Expression of Fluorescent Proteins in Branchiostoma lanceolatum by mRNA Injection into Unfertilized Oocytes
Summary
We report here the robust and efficient expression of fluorescent proteins after mRNA injection into unfertilized oocytes of Branchiostoma lanceolatum. The development of the microinjection technique in this basal chordate will pave the way for far-reaching technical innovations in this emerging model system, including in vivo imaging and gene-specific manipulations.
Abstract
We report here a robust and efficient protocol for the expression of fluorescent proteins after mRNA injection into unfertilized oocytes of the cephalochordate amphioxus, Branchiostoma lanceolatum. We use constructs for membrane and nuclear targeted mCherry and eGFP that have been modified to accommodate amphioxus codon usage and Kozak consensus sequences. We describe the type of injection needles to be used, the immobilization protocol for the unfertilized oocytes, and the overall injection set-up. This technique generates fluorescently labeled embryos, in which the dynamics of cell behaviors during early development can be analyzed using the latest in vivo imaging strategies. The development of a microinjection technique in this amphioxus species will allow live imaging analyses of cell behaviors in the embryo as well as gene-specific manipulations, including gene overexpression and knockdown. Altogether, this protocol will further consolidate the basal chordate amphioxus as an animal model for addressing questions related to the mechanisms of embryonic development and, more importantly, to their evolution.
Introduction
During development, a single cell gives rise to an entire organism in a highly complex process that involves both cell divisions and movements. To better understand the biological principles underlying the dynamics of cell behavior, developmental biologists have started to use fluorescence-based in vivo imaging techniques. Specific compartments of cells, such as cell membranes, can either be labeled by treatments with fluorescent dyes, an approach hampered by a lack of specificity and of tissue penetration1, or by the specific introduction into the embryo of exogenous mRNAs encoding fluorescent proteins2. Different techniques can be used for the efficient delivery of exogenous compounds, such as mRNAs. These include, but are not limited to, microinjection, electroporation, bombardment with microparticles, lipofection and transduction3,4. Although all of these approaches can be used to introduce exogenous compounds into a developing embryo, only microinjection allows the application of predefined and precise quantities into each cell3. Microinjection techniques have been described for all major developmental model systems4 (e.g., fruit flies, nematode worms, zebrafish, frogs, mice) as well as for some alternative models4, including those used for comparative studies aimed at understanding the evolution of developmental mechanisms (e.g., sea anemones, annelid worms, sea urchins, ascidian tunicates, the cephalochordate amphioxus).
Cephalochordates, which together with tunicates and vertebrates establish the chordate phylum, are particularly well-suited models to study the evolution of chordates and the diversification of vertebrates from an invertebrate ancestor5-8. The cephalochordate lineage diverged very early during chordate evolution; and extant cephalochordates, which are subdivided into three genera (Branchiostoma, Asymmetron and Epigonichthys), resemble vertebrates both in terms of overall anatomy and genome architecture5-8. Of the about 30 species of cephalochordates that have been described so far, five are available for embryological and developmental studies6,9: Asymmetron lucayanum (the Bahama lancelet), Branchiostoma floridae (the Florida amphioxus), Branchiostoma lanceolatum (the European amphioxus), Branchiostoma belcheri (the Chinese amphioxus) and Branchiostoma japonicum (the Japanese amphioxus). Ripe adults of three of these species (B. lanceolatum, B. belcheri and B. japonicum) can be induced to spawn on-demand during the breeding season10,11. In addition, at least for B. lanceolatum, efficient spawning can also be induced in artificial sea water12, thereby making this particular cephalochordate species accessible for laboratories that do not have access to natural seawater. The combination, in B. lanceolatum, of a convenient and reliable access to embryos with an efficient delivery method, such as microinjection, so far the only delivery technique developed in amphioxus (in both B. floridae and B. belcheri)13-15, will enable the development of a novel suite of manipulative techniques, including lineage tracing- and dynamic cell behavior-based approaches.
