Co-analysis of Brain Structure and Function using fMRI and Diffusion-weighted Imaging
Summary
We describe a novel approach for simultaneous analysis of brain function and structure using magnetic resonance imaging (MRI). We assess brain structure with high-resolution diffusion-weighted imaging and white-matter fiber tractography. Unlike standard structural MRI, these techniques allow us to directly relate anatomical connectivity to functional properties of brain networks.
Abstract
The study of complex computational systems is facilitated by network maps, such as circuit diagrams. Such mapping is particularly informative when studying the brain, as the functional role that a brain area fulfills may be largely defined by its connections to other brain areas. In this report, we describe a novel, non-invasive approach for relating brain structure and function using magnetic resonance imaging (MRI). This approach, a combination of structural imaging of long-range fiber connections and functional imaging data, is illustrated in two distinct cognitive domains, visual attention and face perception. Structural imaging is performed with diffusion-weighted imaging (DWI) and fiber tractography, which track the diffusion of water molecules along white-matter fiber tracts in the brain (Figure 1). By visualizing these fiber tracts, we are able to investigate the long-range connective architecture of the brain. The results compare favorably with one of the most widely-used techniques in DWI, diffusion tensor imaging (DTI). DTI is unable to resolve complex configurations of fiber tracts, limiting its utility for constructing detailed, anatomically-informed models of brain function. In contrast, our analyses reproduce known neuroanatomy with precision and accuracy. This advantage is partly due to data acquisition procedures: while many DTI protocols measure diffusion in a small number of directions (e.g., 6 or 12), we employ a diffusion spectrum imaging (DSI)1, 2 protocol which assesses diffusion in 257 directions and at a range of magnetic gradient strengths. Moreover, DSI data allow us to use more sophisticated methods for reconstructing acquired data. In two experiments (visual attention and face perception), tractography reveals that co-active areas of the human brain are anatomically connected, supporting extant hypotheses that they form functional networks. DWI allows us to create a “circuit diagram” and reproduce it on an individual-subject basis, for the purpose of monitoring task-relevant brain activity in networks of interest.
Protocol
1. Equipment for MR Data Acquisition
Figures 2 and 3 summarize a number of choices to be made in diffusion MRI acquisition, data reconstruction, and fiber tracking. Keep in mind that these choices typically involve trade-offs, and the best choice may depend upon one’s research objectives. For example, DSI and multi-shell HARDI (see Figure 2) typically use higher “b-values” (i.e., stronger diffusion weighting) than DTI. As a result, these methods have better angular resolution, which is necessary for resolving crossing or “kissing” fibers (i.e., fibers which curve toward one another, making contact at a single tangent before curving away again). However, this gain in angular resolution is often achieved at the cost of lower signal-to-noise ratio (SNR) in EPI data (Figure 3). Researchers may wish to consider the relevance of this trade-off for their specific objectives: if a study focuses on a few major fiber tracts whose trajectories do not cross or run parallel to other tracts, then a low-direction DTI scan with high SNR may be ideal. Imaging of the inferior longitudinal fasciculus might represent such a case. In contrast, the loss of SNR may be an acceptable consequence if a researcher wishes to follow a tract through complex crossings.
A similar trade-off involves the correction of head motion, eddy currents, and non-linear image distortions. DWI protocols use echo-planar imaging (EPI; see Table 1), which is susceptible to magnetic field inhomogeneities caused by air pockets in the sinuses, physiological noise, and other factors3. These inhomogeneities result in undesirable image distortion, particularly in the inferior temporal lobe and orbito-frontal cortex, which reduces the validity and reliability of fiber tracking results in these areas. Additional distortions are created by eddy currents, a product of rapid MR gradient switching4. Participants’ head motion is another factor which degrades image quality and can negatively affect tractography. Current methods can correct both head motion and image distortions in low b-value data, such as DTI; however, these methods have not been extended to higher-resolution methods such as DSI. The difficulty in applying image correction methods to DSI data stems from the low SNR described above (Figure 3). For fiber tracking in brain areas which are susceptible to EPI distortion, it may be best to use low-directionality DTI or another technique for which image distortions can be corrected. On the other hand, if high angular resolution throughout the brain is desired, researchers may opt to use DSI, HARDI, or similar techniques. Tuch (2004)5 suggests that researchers interleave T2 images with no diffusion weighting throughout a DSI scan, providing benchmarks for motion correction (for example, see ref. 6). In all cases, researchers should be aware of the adverse impact of head motion during acquisition: it is advisable to use highly-trained participants and to minimize movement through the use of bite bars, nose guards, padding, or other safeguards.
The results presented here use a 257-direction diffusion spectrum imaging (DSI) protocol, with gradient strengths ranging from b=300 to 7,000 (see parameters in Table 1). The diffusion spectrum imaging (DSI) sequence requires modern MR scanning equipment with certain features necessary for collecting this high-resolution diffusion data. We note that the time requirements of this sequence are considerable: approximately 43 minutes on a Siemens Tim Trio scanner. After extensive empirical testing, we feel that the quality of these data justify the duration and scanning cost; however, in choosing the acquisition protocol, users should carefully weigh their research objectives against the capacities and comfort of participants. We also note that good-quality DSI data has been collected in as few as 10 minutes with advanced acquisition techniques7.
- 3 Tesla field strength MR scanner: 3T is necessary to achieve the signal required for the high-angular direction DSI scan.
- 32-channel phased-array head coil: A head-coil with high sensitivity and excellent signal-to-noise ratio is needed to collect the DSI data. Eight- and 12-channel coils provide less signal at the surface of the brain; consequently, these coils may require increased scanning time to support accurate mapping of projection fields.
- Head stabilization: Due to the long duration of the DSI scanning sequence, and because motion correction cannot be applied to the DSI data, excellent head stabilization is necessary to control subject movement. Movement controls ranging from padding and tape to a bite-bar, vacuum bag, or thermoplastic mask are recommended to stabilize subjects’ heads. More than 2 mm of translational movement or 2° of rotational movement in any direction is excessive and may be considered grounds for excluding data.
- FMRI presentation equipment: For analyses using functional seeds, additional equipment for fMRI scanning is required. Depending on the type of regions to be localized, this usually includes a MR compatible display (such as projector system of MR compatible LCD), a button response system, audio system, and experiment presentation computer synchronized with the scanner acquisition.
