite (negative) direction to a complete reservoir length of 15.24 mm (0.600″). Offset the bit to 3.03 mm (0.119″) along the x-axis (negative direction) and repeat the same process for the second reservoir.
- Position the chamber on its edge and drill a hole using a 1.09 mm (0.043 in.) drill bit on the end of each reservoir (4 total) that connects the end of the reservoir to the side of the chamber for loading medium during experimentation.
- Soak the chamber well in warm soapy water to help remove any chemical contaminants.
- Soak and rinse the chamber well in double-distilled water to remove any soap. The chambers are now ready to use as described above.
Discussion
Conducting chemotaxis research on trunk NCCs has proven challenging for a series of reasons. Trunk NCCs constitute a heterogeneous stem cell population that will differentiate if cultured long-term; therefore, trunk NCCs must be obtained from primary explantation of the trunk-level NT. Conventional methods to study the chemotactic response of homogenously distributed cell populations in vitro are difficult to test on trunk NCCs since they first require that cells are isolated and homogenously replated in the chemotaxis chamber (e.g., a Boyden chamber 12). Isolating enough NCCs can be tedious as the explantation of dozens of NTs may be required to create a homogenous distribution of trunk NCCs sufficient for even one experiment. Additionally, NCCs are never lifted, mixed, and homogenously redistributed in their natural context. Therefore, NCCs that are replated in order to perform certain conventional chemotaxis assays may behave quite differently from in vivo conditions in regards to migratory behavior.
Here, we describe a modified technique for evaluating trunk NCC chemotaxis in a more natural context. A major challenge when evaluating the chemotactic response of trunk NCCs without a homogenous distribution is the strong inherent polarity of NCCs within the original explant culture in addition to their innate heterogeneous identity. For instance, trunk NCCs tend to migrate largely perpendicular (away) from the NT when in culture 13. The various factors that influence trunk NCC polarity within the explant culture can be so powerful that it is difficult to conclusively identify changes in cell polarity induced by a chemotactant gradient. Using our new modified Zigmond assay it is possible to distinguish between natural cell polarity inherent to explant cultures and changes induced by a chemotactant gradient, while also monitoring other migratory parameters such as persistence and speed. We have observed that the angle each border of an explanted trunk NCC culture faces largely determines the inherent directionality of the leading-edge trunk NCCs at that border. For instance, if a border of a NCC culture faces north, then the mean directionality of NCCs from that border would be northerly. Our modified assay minimizes variability caused by the inherent directionality of leading-edge NCCs by standardizing the angle of the NCC border from which they migrate relative to the applied chemotactant gradient (Fig. 2A). By selecting leading-edge NCCs on a relatively straight border of the trunk NCC culture, always positioning that straight NCC border perpendicular to the applied gradient, and then only tracking the migration of leading-edge cells on that border, one is able to detect shifts in the mean directionality of trunk NCCs toward or away from a chemotactant source that would otherwise go undetected.
Below is a summary of the numerous benefits derived from the above protocol:
- It offers the ability to track live cell migration.
- The modified Zigmond chamber can produce a relatively defined gradient that can be visualized with a fluorescent molecule (Fig. 3).
- Zigmond chambers avoid certain limitations of the standard Boyden assay previously described by others 10,14.
- Our modified Zigmond chambers can be home-made at a very low cost (materials are estimated at less than 1 USD per reusable chamber).
- Because neural tubes are cultured on coverslips and later sealed with petroleum jelly, one can readily choose the orientation of the culture relative to the future applied gradient.
- Trypsinization or cell scraping is not required.
- Only a small amount of NT isolations are necessary.
- The NCC culture is more similar to the distribution of trunk NCCs found in vivo than when cells are lifted and replated with a homogenous distribution.
These advantages would also likely apply to analogous primary explant cultures of other cell types, and therefore, this approach may have a broader desirability for use in the evaluation of the natural migratory behavior of other explanted stem cells.
Although the advantages of the modified Zigmond chamber assay are many, it does have some limitations. Some disadvantages of the standard Zigmond chamber have already been discussed by past researchers 14. One limitation of our modified Zigmond assay is the use of petroleum jelly for mounting the CS and plugging each reservoir hole. Petroleum jelly allows one the flexibility to precisely orient a NCC culture relative to the chemotactant gradient, but leads to variability in the distance between the CS and the bridge which could cause variability in the applied gradient 14. Additionally, mounting the CS and sealing the chamber with petroleum jelly often traps small pockets of air in the chamber which could compress leading to small amounts of flow across the bridge 14. Such flow could potentially disrupt the molecular gradient during some experiments and lead to increased variability in one′s results. While this is possible, a severe gradient disruption was not observed when visualized using an Alexa Fluor 488 IgM conjugate (Fig. 3), even while air pockets were present toward the ends of the bridge (away from the area where fluorescent intensity was measured). Therefore, even under conditions that could have potentially disrupted the Alexa Fluor 488 IgM gradient, the gradient was not compromised in the area of the bridge measured (closer to the middle of the bridge). If one requires a long-term gradient that is both highly stable and predictable, then this assay will likely not suffice. However, such precision is often not required and in comparison to some other common chemotaxis assays (such as Boyden assays or certain assays using chemotactant-soaked beads), the gradient is likely to be more stable and predictable.