A protocol for the efficient microinjection of mRNAs to express fluorescent proteins in the B. lanceolatum embryo was hence developed. Furthermore, to provide a basic toolkit for live imaging of B. lanceolatum embryos, vector systems were developed that allow membrane-associated and nuclear expression of fluorescent proteins. For membrane targeting, enhanced green fluorescent protein (eGFP) was fused to the human HRAS CAAX box and nuclear localization of mCherry and eGFP was obtained by fusion to the zebrafish histone 2B (H2B) exon (Figure 1, Supplementary File 1). Furthermore, with the goal to optimize protein translation, the Kozak sequences and codons of the constructs have been modified and adapted to usage in B. lanceolatum. Taken together, the injection method and expression vectors presented here will serve as a basis for the generation of new experimental approaches for cephalochordates, notably analyses using the latest fluorescence-based in vivo imaging techniques.
Protocol
1. Preparation of Instruments and Reagents
- Transfer Pasteur pipettes
- Generate a series of transfer Pasteur pipettes with different tip diameters by pulling 230 mm long Pasteur pipettes above a flame at different speeds. Ensure that the taper is as long as possible for smooth and fine control of aspiration.
- With a diamond scribe, scratch the pipette along a line perpendicular to the length of the pipette. With both hands, pull the pipette parallel to its length to generate a blunt cut. Swiftly flame-polish the pipette without sealing the tip.
- Vary the diameter of the tip with the stage of the oocytes/embryos to be pipetted: 300-400 µm for unfertilized oocytes (around 150 µm in diameter), 600 µm for fertilized eggs (around 500 µm in diameter), 200-400 µm for hatched neurulae. Use pipettes with a mouth-monitored aspiration tube to transfer oocytes/embryos from one dish to another.
- Injection needles
- Manipulate the capillaries with gloves to ensure RNase-free conditions. Use borosilicate glass with filament capillaries with dimensions of OD 1.20 mm, ID 0.94 mm, length 10 mm.
- If capillaries are pulled on the type of heating-filament needle puller described in the Materials List, use the following settings: Heat 600, Pull 50, Velocity 80, Time 60, Pressure 200 or 300. Otherwise, ensure that the shape, which is crucial for successful injections, is as shown in Figure 2 with the following properties: 4-8 µm outer tip diameter, 2 cm taper length.
NOTE: Needles can be pulled before the spawning season and used throughout the season.
- 5% Phenol Red stock solution (4x)
- Prepare fresh before each spawning season.
- In a 0.22 µm-filtration tube, weigh out 25 mg of Phenol Red powder.
- Add 0.5 ml of DNase- and RNase-free water to the tube containing the powder.
- Spin for 3-5 min at 18,000 x g at room temperature to filter-sterilize the solution and remove crystals that could clog the injection needle.
- Store at 4 °C or store 250 µl aliquots at -20 °C.
- 0.25 mg/ml poly-lysine solution
- Prepare fresh before each spawning season.
- Dissolve 5 mg of poly-L-lysine in 20 ml distilled water. Store 5 ml aliquots at -20 °C.
- Use a defrosted aliquot immediately and only once to ensure reproducible and robust adhesion of the oocytes to the poly-lysine-coated dish.
- mRNA synthesis
- Prepare fresh before each spawning season.
- Linearize 5 µg of DNA of interest with the adequate enzyme (usually for 2 hr, at 37 °C). To check the completeness of the digestion, run 2% in volume of the digestion mix on a 1% agarose-TBE gel in TBE buffer at 150 W for 20 min.
- Extract the linearized DNA with 25:24:1 phenol (pH 8.0):chloroform:isoamyl alcohol. Vortex for 20 sec, centrifuge 10 min at 18,000 x g and collect the aqueous (upper) phase.
- Extract the aqueous phase again with 24:1 chloroform:isoamyl alcohol. Vortex for 20 sec, centrifuge 10 min at 18,000 x g and collect the aqueous phase.
- Precipitate the linearized DNA with 100:10:300 linearized DNA:3 M sodium acetate (pH 5.2):100% ethanol overnight at -20 °C.
- Centrifuge 20 min at 18,000 x g at 4 °C. Rinse in 70% ethanol.
- Centrifuge 10 min at 18,000 x g at 4 °C. Let dry and resuspend in RNase-free water at a final concentration of 0.5 µg/µl.
- Transcribe 1 µg of linearized DNA using an mRNA synthesis kit with the appropriate polymerase, according to manufacturer’s instructions.