2. Scanning Procedure
- Brief participants on the nature of the scans to be performed and obtain informed consent. Emphasize the necessity to minimize head motion (especially during the long DSI scan). Offer participants the choice of a movie or other video for entertainment during the DSI scan. For functional scanning of behavioral tasks, instruct subjects to monitor the screen for task-relevant stimuli and to respond as required.
- After screening for MR contraindications, comfortably stabilize participants’ head using one of the methods described above, and slide patient bed into the scanner.
- Perform initial scout scans and calibration.
- Set the slice prescription for the DSI scan parallel to an imaginary line connecting the anterior and posterior commisures. Ensure that the slices for the DSI scan cover the whole brain.
- Run the DSI scan while the subject relaxes in the scanner or watches entertainment on the presentation system.
- Immediately after the completion of the DSI scan, collect a T1-weighted anatomical scan (e.g., MPRAGE) for later use in co-registering (i.e., aligning) the DSI data with other anatomical or functional data.
- Optionally, collect fMRI data in the same session using standard EPI pulse sequences.
- If necessary, conduct fMRI scanning in a separate scanning session. Collect an MPRAGE in both sessions to facilitate co-registration of the datasets.
3. Anatomical MRI Processing
For surface analysis of fMRI data and automatic segmentation using FreeSurfer, as described below, a high-resolution T1-weighted anatomical image with excellent white-gray matter contrast is required. This image provides a common reference space for analyzing functional and diffusion-weighted imaging data. In most modern MRI scanners, this image will be referred to as an MPRAGE (Magnetization Prepared RApid Gradient Echo) image. Most modern MPRAGE sequences can provide sufficient quality data in a single scan (parameters in Table 1). If necessary, two or more scans can be averaged to improve gray-white matter contrast for segmentation. Below, we explain how DWI and fMRI data, which are usually collected with different voxel sizes and different origin points, can be automatically aligned and resampled for simultaneous viewing with the MPRAGE.
Detailed descriptions of the FreeSurfer anatomical MRI processing stream can be found on the FreeSurfer wiki (http://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferWiki); FreeSurfer output includes multiple cortical surface representations, as well as parcellation of cortical anatomical features and segmentation of subcortical structures. We recommend running the AFNI/SUMA script @SUMA_Make_Spec_FS on FreeSurfer output, which converts this output into file formats that can be easily processed using tools from AFNI/SUMA, FSL, SPM, and other neuroimaging software packages. For example, co-registration of images can be performed with one of several programs, such as 3dAllineate (AFNI/SUMA), FLIRT (FSL), bbregister (FreeSurfer), or the SPM Coregister function.
- Perform anatomical segmentation and cortical surface reconstruction by submitting T1-weighted anatomical image to FreeSurfer’s automated algorithm (recon-all).
- Import FreeSurfer processing results into SUMA using @SUMA_Make_Spec_FS script. This step creates NIFTI-format versions of all volumes in the FreeSurfer output, including an intensity-normalized, skull-stripped version of the input anatomical image. We refer to this processed anatomical image as the Surface Volume, according to AFNI/SUMA terminology; the NIFTI version of this image created by @SUMA_Make_Spec_FS is named brain.nii.
- Align the DSI B0 image to the resulting Surface Volume (use the NIFTI-format version of this image, named brain.nii, in the SUMA directory).
- Save the 12-point affine transformation matrix for use in subsequent co-registrations.
4. Functional MRI (fMRI) Processing
Functional MRI analysis can define regions of interest (ROIs) for generation or post-hoc selection of fibers. Any echo-planar imaging (EPI) pulse sequences with parameters optimized for the specific fMRI experiments can be used. Likewise, a large number of software packages for fMRI processing and analysis exist, such as AFNI/SUMA (NIMH, NIH) 8, 9, BrainVoyager (Brain Innovation) 10, FSL (FMRIB, Oxford University) 11, 12, and SPM (Wellcome Trust Center for Neuroimaging, University College London) 13. The “fMRI processing and analysis” section of Figure 4 outlines an analysis pathway based on the AFNI/SUMA software package. For more detailed usage instructions, we refer readers to the excellent tutorials and other educational materials on the AFNI/SUMA website (http://afni.nimh.nih.gov).
The end goal of fMRI analysis for fiber-tracking differs from standard functional localization analyses, in which the emphasis is often to find the locus of maximum activation. Good statistical procedure requires researchers to specify alpha levels for statistical contrasts beforehand; however, researchers should consider the fact that the choice of statistical thresholds will influence the spatial extent of functional activation, and thus the extent of fiber termination fields.
- Correct for subject head motion in each individual’s fMRI data, using the mean image of the first scanner run as the reference image.
- Optionally, perform slice acquisition time correction, particularly if using a rapid event-related task design.
- Correct between-run differences in signal baseline by normalizing the time-series for each voxel, within each run.
- Express each voxel’s time-series as percent change from that voxel’s mean over time for a given run, using a program such as 3dcalc (AFNI/SUMA) or fslmaths (FSL).
- Alternatively, z-transform each voxel’s time-series for each run, based on the signal mean and standard deviation over time.
- Co-register each processed EPI run to the Surface Volume (brain.nii) dataset generated by FreeSurfer (see above).
- Concatenate all EPI runs in time for a given individual.
- Map each EPI run to the surface using 3dVol2Surf (AFNI/SUMA), producing a NIML surface dataset for each run.
- Use the smooth white matter and pial surfaces as the reference surfaces for the mapping; these may be thought of as representing the gray/white-matter boundary and the surface of cortex, respectively.
- Average the signal over the distance between these two surfaces.
- Optionally smooth EPI data on the cortical surface using SurfSmooth (AFNI/SUMA).
- Create separate regressors for epochs of time corresponding to each of the experimental conditions presented to the subject.
- Submit these regressors (along with regressors of no interest) to a general linear model (GLM) analysis of the functional data.
- Contrast beta weights for different regressors to produce a “functional map” of statistical values across the surface
- Optionally enter beta-weight values for multiple participants into a group level analysis of variance (ANOVA), if using a factorial design.
- Derive thresholded functional maps to view statistically significant effects, employing a familywise error rate (Gaussian random field theory) 14 or false discovery rate (FDR) 15, 16 adjustment to correct for multiple comparisons.
- Create regions of interest (ROIs), which will be used later for tractography seeding, from contiguous regions of significant functional activation on the surface by labeling each separable area.
- Automatically segment and label ROIs using a spatial clustering algorithm, such as SurfClust (AFNI/SUMA).
