Colonization of Euprymna scolopes Squid by Vibrio fischeri
Summary
The method outlines the procedure by which the Hawaiian bobtail squid, Euprymna scolopes and its bacterial symbiont, Vibrio fischeri, are raised separately and then introduced to allow for specific colonization of the squid light organ by the bacteria. Colonization detection by bacterially-derived luminescence and by direct colony counting are described.
Abstract
Specific bacteria are found in association with animal tissue1-5. Such host-bacterial associations (symbioses) can be detrimental (pathogenic), have no fitness consequence (commensal), or be beneficial (mutualistic). While much attention has been given to pathogenic interactions, little is known about the processes that dictate the reproducible acquisition of beneficial/commensal bacteria from the environment. The light-organ mutualism between the marine Gram-negative bacterium V. fischeri and the Hawaiian bobtail squid, E. scolopes, represents a highly specific interaction in which one host (E. scolopes) establishes a symbiotic relationship with only one bacterial species (V. fischeri) throughout the course of its lifetime6,7. Bioluminescence produced by V. fischeri during this interaction provides an anti-predatory benefit to E. scolopes during nocturnal activities8,9, while the nutrient-rich host tissue provides V. fischeri with a protected niche10. During each host generation, this relationship is recapitulated, thus representing a predictable process that can be assessed in detail at various stages of symbiotic development. In the laboratory, the juvenile squid hatch aposymbiotically (uncolonized), and, if collected within the first 30-60 minutes and transferred to symbiont-free water, cannot be colonized except by the experimental inoculum6. This interaction thus provides a useful model system in which to assess the individual steps that lead to specific acquisition of a symbiotic microbe from the environment11,12.
Here we describe a method to assess the degree of colonization that occurs when newly hatched aposymbiotic E. scolopes are exposed to (artificial) seawater containing V. fischeri. This simple assay describes inoculation, natural infection, and recovery of the bacterial symbiont from the nascent light organ of E. scolopes. Care is taken to provide a consistent environment for the animals during symbiotic development, especially with regard to water quality and light cues. Methods to characterize the symbiotic population described include (1) measurement of bacterially-derived bioluminescence, and (2) direct colony counting of recovered symbionts.
Protocol
1. Preparation of Bacterial Inocula
- Day 0
Two days prior to squid inoculation, plate the relevant bacterial strains on LBS13 agar. - Incubate bacteria at 25-28°C overnight.
- Day 1
Inoculate 3 ml LBS medium in a glass culture tube with one colony of each V. fischeri strain for infection. Prepare duplicate tubes as backup. - Day 2
(Coordinate bacterial steps 1.4-1.6 with squid steps 3.7-3.10)
1 h prior to inoculation, subculture bacteria 1:80 (37.5 μl) into 3 ml LBS in a glass culture tube and grow for 1 h with aeration. - Measure the OD600 of the sample prior to inoculation. Typical measurements are 0.3-0.6 depending on the strain.
- For a target inoculum of 3-5 x 103 CFU/ml calculate the inoculum volume as follows: Inoculum volume (μl) = 1.25 / OD600 (e.g., For OD600 = 0.5, the calculated inoculum volume = 1.25/0.5 = 2.5 μl).This amount is added directly to the seawater containing squid in Step 4.1. This calculation may need to be adjusted for different strains of V. fischeri or for inoculation at lower or higher levels than those specified here.
2. Preparation of Agar Plates for Enumeration of the Inocula
- For each treatment, label LBS plates (2 per treatment) to plate samples of the inoculum in Step 4.1.
- Add 5 sterile plating beads per plate.
3. Collection of Squid Juveniles
- Measure the salinity of Instant Ocean using the refractometer and adjust to 35‰.
- Filter 1 L of Instant Ocean using the filtration unit and an attached vacuum line or vacuum pump, to generate filter-sterilized Instant Ocean (FSIO). Oxygenate the water by swirling vigorously prior to each dispensing. The filter unit can be reused for 2 days.
- Aliquot 40-50 ml of FSIO into each of two (2) disposable sample bowls. Label one as earlies and one as timelies.
- Prepare an excess of plastic transfer pipettes for acquiring juvenile squid by cutting the pipette approximately 1 cm from the tip, above the lowest ridges (see Figure 3). This facilitates a wider area through which the squid can pass upon collection. Discard any transfer pipettes in which there is a rough exposed surface.
- Using prepared transfer pipettes, collect E. scolopes that hatched overnight and transfer to the earlies bowl of FSIO. Early hatchlings have been in the egg system for over 1 h and are susceptible to colonization by contaminating V. fischeri in the egg system. Do not use earlies for sensitive colonization experiments.