- Extract the mRNA with 5:1 phenol (pH 4.7):chloroform and ammonium acetate stop solution provided with the kit.
- Vortex for 20 sec, centrifuge 10 min at 18,000 x g and collect the aqueous phase.
- Extract the aqueous phase again with 24:1 chloroform:isoamyl alcohol. Vortex for 20 sec, centrifuge 10 min at 18,000 x g and collect the aqueous phase.
- Precipitate the mRNA with 100% isopropanol overnight at -20 °C.
- Centrifuge 20 min at 18,000 x g at 4 °C. Rinse in 80% ethanol.
- Centrifuge 10 min at 18,000 x g at 4 °C. Let the pellet dry at room temperature for no longer than 5-10 min as it will then be difficult to resuspend. Resuspend in DNase- and RNase-free water to a final concentration of at least 2 µg/µl to ensure a decent final mRNA concentration in the injection mix.
- To check the quality and size of the transcription product, run 0.5 µl of the mRNA on a RNase-free 1% agarose-TBE gel in TBE buffer at 150 W for 20 min. Store 2 µl aliquots at -80 °C.
- Poly-lysine-coated dishes
NOTE: Poly-lysine coated dishes are used to immobilize the oocytes during injection.- For each 35 mm cell-culture Petri dish (5 in total), cover the bottom of the Petri dish with 1 ml of the thawed 0.25 mg/ml poly-lysine solution. Incubate at room temperature for 5 min.
- For each 35 mm cell-culture Petri dish (5 in total), transfer the 0.25 mg/ml poly-lysine solution into another 35 mm cell-culture Petri dish. Incubate at room temperature for 5 min.
- Discard the 0.25 mg/ml poly-lysine solution.
- Let the Petri dishes dry, upside-down at room temperature for 2 hr.
- Store the poly-lysine-coated dishes wrapped in a plastic wrap at 4 °C to avoid contamination for one week maximum.
- Agarose-coated dishes
NOTE: They are used to culture injected embryos. The agarose provides a cushion for the injected embryos and prevents them from sticking to the bottom of the dish.- Make artificial seawater (ASW) using 37-38 g/L commercial salts + 0.25 mM NaHCO3 in reverse osmosis water.
- Dissolve agarose to a 1% concentration in 0.22 µm-filtered ASW by heating the solution in a microwave.
- Swiftly pour the warm agarose solution from one 35 mm Petri dish into another one in order to ensure a very thin agarose coating of the dish.
- Store the agarose-coated dishes wrapped in Saran wrap at 4 °C to avoid contamination for one week maximum.
- Injection mix and loading of the injection needles
- About 2 hr before starting the injections, make a 2 µl injection mix in RNase- and DNase-free water with final concentrations of 1-1.8 µg/µl of mRNA, 15% glycerol, 1.25% Phenol Red.
NOTE: The Phenol Red colors the solution, which allows monitoring of the injection efficiency and the identification of successfully injected embryos. Glycerol favors mRNA diffusion within the oocyte. - Centrifuge 4 min at 18,000 x g to pellet crystals. Keep on ice until use.
- With a 10 µl pipette, collect 0.5 µl of the injection mix, avoiding the bottom of the tube, where the crystals have been pelleted.
- Backfill at least two injection needles (in case one breaks during the injection) by pipetting the 0.5 µl drop of injection mix at the large opening of the needle.
- Install the needles in a storage jar with liquid at the bottom at 4 °C to prevent evaporation of the injection mix. Let the injection mix slowly travel to the tip of the injection needle for at least 1 hr to minimize the creation of bubbles.
- Store additional injection mix at -80 °C for maximum of three additional uses, after which the mRNA quality deteriorates (data not shown).
- About 2 hr before starting the injections, make a 2 µl injection mix in RNase- and DNase-free water with final concentrations of 1-1.8 µg/µl of mRNA, 15% glycerol, 1.25% Phenol Red.
2. Collection of Biological Material, Microinjection and Embryo Culture
- Oocyte and sperm collection
NOTE: See Theodosiou et al.12 for a detailed protocol for inducing spawning and for gamete collection.- Shock males and females in ASW at 23 °C for 24 hr.
- One to two hours before sunset, transfer the adults into individual cups in ASW at 19 °C because most adults will spawn 1-2 hr after sunset.