- Alternatively, hand-draw ROIs using SUMA’s Draw ROI feature.
- Expand ROIs into the white matter using 3dSurf2Vol (AFNI/SUMA), to maximize contact with streamlines during tractography.
- As in step 6, use the smooth white matter and pial surfaces for the mapping.
- Set f_p1_fr = -0.5 in order to grow the ROI below the gray/white matter boundary by 50% of the gray matter thickness at each surface node.
- Set f_pn_fr=1 in order to grow the ROI in the opposite direction to the pial surface.
- Use the AFNI program cat_matvec to find the inverse of the 12-point affine transformation matrix generated when aligning the B0 image to the Surface Volume (brain.nii).
- niiApply the inverted matrix to functional ROIs in order to align them with DSI data.
5. Processing of Diffusion Weighted Imaging Data
Diffusion-weighted imaging is a general term for white matter imaging techniques, encompassing many different combinations of data acquisition and reconstruction methods. Perhaps the most frequently used method, referred to as diffusion tensor imaging (DTI) 17, 18, is based on 5-10 minutes of data acquisition, measuring diffusion in 6 or 12 directions. Based on these data, diffusion patterns are typically modeled with a simple tensor model, which is best suited for detecting a single dominant diffusion direction. This limitation means that DTI does not perform well for imaging fibers which cross one another or “kiss” at a single point. Crossing and kissing fibers are better detected with a combination of high-resolution acquisition and reconstruction methods, such as high angular resolution diffusion imaging (HARDI) 19-21, diffusion spectrum imaging (DSI) 1, 2, and generalized q-ball imaging (GQI) 22-24.
A 257-direction multi-shell DSI sequence run on Siemens 3T scanners was used for acquisition of the results presented here (parameters in Table 1). Acquired data were reconstructed with the GQI method24, which models diffusion patterns in each voxel with an orientation distribution function (ODF) that can detect simultaneous diffusion in multiple directions. Other high-angular resolution diffusion sequences should produce similar results. Note that correct reconstruction of ODFs requires the researcher to input a gradient table (also referred to as a b-table) to DSI Studio, the DWI processing and tractography program used here. (Detailed usage instructions for DSI Studio can be found on the software’s website, http://dsi-studio.labsolver.org.) This table lists the gradient direction and magnetic field strength for each of the acquired DWI volumes. The gradient table depends on the MR acquisition protocol and is automatically extracted from DICOM images by DSI Studio. However, we recommend that researchers compare this automatically-extracted gradient table with the standard table for their scanner’s DWI protocol.
- If necessary, convert MR images to .dcm (DICOM) format using mri_convert (FreeSurfer).
- Identify which image(s) in the dataset are B0 images (i.e., echo-planar images collected with no diffusion weighting).
- Convert the B0 image(s) to NIFTI format using AFNI program to3d.
- In DSI Studio, open DICOM images and combine to create a source (.src) file.
- Supply a gradient table (see above).
- Check that the default reconstruction mask encompasses all gray matter, without including empty space, skull, or non-brain tissue. Edit the mask as necessary.
- Alternatively, create a reconstruction mask by running AFNI program 3dAutomask on the B0 image.
- Choose a high-resolution reconstruction model: DSI, GQI, or GQI variant.
- Create a fiber information file (.fib) to represent the principal diffusion direction(s) in each voxel.
6. Evaluating Data Quality and Tracking Parameters through Whole-brain Tractography
Tracking fibers with a whole-brain seed is a fast and effective way to assess overall data quality. It also presents an opportunity to decide appropriate values for global parameters, especially the anisotropy threshold used as a stopping criterion in tractography. This procedure is necessary to strike a balance between improving coverage in the fiber tracking process and reducing noise. Special care should be taken in setting key tracking parameters, such as angle threshold and tracking thresholds.
It is important to note that the relative anisotropy of different tracts may vary between individuals, depending on biological factors such as age and white matter integrity, as well as extraneous factors like hardware calibration between sessions. Below, we suggest multiple methods for equilibrating tracking thresholds between datasets. At all times, check the quality of tracking results by comparing them to known neuroanatomy. For example, fibers which cross the interhemispheric fissure outside of known interhemispheric connections (i.e., corpus callosum, anterior & posterior commissures) may indicate that the tracking threshold is too low and should be raised, or may be evidence of head movement artifacts.
In contrast to tracking threshold, angle threshold should be invariant for a given individual across sessions, since fiber tracts do not change in curvature over the short term, if ever. Likewise, tract curvature should be relatively constant across individuals, in the absence of major differences in brain size or morphology. Nevertheless, care should be taken in setting initial values of this parameter. Fibers which follow improbable trajectories, such as hairpin curves, may indicate that the angle threshold is too high.
- Create a whole-brain seed region.
- Set an initial tracking threshold value to mask out low-signal voxels.
- Set angle threshold to allow fibers to curve up to n degrees in a single step.
- Set tracking step size in mm.
- Set desired number of fibers or seed points.
- Perform whole-brain tractography to check overall ODF reconstruction quality.
- To begin, choose a median (across datasets) tracking threshold.
- Load a whole-brain trk file into TrackVis, a fiber tract visualization and analysis program (Martinos Center for Biomedical Imaging, Massachusetts General Hospital) 25.
- Load the gray matter (GM) volumes in the SUMA directory (lh/rh.ribbon.nii) as ROIs.
- Set the GM ROIs as filters on the track group, accepting only fibers which have either end in one of the ROIs.
- Verify that most of the fibers (90-100%) in the trk file remain in the filtered track group.
- Repeat as necessary, adjusting tracking threshold in DSI Studio each time.
- Further check that the tracking threshold masks out voxels in empty space (i.e., around the edges of the brain and in intra-gyral space) without removing voxels, which clearly lie in white matter.
- Optionally, equilibrate the tracking threshold across datasets (i.e., different sessions and/or participants).
- Set a tracking threshold as desired in one dataset via the DSI Studio interface.
- Rename the .fib file created by DSI Studio with a .mat extension and import into MATLAB, per instructions on the DSI Studio website (http://dsi-studio.labsolver.org).
- Create a histogram of the values that you wish to threshold.
- Convert the map values into z-scores.
- Find the z-score of the tracking threshold which you initially set in the DSI Studio interface.
- Perform steps b-d for all other datasets, finding the tracking threshold that corresponds to the z-score found in step e.
- As a cross-check for steps a-f, track a set of control fibers from an anatomical ROI at the occipital pole with 500,000 seeds.