- Check egg tanks every 30-45 min for new hatchlings. Ensure that all hatchlings are cleared during each check. Remove hatchlings with a transfer pipette, and deposit into the timelies bowl of FSIO. Animals collected in a timely fashion are available for colonization experiments.
- When the collection has finished (~ 45 min after dusk), transfer the squid to the main laboratory. Empirically it is advantageous to colonize the animals under uninterrupted laboratory light conditions for 3 h inoculations.
- For each treatment, prepare a bowl with 40 ml FSIO. Add squid to the bowls for the assay (maximum n=40 per bowl).
- Prepare an additional bowl as an aposymbiotic (negative) control.
- Prepare a dedicated transfer pipette for each treatment.
- Euthanize extra squid in 2% ethanol.
4. Squid Colonization
- Day 2 – Using a P10 Pipetman, dispense the calculated aliquot of bacteria (Step 1.5) into each squid bowl (Step 3.8) for each treatment. Start a 3 h timer immediately after the first inoculation.
- For each treatment, create a “vortex” in the bowl with the dedicated transfer pipette by placing the pipette near the edge of the bowl and pipetting up and down repeatedly to mix the water and squid for approximately 10 sec. Thorough mixing is critical.
- Plate 50 μl from each bowl onto an LBS agar plate from Step 2.2 (for technical replicates, plate two 50 μl plates per treatment). Incubate at 25-28°C overnight.
- Prepare wash bowls (100 ml FSIO/ea) for each treatment.
- Prepare Drosophila vials (4 ml FSIO/ea) for each squid.
- After exactly 3 h, transfer the squid to their respective wash bowls (complete for all treatment). This stops the inoculation.
- Proceed to transfer each individual squid to its own Drosophila vial with FSIO. Use a designated transfer pipette for each treatment.
- Move trays of Drosophila vials to the squid facility to return to the day/night light cycle the animals experienced during embryogenesis.
- Day 3 – Prepare Drosophila vials (4 ml FSIO/ea) for each squid.
- Prior to dusk at 22-24 h post-inoculation, transfer each squid to a new Drosophila vial. Use a designated transfer pipette for each treatment.
- Day 4 – Prepare labelled 1.5 ml microcentrifuge tubes (1/squid).
- Prior to dusk at 46-48 h post-inoculation, measure and record the luminescence of each squid in the Drosophila vial (luminometer set for 6 s integration and auto-read on lid closure).
- As a negative control for background luminescence, measure a vial with FSIO that does not contain any squid.
- Transfer each squid in a volume of approximately 700 μl to a 1.5 ml microcentrifuge tube from Step 4.12. Move to a cardboard freezer box. Once the lid is placed on the box, do not remove it as the light cues for bacteria expulsion are not well-understood.
- Freeze microcentrifuge tubes at -80°C overnight.
5. Determination of Colonization Levels
- For each squid, prepare two (2) microcentrifuge tubes, each with 475 μl FSIO (or autoclaved 70% Instant Ocean).
- Prepare pestles by first using a Kimwipe to clean the pestle and remove gross debris and/or tissue.
- Place pestles tip-down in a 50 ml beaker containing 95% ethanol. Ethanol should be added to a height of approximately 3 cm.
- For each pestle, remove from the beaker and wipe the tip with a Kimwipe.
- Dip pestle back into the ethanol bath, remove and insert (tip up) into an microcentrifuge tube rack and allow to air dry completely for approximately 15 minutes.
- Thaw squid in a microcentrifuge tube rack (maximum n=8).
- If necessary, adjust the volume to 700 μl.
- Using a pestle from Step 5.5, disrupt the animal tissue until the ink sac ruptures (the water will turn a murky grey color).
- Remove the pestle and ensure all tissue remains in the tube.
- Vortex the tissue briefly for exactly 10 seconds (use a timer).
- Allow the tissue to rest for 10 min. The tissue will settle and the bacteria and ink remain in solution. For the calculations that follow, the bacteria/ink solution is the [A] dilution (i.e. the E. scolopes light organ homogenate in 700 μl). Serial 1:20 dilutions ([B], [C]) are described below.
- For the [B] dilution, add 25 μl [A] to one of the microcentrifuge tubes prepared in Step 5.1. Vortex.
- For the [C] dilution, add 25 μl [B] to one of the microcentrifuge tubes prepared in Step 5.1. Vortex.
- Plate 50 μl of each dilution onto LBS agar, 2 replicates per treatment.
- Incubate the plates at 25-28°C overnight.