- Rinse 35 mm Petri dishes in filtered ASW and let them dry upside-down to prevent the oocytes from sticking to the bottom of the dish.
- Upon spawning, immediately collect sperm and oocytes with a 1,000 µl pipette.
- Keep sperm and oocytes separate from adult amphioxus because the contact is detrimental for gamete health (data not shown).
- Furthermore, avoid startling the adults, which leads to movements that dissipate, and hence dilute, both sperm and oocytes. To keep the sperm active as long as possible and to optimize fertilization rate, collect the sperm as concentrated as possible.
- Keep sperm on ice in a 1.5 ml tube.
- Transfer oocytes in filtered ASW into the pre-rinsed 35 mm Petri dishes.
- Transfer 100-500 oocytes with a previously pulled 300-400 µm transfer Pasteur pipette to another 35 mm Petri dish to perform the injections.
- Fertilize the remainder of the clutch as a control for sperm and oocyte quality or for other experiments.
- Oocyte injection
- Install the injection needle on the micromanipulator at a 50° angle relative to the horizontal plane.
NOTE: Angles of less than 50° will push the oocytes around on the dish, while angles of more than 50° will not allow an appropriate monitoring of the needle position relative to the oocyte. - Under a fluorescent dissecting scope with 25X oculars, transfer 30 oocytes with the 300-400 µm transfer Pasteur pipette on a poly-lysine-coated dish containing filtered ASW.
- Deposit the oocytes along a line to carry out injections in an ordered way and to distinguish injected from non-injected oocytes. Inject small numbers (30 oocytes) to minimize the exposure time of oocytes to poly-lysine, which tends to deform developing embryos (data not shown).
- Use the dark field illumination to render the oocytes as translucent as possible.
- With the coarse movement knob of the micromanipulator, bring the injection needle close to an oocyte.
- With fine forceps, cut open the needle at the level where the tip starts to be curved. By pulsing with the injector, verify that red injection mix is actually flowing out of the needle.
- At 200X magnification and with the fine movement knob of the micromanipulator, gently move the injection needle inside the core of the oocyte.
NOTE: If inserted too superficially, the injected solution will not remain inside the oocyte. If inserted too far, the oocyte will be destroyed. - Inject with 1-3 pulses of 120 msec duration and 1-10 psi pressure. If the needle is fine enough, inject with continuous flow at constant pressure. Ensure that the injection volume corresponds to 1/5 to 1/3 of the volume of a single oocyte.
- Following injection, pull the needle out swiftly to avoid leakage of the oocyte.
- Verify that the injected solution remains within the oocyte and that after a few sec, the injected solution spreads throughout the oocyte.
- Move on to the next oocyte in line.
- Keep some uninjected embryos of each series as negative control to estimate the background fluorescence when scanning for injected embryos.
- Install the injection needle on the micromanipulator at a 50° angle relative to the horizontal plane.
- Fertilization, selection of injected embryos and embryo culture
- Fertilize the oocytes as soon as a series has been injected. As oocyte quality declines with time, inject and fertilize oocytes within 1 hr after spawning12.
- Depending on the sperm concentration, add 1-5 drops of sperm to the oocytes and swirl the dish.
NOTE: The fertilization envelope should become apparent on the embryos after about 1 min. - Allow the embryos to detach from the poly-lysine-coated dish, while injecting another series of oocytes.
- Transfer the embryos with the 600 µm transfer Pasteur pipette into an agarose-coated Petri dish. Remove the embryos from the poly-lysine-coated dish as soon as possible, if at all possible before the 2-cell stage.
NOTE: In case of prolonged exposure to poly-lysine, the embryos tend to become densely-packed blastulae, flattened on the side touching the bottom of the dish. - At the 2-cell to 4-cell stage, select with a fluorescent dissecting scope with DSR filter the successfully-injected embryos, i.e., those with a normal morphology that exhibit a Phenol Red-derived red fluorescent signal.
- Keep the embryos in culture in filtered ASW in agarose-coated Petri dishes at 19 °C until the desired stage for in vivo imaging.