- Check that this procedure produces approximately the same number of fibers across datasets (+- 100 fibers).
7. Locally-constrained Tractography
In contrast to whole-brain tractography, locally-constrained tractography makes use of ROI-based Boolean operations, such as specifying volumes through which fibers must or may not pass. As a result, locally-constrained tractography offers higher sensitivity and greater control for tracking select fibers of interest. Whole-brain tractography undersamples the space of possible seed points, due to the high computational cost of seeding operations and limited computer graphics memory. (It is possible that these constraints will be alleviated in the future, due to changes in tractography algorithms, increased memory capacity, or other factors.) As a result of undersampling, whole-brain tractography often produces results which are biased toward the dominant diffusion pathways in the brain. User-supplied ROIs address this problem by providing limited target regions with a high density of seed points, making it easier to capture difficult-to-detect fiber tracts.
- Create a whole-brain seed region in DSI Studio.
- Load one or more NIFTI region-of-interest (ROI) files.
- Optionally load a region of avoidance (ROA) file to indicate voxels which fibers should NOT pass through.
- Set anisotropy threshold and angle threshold as described above.
- Perform tracking.
- Check quality by comparing fiber tracks to anatomical detail.
8. Endpoint Density Analysis
- Load NIfTI ROIs and trk files into TrackVis.
- Perform Boolean operations between regions.
- Save the results of each operation as a new trk file.
- Use the track_transform (Diffusion ToolKit) function to spatially transform trk files to the Surface Volume (brain.nii) file.
- Load the transformed trk file and Surface Volume (brain.nii) in TrackVis to inspect.
- Load trk and ROI files into MATLAB to make quantitative connectivity estimates.
- Find the {x/y/z} centroid of an ROI.
- As one measure of connectivity, calculate the total number of fiber endpoints in an ROI, normalized by ROI volume.
- Alternatively, calculate the Euclidean distance between fiber endpoints and ROI centroid, as a measure of the specificity and consistency of the tract’s connectivity to that ROI.
9. Representative Results
High-resolution diffusion-weighted imaging and fiber tractography can be applied to a wide range of neuroscientific questions. Our focus in this paper is to detail the coupling of structural connectivity methods with functional neuroimaging. However, we note that any application of DWI requires careful evaluation of tractography results, given that the data acquisition protocol, reconstruction method, and tractography parameters can exert significant, independent effects on the final product. Figure 5 illustrates optimal and sub-optimal results using whole-brain tractography. All three images are based on the same 257-direction DWI dataset from a single participant; optimal results are shown in the left panel. In contrast, the middle panel shows the effect of excessively lenient tractography parameters (FA and angle thresholds). The right panel shows the reduction in quality that results from using a single-tensor model to reconstruct the DWI data.
We include two examples of how tractography results can corroborate and inform interpretation of functional imaging data. These experiments assess cognitive processes that allowed the creation of functional seed regions: namely, face perception and visual attention. These seed regions can be used to test questions of white-matter connectivity within a cognitive network. Figure 6 shows an example of regions activated during a face perception task. Subjects viewed pictures of faces and everyday objects while undergoing fMRI scanning. Two ventro-temporal regions, in the middle fusiform gyrus (mFG) and inferior occipital gyrus (IOG), showed significantly greater BOLD responses for faces than for objects. These two functionally defined regions were then used as seed regions during tractography (as outlined in Sections 6-7 above). Figure 6A shows the large bundle of fiber streamlines (shown in red) that connect these two regions of interest within the temporal lobe, over a distance of approximately 12 cm. Note the tight packing of the fibers and small degree of fiber curvature over this distance. This pattern is typical of one-to-one connections within functional networks over long distances (for example, see ref. 26). Figure 6B shows the IOG functional seed region (shown in yellow) along with the individual fiber endpoints (red dots). The fiber endpoints are located throughout the ROI. This connectivity pattern suggests that these regions have direct, long-distance connections that may underlie fast communication within the face perception network.
Our second example (Figure 7) shows the connections between visual cortex sensory regions and a region of attentional control in the posterior parietal cortex (PPC). In this case, the two sets of functional activations (occipital and parietal regions) were produced via independent sets of fMRI data from the same individuals. Parietal activations were generated via an attention shifting task between 6 locations in the visual field (for details, see ref. 27), whereas the occipital regions were defined using standard visual field meridian mapping 28, which was used to mark the borders between functional seed regions of visual cortex (V1 – V3). Figure 7A shows the approximate locations of V1, V2, & V3 seed regions (red, green, & blue, respectively), the PPC seed region labeled IPS-1, and the fiber tracts that connect these regions. Tracts are colored by the occipital ROI from which they were seeded. Unlike the long, straight fibers in the temporal lobe (Figure 6), these white-matter tracts cover a shorter distance (range 3 – 5 cm) and are, therefore, more U-shaped and less tightly packed as they travel from the occipital lobe to the parietal lobe. Figure 7B shows the functionally-defined regions in IPS (brown), V1 (red), V2 (green), & V3 (blue) on the cortical surface along with the fiber endpoints in each region. Note the segregation of tracts in the occipital lobe by seed region, contrasted with the large degree of endpoint interdigitation in IPS-1. This suggests that our PPC region (identified through fMRI activity during a selective attention task) may be a convergence area of the brain, with structural connections to many different nodes of sensory cortex. This connectivity pattern may allow for the transmission of attentional biasing signals from higher cortical regions to modulate activity in early cortices; these signals help to enhance target representations in visual cortex29, 30.
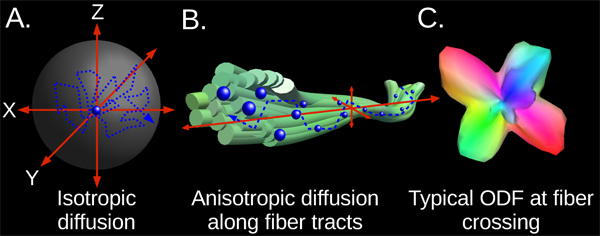
Figure 1. Schematic illustration of key concepts in diffusion-weighted imaging (DWI). Panel A: in a homogenous medium, diffusion occurs randomly as a result of Brownian motion. For large numbers of water molecules, diffusion is isotropic-that is, the aggregate diffusion pattern is spherical. Panel B: diffusion of water molecules within axons and in the interstices of axonal bundles is constrained by axonal walls and other support structures. Thus, diffusion along fiber tracts is anisotropic: it is much greater along the fiber tract’s trajectory than in other directions. Panel C: high-resolution DWI methods use models such as the orientation distribution function (ODF) to model anisotropic diffusion in complex configurations of white matter tracts. As seen in this example, ODFs can distinguish separate diffusion pathways for multiple fiber tracts crossing at a single point. Crossings involving two or three different fiber tracts are common in the brain.