6. Data Analysis
- To calculate the CFU/light organ (LO), count colonies on the plate for each treatment in which 10-400 colonies are present, and use the appropriate formula:
CFU/LO = (colonies on [A] plate) x 14; or
CFU/LO = (colonies on [B] plate) x 280; or
CFU/LO = (colonies on [C] plate) x 5600. - Plot individual data points and medians on a logarithmic scale.
- The data are often not normally distributed, with different variances, and the outliers may contain biologically meaningfully information. Therefore, non-parametric tests provide a useful method to determine whether the treatments differ significantly.
- Use GraphPad Prism software for statistical analysis. For two treatments, use the Wilcoxon Rank Sum test. For comparisons among greater than two treatments, use the Kruskal-Wallis test with appropriate post-tests.
7. Representative Results
Results from a sample colonization assay are shown in Figure 4. Two strains of V. fischeri that exhibit different relative levels of luminescence were each inoculated into six squid, along with six squid that served as aposymbiotic (uncolonized) controls. The E. scolopes symbiont, ES11414, and the brighter Sepiola robusta symbiont, SR515,16. Similar inoculum levels (Fig. 4A) lead to 100% colonization within 3 h. At 48 h, the luminescence levels (Fig. 4B) and CFU counts (Fig. 4C) were determined to assess the colonization proficiency of the strain. Determination of the specific luminescence (Fig. 4D; per bacterium) allows for determination of the brightness of each bacterial strain during the symbiosis.

Figure 1. Flow chart of the colonization procedure. Bacteria and squid are harvested separately, then mixed at the specified inoculum. Squid are washed, then transferred to new water at 3 h, 24 h, and 48 h post inoculation. At 48 h the luminescence is measured and the animals are frozen, which serves to surface-sterilize the animals. Light-organ colonized bacteria remain viable at -80°C through one thaw (no additional freeze-thaw cycles).
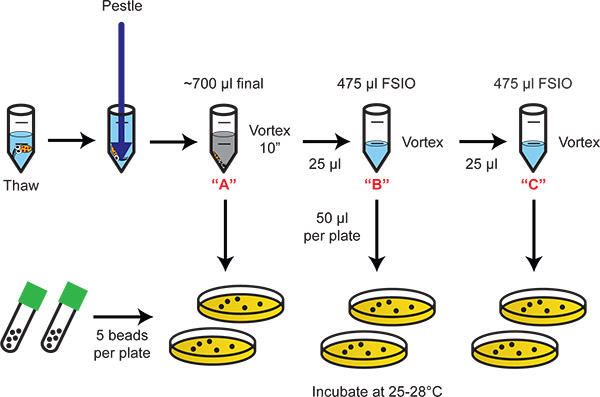
Figure 2. Flow chart illustrating homogenization and dilution plating of the bacteria. Serial 20-fold dilutions provide the appropriate dynamic range for enumeration of colonized bacteria.

Figure 3. Transfer pipettes with an appropriately narrow shaft taper to a narrow bore (A) that would damage juvenile squid. Pretreatment by cutting off the narrowest section with scissors or a razor blade yields an appropriate tool (B) for transferring juvenile squid.

Figure 4. Sample data for a colonization assay. (A) Levels of the bacteria in the inoculum bowls. (B) Luminescence of individual squid. (C) Colony counts of individual squid. (D) Specific luminescence of individual squid. Apo, Aposymbiotic (uncolonized negative control).
Discussion
The colonization assay described allows for analysis of a natural symbiotic process in a controlled laboratory environment. As such, it can be used to assess colonization by mutant strains, by different natural isolates, and under different chemical regimes. Variations on the experiments described are commonly used to assess different aspects of the symbiosis. The kinetics of colonization can be measured by examining luminescence during the first 24 h, which can be detected automatically in a scintillation counter in which the coincidence detector has been removed. Furthermore, the relative colonization ability of one strain relative to another can be measured by a competitive colonization assay, in which the output ratio of the two strains in a set of animals are normalized to the input ratio (differential detection by distinct antibiotic resistance, fluorescence, or chromogenic [LacZ/Xgal] markers). Finally, colonization can be imaged directly by confocal microscopy.
It is critical to use healthy juvenile squid for the experiments. Behavioral indicators of poor squid health include swimming in circles (euthanize the animal), or animals that remain white and do not alter their chromatophores to turn brown on a dark surface (use with care). As there exists variation in the host populations, a greater number of replicate experiments with smaller numbers of animals is often more valuable than a smaller number of replicate experiments with large sample sizes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Mattias Gyllborg for squid facility support and for comments on this manuscript, Michael Hadfield and the Kewalo Marine Laboratory for assistance during field collection, and members of the Ruby and McFall-Ngai Laboratory for contributions to this protocol. Work in the Mandel Laboratory is supported by NSF IOS-0843633.