Representative Results
The protocol detailed above provides the basis for the microinjection of B. lanceolatum oocytes and hence for the introduction into developing B. lanceolatum embryos of mRNA encoding fluorescent proteins for in vivo imaging. Although the technique is certainly robust and reliable, the rate of successful injections using this protocol remains variable (Table 1). The very likely explanation for this intriguing fact is the extreme variability of oocyte clutches: different egg batches do indeed behave very differently, when subjected to the injection pressure. Some oocytes are rather elastic and tend to flatten at the bottom of the dish, while others are quite stiff and remain round upon injection. Furthermore, some oocyte batches tend to inflate their chorion membranes upon injection, while others simply lyse (data not shown). No obvious correlation could be established between oocyte behavior upon injection and embryonic development after fertilization. It is thus difficult to anticipate which category of oocytes is best suited for microinjection. However after fertilization, embryos undergoing normal development can be identified as early as cleavage stages: 2-cell to 4-cell stage embryos can be considered normal when the contact surface between blastomeres is small, while a compacted cleavage-stage embryo, where individual cells are difficult to discern, is definitely abnormal.
In our hands, about half of the oocytes do not survive the trauma caused by the injection and about half of the embryos that do survive the injection exhibit a specific fluorescent label. Thus, we estimate that with this injection protocol, between 50 and 120 embryos can be successfully injected on a given spawning day yielding between 15 and 60 labeled embryos.
The red fluorescence of the Phenol Red in 2-cell to 4-cell stage embryos very reliably marks embryos that have been properly injected, as 100% of embryos selected in this manner subsequently produce the fluorescent proteins encoded by the injected mRNA. Additionally, there is a very clear positive correlation between the intensity of Phenol Red fluorescence at the 2-cell to 4-cell stage and the time of onset of the fluorescence produced by the proteins encoded by the injected mRNA. This correlation thus allows the identification of those embryos that have received the highest quantity of mRNA during the injection process.
If the signal of the fluorescent protein encoded by the injected mRNA is not detected after selection of Phenol Red-positive embryos, we recommend to run the injection mix on an RNAse-free gel in order to check for mRNA degradation or trapping (Figure 3). In lane 1, an intact mRNA band is detected. Injection of this mix led to strong fluorescent signal in the embryo. On the contrary, in lane 2 which corresponds to another experiment where Phenol Red was replaced with Dextran dye in the mix (see discussion for further details), the mRNA seems to have been trapped by the Dextran dye and injection of this mix never led to fluorescent signal in the embryo.
Using this microinjection technique and our constructs (Tables 2 and 3, Supplementary File 1) (see discussion), we can thus reproducibly produce homogenous fluorescent labeling throughout the embryo with mCherry or eGFP in the nucleus and with eGFP at the membrane (Figure 4). The imaging protocol used to generate Figure 4 will be described elsewhere (Faure et al., currently in preparation). Depending on the amount of mRNA injected, fluorescent protein expression typically becomes detectable between 16-cell (Figure 4A) and 64-cell stages and stays detectable at least up to the late neurula stage (data not shown). Later stages have not been tested. Nuclear signal typically appears earlier than membrane signal, potentially because of the intrinsically diffuse nature of the membrane compared to the compact nature of the nucleus (data not shown). Although it has not been monitored systematically, embryos injected with both a nuclear and a membrane label appear to be less healthy than embryos injected exclusively with a nuclear eGFP label. Intriguingly, this effect seems to be independent of the total amount of mRNA injected as the total amount of injected mRNA is the same, whether one or two mRNA species are included to the mix (data not shown).
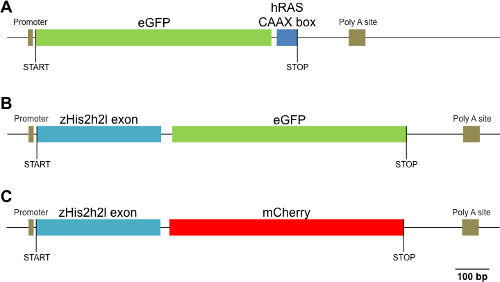
Figure 1: Construct details. Sketches of (A) the membrane-targeted eGFP (eGFP:CAAX box), (B) the nuclear-targeted eGFP (H2B:eGFP) and (C) the nuclear-targeted mCherry (H2B:mCherry) constructs are shown.