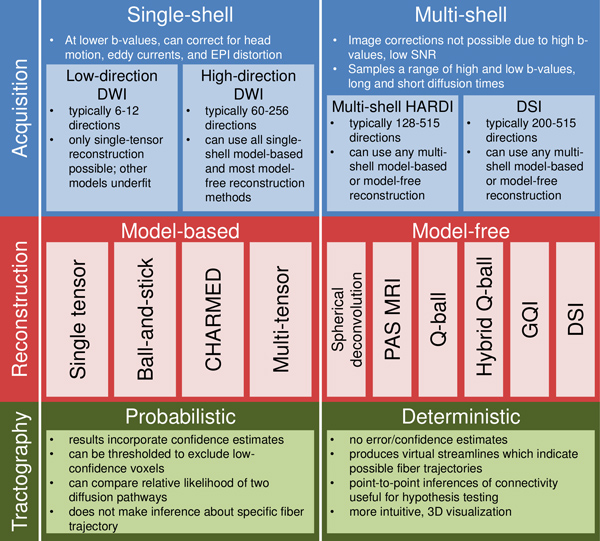
Figure 2. Fiber-tracking research can be performed in a number of ways. The most important choices involve acquisition protocol, reconstruction technique, and tractography method. In the current paper, we use a diffusion spectrum imaging (DSI) 1, 2 protocol for acquisition; generalized Q-sampling imaging (GQI) 24 for reconstruction; and FACT deterministic tractography40, 41 . We especially highlight model-free and hybrid reconstruction techniques, which generate orientation distribution functions (ODFs; see Figure 1) to represent diffusion in each voxel. Researchers may choose different pipelines based on budget, time available, the need for high angular resolution, and the importance of correcting for head motion and non-linear image distortions. This figure is not a comprehensive list of all viable acquisition, reconstruction, and tractography methods. See Seunarine & Alexander42 for an excellent review of reconstruction techniques.
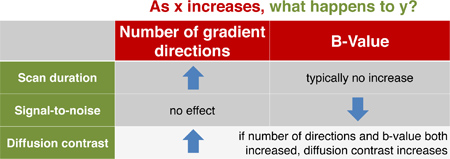
Figure 3. Interactions of diffusion MRI acquisition variables, scan duration, and ability to resolve fiber crossings. High diffusion contrast is necessary for resolving fibers in complex crossing configurations. This contrast depends upon several factors, including the number of gradient directions (i.e., the number of possible fiber orientations) and b-value (which indicates the degree of diffusion weighting). Here we present typical effects of increasing b-values and the number of gradient directions. Note that this table only indicates trends, and individual techniques may have different effects upon scan duration, signal-to-noise ratio (SNR), and diffusion contrast. Generally, contrast can be improved by increasing both the number of gradient directions and the magnitude of b-values. At higher b-values, however, the signal-to-noise ratio of diffusion-weighted images is diminished, and scan time is often increased.
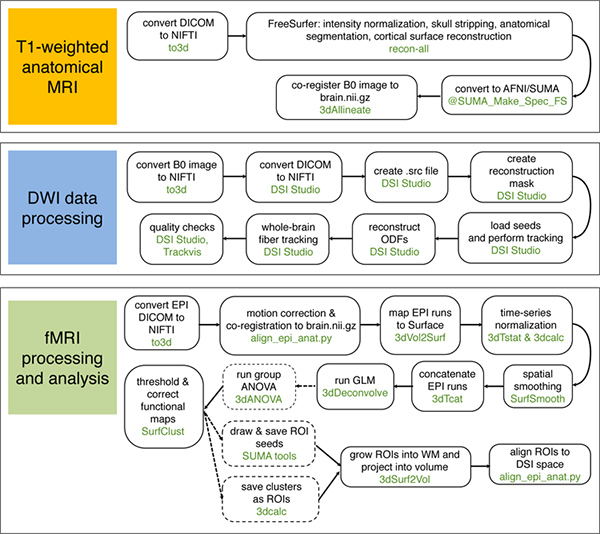
Figure 4. Graphical summary of anatomical MRI, DWI-MRI, and fMRI processing streams. Text in black describes the nature of each processing step, while text in green indicates software which may be used. Dashed lines and boxes indicate optional steps, which may not be applicable to all projects. In this example, processing is performed in the AFNI/SUMA package (except where DSI Studio or TrackVis is indicated). Comparable functions in other neuroimaging analysis packages may often be substituted. Many of the steps illustrated in these diagrams have been partially consolidated by the software developers into convenient scripts: we particularly refer readers to the FreeSurfer recon-all pipeline (http://surfer.nmr.mgh.harvard.edu/fswiki/ReconAllDevTable). We note additionally that several software packages provide complete processing pipelines for DWI data;however, these packages vary in their strengths and weaknesses, and some do not include tools for working with high angular resolution diffusion MRI data. Click here to view larger figure.
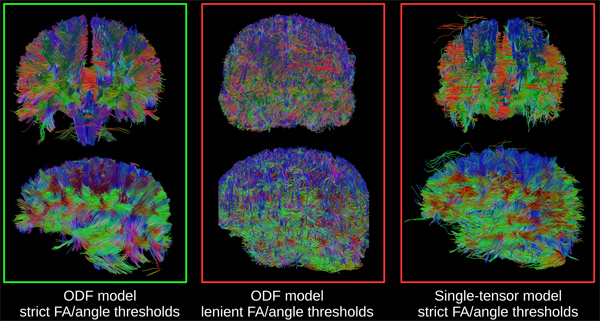
Figure 5. Illustration of whole-brain tractography with different reconstruction methods and tractography parameters. All images were derived from the same dataset, a 257-direction diffusion spectrum imaging (DSI) sequence with multiple b-values (7,000 s/mm2, 5 shells). Panel A: optimal results, achieved by using a high-resolution, ODF-based reconstruction method. A relatively high tracking threshold of 0.06 was selected, in order to generate fibers only from strongly anisotropic voxels; and an angle threshold of 55° was selected to preclude the generation of fibers with biologically unrealistic curvature (i.e., “looping” fibers). Note the clear delineation of the hemispheres, separated by the longitudinal fissure; also note how fiber bundling follows expected sulcal/gyral contours. Panel B: the same reconstruction method was used as in (A), but FA and angle thresholds were set more leniently during tractography (0.03 and 85°, respectively). Inappropriate tracking parameters can cause the generation of large numbers of “junk” fibers, which conceal true information about anatomical structure. See Section 5, “Evaluating Data Quality and Tracking Parameters through Whole-Brain Tractography”, for advice on appropriate parameter choices. Panel C: data were reconstructed using a single tensor model, one of the most widely-used methods in DWI. With appropriate tracking parameters (same as A), the single-tensor model reproduces many known major fiber tracts, and gyral contours are somewhat visible in the sagittal view. However, it also produces more false positives than the ODF model: note fibers traveling horizontally across the interhemispheric fissure. Click here to view larger figure.