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We give special thanks to Lino Kim, Steve Guzman and Ujit Satyarthi for technical assistance during the development of this method. Myron Hawthorne, Richard Spengel, and Roberto Rojas machined the chambers used here and provided much-needed technical assistance. Notably, Roberto Rojas produced Figure 4. We are also thankful for Scott Fraser’s invaluable advice prior to the development of the above chemotaxis assay. This work was partly supported by an NIH-MBRS SCORE-5S06GM048680-13 to MEdB and by an award from the CSU, Northridge Graduate Thesis Support Program to CW.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| DMEM | Omega Scientific | DM-22 | |
| Penicillin Streptomycin Solution | Omega Scientific | PS-20 | 100X Stock Concentration |
| L-Glutamine | Omega Scientific | GS-60 | 100X Stock Concentration |
| Fetal Bovine Serum | Omega Scientific | FB-11 | Lot# 105247 (or another that is comparable) |
| Modified Zigmond chamber | Home made | N/A | Reservoir volume: ~ 160 μl ea; for additional specifications, see Fig. 4 and the supplemental fabrication protocol |
| Cell culture dish | Denville | T6040 | 40 x 10 mm |
| Fibronectin | BD | 354008 | 10X Stock prepped by diluting 1 mg FN in 1 ml H2O and 9 ml DMEM |
| Coverslips | Fisher | 12-548-B | Precleaned; 22 x 22 mm |
| L15 medium | Thermo Scientific | SH30525.02 | |
| Petroleum Jelly | Comforts | 011110794642 | 100% |
| Centrifuge tube | Biologix | 10-9152 | 15 ml |
| Dispase | Cell Systems | 4Z0-850 | 10X Stock Concentration |
| Syringe | BD | 309602 | 1 ml |
| Needle | BD | 305127 | 25 G x 1.5 in. |
| Alexa Fluor 488-IgM | Invitrogen | A21042 | Stock is 2 mg/ml; 7 moles dye/mole IgM |
| Dissecting Forceps | FST | Misc. | Dumont #5 or 55; straight tipped; stainless steel or titanium |
| Tungsten Needle | N/A | N/A | Home made; placed in a pin holder |
| Blunt Forceps | Tiemann | 160-18 | Used for transferring embryos to Ringer’s from egg yolk |
Supplemental Protocol: Fabrication of a Modified Zigmond Chamber
Please refer to Figure 4 as a reference for the protocol below:
- Purchase a sheet of 3/16″ thick polished acrylic (4.45 mm actual thickness).
- Using a table saw, cut chamber blanks oversized to the rough dimensions of 33.25 mm x 64.57 mm. This allows 3.175 mm extra material for machining.
- Set the chamber blank on a vise. With a milling machine and a 6.35 mm (1/4″) end mill bit, finish machining the sides of the chamber to their exact dimensions: 30.07 mm x 61.39 mm.
- Position the chamber blank on the milling machine and locate the center of the blank along both the x and y axes with an edge finder; then zero the center location.
- Acquire the chamber height (z-axis) by touching the end mill bit to the top surface and zero the height.
- Using a 3.91 mm (0.154″) end mill bit, offset the bit 3.03 mm along the x-axis (positive direction) for the first reservoir. Begin machining into the chamber to a depth of 2.84 mm while moving along the y-axis (positive direction) to 7.62 mm (0.300″) and then traverse to 7.62 mm (0.300″) in the opposite (negative) direction to a complete reservoir length of 15.24 mm (0.600″). Offset the bit to 3.03 mm (0.119″) along the x-axis (negative direction) and repeat the same process for the second reservoir.
- Position the chamber on its edge and drill a hole using a 1.09 mm (0.043 in.) drill bit on the end of each reservoir (4 total) that connects the end of the reservoir to the side of the chamber for loading medium during experimentation.
- Soak the chamber well in warm soapy water to help remove any chemical contaminants.
- Soak and rinse the chamber well in double-distilled water to remove any soap. The chambers are now ready to use as described above.
References
- Le Douarin, N. M. The avian embryo as a model to study the development of the neural crest: a long and still ongoing story. Mechanisms of Development. 121, 1089-1102 (2004).
- Baker, C. V. . Neural Crest and Cranial Ectodermal Placodes. , (2005).
- Gammill, L. S., Roffers-Agarwal, J. Division of labor during trunk neural crest development. Dev. Biol. 344, 555-565 (2010).
- Kulesa, P. M., Gammill, L. S. Neural crest migration: patterns, phases and signals. Dev. Biol. 344, 566-568 (2010).
- Wang, H. U., Anderson, D. J. Eph family transmembrane ligands can mediate repulsive guidance of trunk neural crest migration and motor axon outgrowth. Neuron. 18, 383-396 (1997).
- Krull, C. E. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr. Biol. 7, 571-580 (1997).
- Gammill, L. S., Gonzalez, C., Gu, C., Bronner-Fraser, M. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development, Cambridge, England. , 133-199 (2006).
- De Bellard, M. E., Rao, Y., Bronner-Fraser, M. Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells. The Journal of cell biology. 162, 269-279 (2003).
- Kasemeier-Kulesa, J. C., McLennan, R., Romine, M. H., Kulesa, P. M., Lefcort, F. CXCR4 controls ventral migration of sympathetic precursor cells. J. Neurosci. 30, 13078-13088 (2010).
- Zigmond, S. H. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. The Journal of Cell Biology. 75, 606-616 (1977).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chicken embryo. J. Morph. 88, 49-52 (1951).
- Boyden, S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. The Journal of Experimental Medicine. 115, 453-466 (1962).
- Davis, E. M., Trinkaus, J. P. Significance of cell-to cell contacts for the directional movement of neural crest cells within a hydrated collagen lattice. Journal of Embryology and Experimental Morphology. 63, 29-51 (1981).
- Zicha, D., Dunn, G. A., Brown, A. F. A new direct-viewing chemotaxis chamber. Journal of Cell Science. 99, 769-775 (1991).