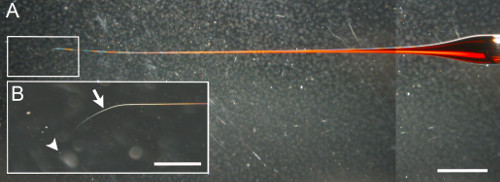
Figure 2: Shape of a representative injection needle. (A) The taper is about 2 cm in length and the needle curves at the tip. Scale bar = 2 mm. (B) Magnification of the boxed area in A. The position for clipping the needle (arrow) is in the curve of needle. The tip of the needle is indicated by the arrowhead. Scale bar = 500 µm. Please click here to view a larger version of this figure.
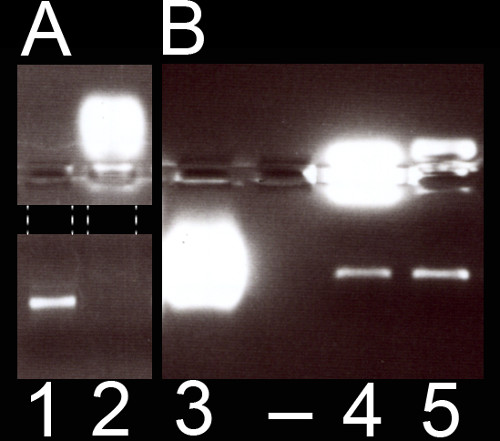
Figure 3: Interactions of mRNA and selected fluorescent dyes. (A) Result of the migration of a mix containing mRNA and Phenol Red in lane 1 and of mRNA and Texas Red dextran (at a final concentration of 3.3 mg/ml) in lane 2. (B) Result of the migration of a mix containing mRNA and Oregon Green dextran (at a final concentration of 3.3 mg/ml) in lane 3, of mRNA and FITC dextran (at a final concentration of 3.3 mg/ml) in lane 4 and of mRNA and Rhodamine dextran (at a final concentration of 3.3 mg/ml) in lane 5. Gel migration time in A and B was different. For the sake of clarity, the black rectangle in A masks a light reflection and the dashed lines delineate the migration lanes. Please click here to view a larger version of this figure.
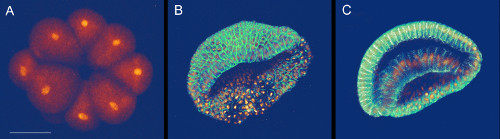
Figure 4: 3D rendering (Amira software) of amphioxus embryos injected with mRNAs coding for fluorescent proteins. Images are extracted from 3D time-lapses acquired on an optimized Leica SP5 2-photon laser-scanning microscope. (A) 16-cell stage embryo expressing nuclear H2B:eGFP. (B) Gastrula stage embryo expressing nuclear H2B:mCherry and the membrane-targeted eGFP:CAAX box. (C) Gastrula stage embryo expressing nuclear H2B:mCherry and the membrane-targeted eGFP:CAAX box. The optical section shows internal cells with nuclear mCherry and membrane-associated eGFP signals. Scale bar = 50 µm.
| Experiment number | Injected construct mRNA | Number of injected embryos | Number of labeled embryos |
| 1 | H2B:eGFP | 53 | 23 |
| 2 | H2B:eGFP | 50 | 31 |
| 3 | H2B:eGFP | 48 | 20 |
| 4 | H2B:eGFP | 54 | 24 |
| 5 | H2B:eGFP | 114 | 47 |
| 6 | H2B:eGFP | 80 | 46 |
| 7 | H2B:eGFP or | 62 | 24 |
| H2B:mCherry + eGFP:CAAX box | |||
| 8 | H2B:eGFP | 87 | 60 |
| 9 | H2B:eGFP | 77 | 45 |
| 10 | eGFP:CAAX box or | 110 | 51 |
| H2B:mCherry + eGFP:CAAX box | |||
| 11 | H2B:mCherry + eGFP:CAAX box | 45 | 26 |
| 12 | H2B:mCherry + eGFP:CAAX box | 52 | 16 |
| 13 | eGFP:CAAX box or | 74 | 35 |
| H2B:mCherry + eGFP:CAAX box | |||
| Total | 906 | 448 | |
| Success rate | 49% |
Table 1: Injection success rates. The injected constructs, the number of injected embryos that survived the injection and the number of labeled (or successfully injected) embryos are indicated, as is the overall percentage of successful injections.