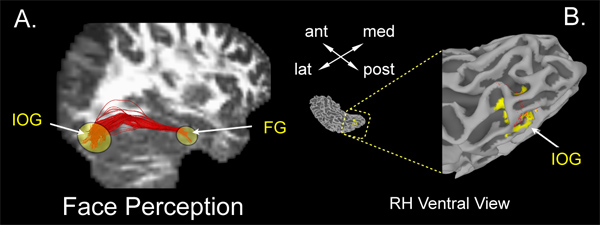
Figure 6. Tractography results from a face perception experiment. Panel (A) shows streamlines resulting from tractography between functional ROIs identified from a face perception experiment. General areas of inferior occipital gyrus (IOG) and mid-fusiform gyrus (mFG) are indicated by yellow ovals. Panel (B) shows the IOG endpoints of the fibers indicated in panel (A) displayed on an enlarged ventral view of the posterior temporal cortical surface. The ROI rendered in yellow resulted from a face perception functional MRI experiment. Note the large agreement between functionally-defined activation and fiber endpoints in IOG. These fibers track from the mFG, a brain region involved face perception. Click here to view larger figure.
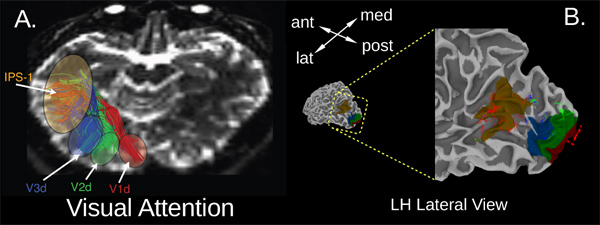
Figure 7. Tractography results from a visual attention experiment. Panel (A) shows the streamlines resulting from tractography between functional ROIs identified from a visual attention experiment27. General areas of posterior parietal cortex (IPS-1) and visual cortex (V1d, V2d, & V3d) are indicated by colored ovals. Fiber tracts are rendered in corresponding colors: red for V1d, green for V2d, and blue for V3d. Panel (B) shows the endpoints of the fibers indicated in panel (A) displayed on an enlarged lateral view of the posterior (parietal and occipital) cortical surface. Color conventions match those of panel (A). Regions of interest resulting from a visual attention functional MRI experiment are displayed on the cortical surface. All three sets of tracts/endpoints converge in the IPS-1 region, which is thought to contain a priority map of visual attention that may be the source of attention biasing signals to targets in visual cortex. Tracts in IPS-1 are largely interdigitated, whereas the occipital ends of these fiber tracts are clearly segregated by region of visual cortex.
| MR Scan | Parameters |
| DSI | 257 direction diffusion spectrum imaging (DSI) scan using a twice-refocused spin-echo EPI sequence and multiple q-values with a 43 min acquisition time (TR = 9,916 ms, TE = 157 ms, voxel size = 2.4 x 2.4 x 2.4 mm, FoV = 231 x 231 mm, b-max = 7,000 s/mm2, 5 shells) |
| Anatomical | T1-weighted MPRAGE sequence (1 mm x 1 mm x 1 mm, 176 sagittal slices, TR = 1,870, TI = 1,100, FA = 8°, GRAPPA = 2) |
| fMRI | T2*-weighted echo-planar imaging (EPI) pulse sequence (31 oblique axial slices, in-plane resolution 2 mm x 2 mm, 3 mm slice thickness, no gap, repetition time [TR] = 2,000 ms, echo time [TE] = 29 ms, flip angle = 90°, GRAPPA = 2, matrix size = 96 x 96, field of view [FOV] = 192 mm) |
Table 1. Neuroimaging acquisition parameters.
Discussion
High-resolution DWI and fiber tractography provide a powerful approach for examining the connective structure of the human brain. Here, we present evidence that this structural architecture is meaningfully related to brain function, assessed by fMRI. By using tractography seeds based on fMRI task activation, we find evidence that brain areas which are co-active during visual attention are anatomically connectedconsistent with prior knowledge of functional neuroanatomy (Figure 7). Similarly, the functional neuroanatomy for face perception is consistent with our present structural connectivity findings (Figure 6). Knowledge of anatomical connectivity is necessary, although not sufficient, for inferring a direct functional connection between brain areas in a given task (and vice versa). In many neuroimaging studies, direct structural and functional connections are inferred-problematically-on the basis of concurrent functional activation alone. Such inferences neglect other interpretations: for example, two brain areas may appear co-active because they share a common input, because of global neuromodulatory influences for which the experimental design does not control, or even because of a common noise source, such as head motion. MR diffusion tractography provides converging evidence for dynamic functional relationships between distal brain areas, by confirming that a possible connective substrate exists between them.
Users should consider several limitations and caveats of fiber-tracking research. The most fundamental of these is that fiber streamlines generated in deterministic tractography represent possible diffusion pathways, and not real fiber bundles. Tractography results can be affected by both false positive and false negative results, and interpretation of tractography should be guided by existing neuroanatomical knowledge. The best prior evidence of white matter connectivity comes from “gold-standard” techniques such as microdissection or tracer labeling. Especially useful are probabilistic maps of fiber tract contours derived from postmortem human brains, such as that created by Buergel, Amunts, and colleagues31; free online resources such as the Digital Anatomist project (http://www9.biostr.washington.edu/da.html) can also provide useful guidance. We note that functional connectivity analysis of EEG, MEG, and BOLD fMRI data provides only weak evidence, if any, for anatomical connectivity between brain areas.