Materials
| Name of reagent | Company | Catalogue Number | Comments |
| Glass Culture Tubes, 16 mm Diameter | VWR | 47729-580 | |
| Caps for Glass Culture Tubes | Fisher | NC9807998 | |
| Visible Spectrophotometer for Determination of OD600 | Biowave | CO8000 | Any spectrophotometer capable of measuring OD600 will work. This unit can measure the OD600 of liquid directly in the glass culture tubes. Some adjustment of the inoculum calculation may be necessary depending on the instrument used. |
| GloMax 20/20 Single-Tube Luminometer | Promega | E5311 | Equivalent to the Turner BioSystems 20/20n Luminometer. Includes the microcentrifuge tube holder. |
| GloMax 20/20 Light Standard | Promega | E5341 | For luminometer calibration. |
| Refractometer, Handheld | Foster and Smith Aquatics | CD-14035 | Calibrate before each use with deionized water. Rinse after every use with deionized water to prevent salt build-up. |
| Instant Ocean (artificial seawater concentrate) | Foster & Smith Aquatics | CD-16881 | Prepare at 35 ‰ in deionized water, using the refractometer, then filter through a 0.2 μm SFCA filter. |
| Filtration Unit | Nalgene | 158-0020 | Surfactant-free cellulose acetate (SFCA) membrane, 0.2 μm. We have observed variable results with some surfactant-containing PES filters. |
| Transfer Pipettes | Fisher | 13-711-9AM | Using scissors or razor blade, cut the tip cleanly above the first ridge to increase the diameter of the pipette tip and avoid squeezing the squid hatchlings. |
| Disposable Sample Bowls (plastic tumblers) | Comet | T9S (9 oz.) | Bowls for inoculation, with upper diameter 3 ¼”, lower diameter 2 ¼”, height 3″. Bowls create a homogenous environment as they have no bottom rim, in which squid can get trapped in a low-oxygen niche. The size is optimized for 40-ml inoculum. Available at webstaurantstore.com, #619PI9. |
| Drosophila Vials | VWR | 89092-720 | Vial diameter matches the opening on the luminometer PMT. |
| 1.5 ml Microcentrifuge Tubes | ISC Bioexpress | C-3217-1CS | Tubes must fit the shape of the pestles. |
| Ethanol, 200 Proof | Fisher | BP2818-100 | |
| Pestles | Kimble Chase/Kontes | 749521-1500 | |
| Plating Beads, 5 mm diameter | Kimble Chase | 13500 5 | Prepare 5 per tube and autoclave. |
References
- Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., Dewhirst, F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721-5732 (2005).
- Mandel, M. J., Wollenberg, M. S., Stabb, E. V., Visick, K. L., Ruby, E. G. A single regulatory gene is sufficient to alter bacterial host range. Nature. 458, 215-218 (2009).
- Grice, E. A., Segre, J. A. The skin microbiome. Nat. Rev. Microbiol. 9, 244-253 (2011).
- Malic, S. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology. 155, 2603-2611 (2009).
- Turnbaugh, P. J. The human microbiome project. Nature. 449, 804-810 (2007).
- Nyholm, S. V., McFall-Ngai, M. J. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632-642 (2004).
- Ruby, E. G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50, 591-624 (1996).
- McFall-Ngai, M. J., Ruby, E. G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 254, 1491-1494 (1991).
- Jones, B., Nishiguchi, M. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Marine Biology. 144, 1151-1155 (2004).
- Graf, J., Ruby, E. G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 95, 1818-1822 (1998).
- Ruby, E. G., McFall-Ngai, M. J. A squid that glows in the night: development of an animal-bacterial mutualism. J. Bacteriol. 174, 4865-4870 (1992).
- Lee, P. N., McFall-Ngai, M. J., Callaerts, P., de Couet, H. G. The Hawaiian bobtail squid (Euprymna scolopes): a model to study the molecular basis of eukaryote-prokaryote mutualism and the development and evolution of morphological novelties in cephalopods. Cold Spring Harbor Protocols. , (2009).
- Stabb, E., Visick, K., Millikan, D., Corcoran, A. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction. Recent Advances in Marine Science and Technology. , (2001).
- Boettcher, K. J., Ruby, E. G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172, 3701-3706 (1990).
- Fidopiastis, P. M., von Boletzky, S., Ruby, E. G. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180, 59-64 (1998).
- Bose, J. L. Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl. Environ. Microbiol. 77, 2445-2457 (2011).