| Construct | Position in the pCS2+ vector | Original sequence | Mutated sequence |
| pCS2+ eGFP:CAAX box | 79..87 | GGA TCC ACC | ACC GTC AAC |
| pCS2+ H2B:eGFP | 82..90 | TCC GAC ACG | ACC GTC AAC |
| pCS2+ H2B:mCherry | 82..90 | TCC GAC ACG | ACC GTC AAC |
Table 2: Tentative optimization of Kozak sequences. Original and mutated Kozak sequences are indicated for each construct. The introduced mutations recover the Kozak sequence of the B. lanceolatum actin gene, which very closely resembles that of the inferred theoretical preferred Kozak sequence of amphioxus (A/CGA/C G/TTC A/GAC atg TG/CT).
| pCS2+ eGFP:CAAX box | |||
| Original codon | Mutated codon | Position in the pCS2+ vector | |
| (usage level) | (usage level) | ||
| GTA | GTG | 154..156 | eGFP sequence |
| (0.08) | (0.43) | ||
| AGA | AGG | 814..816 | Linker |
| (0.09) | (0.27) | ||
| pCS2+ H2B:eGFP | |||
| Original codon | Mutated codon | Position in the pCS2+ vector | |
| (usage level) | (usage level) | ||
| GTA | GTG | 223..225 | H2B sequence |
| (0.08) | (0.43) | ||
| 289..291 | H2B sequence | ||
| 469..471 | Linker | ||
| 568..570 | eGFP sequence | ||
| CTA | CTG | 226..228 | H2B sequence |
| (0.06) | (0.51) | ||
| pCS2+ H2B:mCherry | |||
| Original codon | Mutated codon | Position in the pCS2+ vector | |
| (usage level) | (usage level) | ||
| GTA | GTG | 223..225 | H2B sequence |
| (0.08) | (0.43) | ||
| 289..291 | H2B sequence | ||
| 469..471 | Linker | ||
| 913..915 | mCherry sequence | ||
| CTA | CTG | 226..228 | H2B sequence |
| (0.06) | (0.51) | ||
Table 3: Tentative optimization of codon usage. Original and mutated codons are indicated for each construct. While this has not been experimentally tested, the introduced mutations are meant to optimize mRNA translation in amphioxus.
Supplementary File 1: Construct sequences. The nucleotide sequences of (A) the membrane-targeted eGFP (eGFP:CAAX box), (B) the nuclear-targeted eGFP (H2B:eGFP) and (C) the nuclear-targeted mCherry (H2B:mCherry) constructs are given in the context of the pCS2+ vector backbone. The main domains encoded by each of the constructs are indicated in color: eGFP (green), mCherry (red), membrane localization signal from human HRAS (CAAX box) (cyan blue), zebrafish histone 2B exon (H2B) (yellow).
Discussion
In this article, we present, for the first time, a detailed and reproducible protocol for the injection of B. lanceolatum oocytes, which, after B. floridae13,14 and B. belcheri15, is thus the third amphioxus species, for which such a technique has been described. Importantly, the protocol described here also includes the description of vector systems suited for the production of fluorescent proteins in B. lanceolatum from injected mRNA produced in vitro (described below). Together, these new tools allow in vivo imaging of the early development of amphioxus and the analysis of dynamic cell behaviors that underlie the earliest morphogenetic events in the embryo, such as cleavage and gastrulation.