An additional caveat concerns false continuations of fiber streamlines, which can occur when two independent diffusion pathways are roughly aligned end-to-end, and they appear to flow into one another. In such cases, the tractography algorithm may continue beyond the true stopping point. For example, thalamic afferents from the brainstem and thalamic efferents to dorsal parts of cortex may have similar orientations. As a result, tractography algorithms may be duped into producing long fibers which rise from the brainstem, pass through the thalamus, and appear to end in cortex. Such false fibers may result from the conjoining of two anatomically correct pathways, each of which is correct in itself. However, they can also result from anatomically invalid fiber trajectories. Other common false continuations include fibers which appear to pass from the temporal poles into the insula and fibers which cross the longitudinal fissure outside of known interhemispheric crossing points (i.e., the corpus callosum and the commissures). These false continuations often occur because 1) partial volume effects obscure the borders of the lobes/hemispheres; or 2) because the tracking threshold has been set too leniently. As we noted above, researchers must evaluate fiber-tracking results in the light of extant neuroanatomical knowledge. As a final caution, we note that diffusion MRI and fiber tractography provide no information about the directionality of connections: that is, they cannot discern feedforward from feedback fibers, or afferents from efferents.
Deterministic tractography can be useful for hypothesis testing, as the fiber streamlines it generates constitute inferences about point-to-point connectivity along specific trajectories, which can be compared against hypotheses. However, users may also wish to consider probabilistic tracking methods (see Figure 2). The major advantage of probabilistic methods is that they yield confidence estimates for diffusion pathways between two points, based on the accumulation of diffusion probabilities in voxels which connect those points32. In contrast, the results of deterministic tractography do not take into account the uncertainty which accumulates at each step of a virtual fiber’s propagation; this uncertainty increases as step size, a user-set parameter, is increased. The confidence estimates produced by probabilistic tractography can be especially useful when trying to determine the relative likelihood of two or more different diffusion pathways; moreover, users can easily mask out voxels with low confidence estimates, a possibility not afforded with deterministic methods. Like deterministic tractography, however, probabilistic methods do not conclusively demonstrate the existence of white-matter fibers; rather, they demonstrate possible diffusion pathways.
Users may find deterministic tractography results more intuitive for visualization, as the results are typically presented as 3-dimensional fiber streamlines, which allow the viewer to quickly apprehend possible fiber trajectories. In contrast, probabilistic tractography results are typically represented as 2-dimensional slices of volumetric data. These images usually show heat maps of contiguous voxels, corresponding to diffusion probability within a thresholded tract volume, without modeling possible fiber trajectories within the tract. Regardless of users’ choice of tractography and visualization methods, they should recognize that fiber-tracking results only demonstrate possible diffusion pathways, and that the results of both methods may include Types I and II statistical error.
Our research group has used the techniques described here to visualize and quantify connections of the corticospinal33 tract, corpus callosum34, and visual attention system27, as well as to map cortical projection circuits in the basal ganglia35. In some cases tractography results may generate novel findings: for example, Wang et al. (submitted) used high-resolution DWI to detail previously undescribed fiber tracts, with cross-validation in cadaver dissection36. Results such as these may provide the impetus for investigations of brain function, in order to assess the functional utility of the newly discovered tracts. Finally, non-invasive, high-resolution DWI holds promise in a number of clinical situations, such as neurosurgical planning37; surgery for tumors, bleeds and cavernovas38; and traumatic brain injury (TBI)39. Our group has applied these techniques in a large number of neurosurgery and traumatic brain injury cases, relating disrupted fiber connectivity to behavioral symptoms.
Ultimately, easy-to-obtain information regarding global brain connectivity will allow researchers to construct better models of the brain. For example, graded measurements of white-matter connectivity could be used to improve source localization in MEG/EEG, or to place constraints on fMRI-based effective connectivity analyses. High-resolution tractography is also likely to improve models of disrupted or pathological brain connection, such as may occur in TBI or autism. Finally, high-resolution tractography may enable researchers to better integrate knowledge of human functional neuroanatomy with invasive studies of non-human brains. We hope and anticipate that a growing number of researchers will explore the potential of combining assessments of brain function with high-resolution diffusion-weighted imaging.
Disclosures
The authors have nothing to disclose.
Acknowledgements
List acknowledgements and funding sources. The work is supported by NIH RO1-MH54246 (M. B.), National Science Foundation BCS0923763 (M.B.), the Defense Advanced Research Projects Agency (DARPA) under contract NBCHZ090439 (W. S.), the Office of Naval Research (ONR) under award N00014-11-1-0399 (W. S.), and the Army Research Lab (ARL) under contract W911NF-10-2-0022 (W. S.). The views, opinions, and/or findings contained in this presentation are those of the authors and should not be interpreted as representing the official views or policies, either expressed or implied, of the above agencies or the United States Department of Defense.
References
- Wedeen, V. a. n. J., Hagmann, P., Tseng, W. I., Reese, T. G., Weisskoff, R. M. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magnetic Resonance in Medicine. 54 (6), 1377-1386 (2005).
- Wedeen, V. J., Wang, R. P., Schmahmann, J. D., Benner, T., Tseng, W. Y. I., Dai, G., Pandya, D. N., et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. NeuroImage. 41 (4), 1267-1277 (2008).
- Pipe, J. Pulse Sequences for Diffusion-weighted MRI. Diffusion MRI: From quantitative measurement to in-vivo neuroanatomy. , 12-35 (2009).
- Le Bihan, D., Poupon, C., Amadon, A., Lethimonnier, F. Artifacts and pitfalls in diffusion MRI. Journal of Magnetic Resonance Imaging: JMRI. 24 (3), 478-488 (2006).
- Tuch, D. S. Q-ball imaging. Magnetic Resonance in Medicine. 52 (6), 1358-1372 (2004).
- Sakaie, K. E., Lowe, M. J. An objective method for regularization of fiber orientation distributions derived from diffusion-weighted MRI. NeuroImage. 34 (1), 169-176 (2007).
- Reese, T. G., Benner, T., Wang, R., Feinberg, D. A., Wedeen, V. J. Halving imaging time of whole brain diffusion spectrum imaging and diffusion tractography using simultaneous image refocusing in EPI. Journal of Magnetic Resonance Imaging. 29 (3), 517-522 (2009).
- Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research. 29 (3), 162-173 (1996).
- Cox, R. W., Hyde, J. S. Software tools for analysis and visualization of fMRI data. NMR in Biomedicine. 10 (4-5), 171-178 (1997).
- Goebel, R. BRAINVOYAGER: a program for analyzing and visualizing functional and structural magnetic resonance data sets. NeuroImage. 3, S604 (1996).
- Smith, S. M., Jenkinson, M., Woolrich, M. W., Beckmann, C. F., Behrens, T. E. J., Johansen-Berg, H., Bannister, P. R., et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 23, S208-S219 (2004).
- Woolrich, M. W., Jbabdi, S., Patenaude, B., Chappell, M., Makni, S., Behrens, T., Beckmann, C., et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 45, S173-S186 (2009).
- Friston, K. J. . Statistical parametric mapping: the analysis of functional brain images. , (2007).
- Nichols, T., Hayasaka, S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Statistical Methods in Medical Research. 12 (5), 419-446 (2003).
- Benjamini, Y., Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological. 57 (1), 289-300 (1995).
- Logan, B. R., Rowe, D. B. An evaluation of thresholding techniques in fMRI analysis. NeuroImage. 22, 95-108 (2004).
- Basser, P. J., Mattiello, J., LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. Journal of Magnetic Resonance, Series B. 103 (3), 247-254 (1994).
- Basser, P. J., Mattiello, J., LeBihan, D. MR diffusion tensor spectroscopy and imaging. Biophysical Journal. 66 (1), 259-267 (1994).
- Frank, L. R. Anisotropy in high angular resolution diffusion-weighted MRI. Magnetic Resonance in Medicine. 45 (6), 935-939 (2001).
- Frank, L. R. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magnetic Resonance in Medicine. 47 (6), 1083-1099 (2002).
- Tuch, D. S., Reese, T. G., Wiegell, M. R., Makris, N., Belliveau, J. W., Wedeen, V. J. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magnetic Resonance in Medicine. 48 (4), 577-582 (2002).
- Descoteaux, M., Angelino, E., Fitzgibbons, S., Deriche, R. Regularized, fast, and robust analytical Q-ball imaging. Magnetic Resonance in Medicine. 58 (3), 497-510 (2007).
- Tuch, D. S. Q-ball imaging. Magnetic Resonance in Medicine. 52 (6), 1358-1372 (2004).
- Yeh, F. C., Wedeen, V. J., Tseng, W. -. Y. I. Generalized Q-sampling imaging. IEEE Transactions on Medical Imaging. 29 (9), 1626-1635 (2010).
- Wang, R., Benner, T., Sorensen, A. G., Wedeen, V. J. Diffusion Toolkit: a software package for diffusion imaging data processing and tractography. Proc. Intl. Soc. Mag. Reson. Med. , 3720 (2007).
- Sundaram, S. K., Kumar, A., Makki, M. I., Behen, M. E., Chugani, H. T., Chugani, D. C. Diffusion tensor imaging of frontal lobe in autism spectrum disorder. Cereb Cortex. 18 (11), 2659-2665 (2008).
- Greenberg, A. S., Verstynen, T., Chiu, Y. -. C., Yantis, S., Schneider, W., Behrmann, M. Visuotopic Cortical Connectivity Underlying Attention Revealed with White-Matter Tractography. The Journal of Neuroscience. 32 (8), 2773-2782 (2012).
- Slotnick, S. D., Yantis, S. Efficient acquisition of human retinotopic maps. Human Brain Mapping. 18 (1), 22-29 (2003).
- Greenberg, A. S., Esterman, M., Wilson, D., Serences, J. T., Yantis, S. Control of spatial and feature-based attention in frontoparietal cortex. The Journal of Neuroscience. 30 (43), 14330-14339 (2010).
- Kastner, S., Ungerleider, L. G. Mechanisms of visual attention in the human cortex. Annual Review of Neuroscience. 23, 315-341 (2000).
- Bürgel, U., Amunts, K., Hoemke, L., Mohlberg, H., Gilsbach, J. M., Zilles, K. White matter fiber tracts of the human brain: Three-dimensional mapping at microscopic resolution, topography and intersubject variability. NeuroImage. 29 (4), 1092-1105 (2006).
- Behrens, T. E. J., Jbabdi, S. . MR Diffusion Tractography. Diffusion MRI: From quantitative measurement to in-vivo neuroanatomy. , 333-352 (2009).
- Verstynen, T., Jarbo, K., Pathak, S., Schneider, W. In vivo mapping of microstructural somatotopies in the human corticospinal pathways. Journal of Neurophysiology. 105 (1), 336-346 (2011).
- Jarbo, K., Verstynen, T., Schneider, W. In vivo quantification of global connectivity in the human corpus callosum. NeuroImage. , (2012).
- Verstynen, T., Badre, D., Jarbo, K., Schneider, W. Microstructural organizational patterns in the human corticostriatal system. , (2012).
- Wang, Y., Fernández-Miranda, J. C., Verstynen, T., Pathak, S., Schneider, W. Identifying human brain tracts with tractography and fiber microdissection: mapping connectivity of the middle longitudinal fascicle as the dorsal auditory pathway. , (2012).
- Fernandez-Miranda, J. C., Engh, J. A., Pathak, S. K., Madhok, R., Boada, F. E., Schneider, W., Kassam, A. B. High-definition fiber tracking guidance for intraparenchymal endoscopic port surgery. Journal of Neurosurgery. 113 (5), 990-999 (2010).
- Fernandez-Miranda, J. C., Engh, J., Pathak, S., Wang, Y., Jarbo, K., Verstynen, T., Boada, F., Schneider, W., Friedlander, R. High-definition fiber tractography of the human brain: neuroanatomical validation and neurosurgical applications. , (2012).
- Shin, S., Verstynen, T., Pathak, S., Jarbo, K., Hricik, A., Maserati, M., Beers, S., Puccio, A. M., Okonkwo, D., Schneider, W. High definition fiber tracking for assessment of neurologic deficit in a case of traumatic brain injury. Journal of Neurosurgery. , (2012).
- Mori, S., Crain, B. J., Chacko, V. P., Van Zijl, P. C. M. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 45 (2), 265-269 (1999).
- Tournier, J., Mori, S., Leemans, A. Diffusion tensor imaging and beyond. Magnetic Resonance in Medicine. 65 (6), 1532-1556 (2011).
- Seunarine, K. K., Alexander, D. C. . Multiple Fibers: Beyond the Diffusion Tensor. Diffusion MRI: From quantitative measurement to in-vivo neuroanatomy. , (2009).