In general, the microinjection technique has the benefit of allowing the delivery, into a target cell, of a pre-defined volume of a specific compound. With this being said, one major limitation of this technique is the limited number of cells that can be injected during a given experiment. For example, the protocol described here for B. lanceolatum allows the injection of 100 to 120 eggs per injection session, of which about half will be labeled (Table 3). For B. floridae, the numbers are very similar with 500 or more eggs that can be injected per day, of which more than 50% will survive the injection13,14. While the protocols for injecting B. lanceolatum and B. floridae eggs closely resemble each other, the B. belcheri technique is slightly different: the eggs are placed on poly-lysine-coated coverslips and injections are performed under an inverted microscope using a microinjector of a different brand. This approach apparently allows the injection of between 200 and 300 B. belcheri eggs per injection session with a survival rate, after hatching at the neurula stage, of 89.39% to 95.83%15. Considering the differences in the protocols, it is difficult to know whether the differences in success rates are due to species-related or protocol-related differences. One would have to test the different species in parallel with the different protocols to answer this question. Nonetheless, to further increase the number of oocytes targeted for the delivery of exogenous compounds, other protocols will have to be developed for amphioxus. Candidate techniques include electroporation, bombardment with microparticles, lipofection, and transduction4. Of note, electroporation has already been established as a standard technique for the introduction of exogenous material into fertilized ascidian tunicate eggs, another invertebrate chordate model used for studying the evolution of developmental mechanisms8.
For successful microinjection and subsequent in vivo imaging of fluorescent proteins, suitable expression vectors are a mandatory prerequisite. Towards this end, three plasmids have been developed and experimentally validated (Figure 1, Supplementary File 1): for membrane targeting, the eGFP gene was fused at its 3’ end to the human HRAS CAAX box (resulting in a eGFP:CAAX box construct)16 and, for nuclear targeting, both the eGFP and the mCherry genes were fused at their 5’ ends to the zebrafish histone 2B (H2B) exon (resulting, respectively, in a H2B:eGFP and a H2B:mCherry construct)17. Each one of these constructs has been cloned into the pCS2+ plasmid containing a SV40 late polyadenylation site and a SP6 promoter allowing in vitro capped mRNA synthesis18,19. Furthermore, with the goal to optimize translation of the constructs in B. lanceolatum, both the Kozak sequences and the codons have been adapted using single nucleotide mutagenesis (Tables 2 and 3, Supplementary File 1). Based on the approach by Nakagawa20, a small pool of B. lanceolatum genes was analyzed to identify a preferred Kozak sequence in this species. The closest known naturally-occurring sequence to this theoretical preferred sequence is that of the B. lanceolatum actin gene. The Kozak sequences of the three constructs were hence modified to match this actin gene sequence. Furthermore, the codon usage of B. lanceolatum was analyzed using the codon usage database21 and codons in the constructs with a low usage probability in B. lanceolatum (<10%) were replaced, wherever possible, by equivalent codons with a higher usage probability.
The results of the injections show that the level of protein expression from these vectors is suitable for in vivo imaging analyses. Given that the expression output of the modified vectors has not been compared with that of unmodified constructs, we cannot conclude on the efficiency of the modifications that were introduced in the Kozak consensus and the coding sequences. It would certainly be interesting to test whether these modifications indeed have an impact on protein expression levels. Along these lines, a recent analysis based on microinjections into B. belcheri suggests that other, unmodified vectors can also be used for the synthesis of mRNA for fluorescent protein production in amphioxus15.
Amphioxus contains endogenous green fluorescent proteins22. The embryos therefore exhibit low levels of green as well as red fluorescence (our unpublished observations). However, these endogenous levels are negligible with respect to the fluorescence levels resulting from the injection of our mRNA constructs. Therefore, the endogenous fluorescence of amphioxus embryos does not disturb experimental observations using fluorescent binoculars or multi-laser scanning microscopes. In addition, the injection experiments reveal that the intensity of the exogenous fluorescence generated by mRNA injection depends on the actual amount of injected material, which is directly correlated with the final concentration of mRNA used in the injection mix. B. lanceolatum embryos tend to tolerate a concentration of up to 1.8 µg/µl mRNA, although, independent of the actual mRNA concentration, some mRNA combinations seem to be less tolerated by B. lanceolatum embryos than others. Thus, the combination of nuclear mCherry and membrane eGFP mRNAs appeared to be more toxic than the injection of nuclear eGFP alone.
Texas Red dextran has previously been used as a tracer for successful injections into amphioxus oocytes13-15. Intriguingly, a fluorescent signal derived from the injected mRNA was never obtained after injection of solutions containing Texas Red dextran and this in both amphioxus oocytes (n = 4 experiments) and zebrafish embryos (n = 3 experiments). When an injection mix containing in vitro transcribed mRNA and Texas Red dextran is loaded on a RNase-free agarose gel (Figure 3, lane 2), the band corresponding to the mRNA is absent. In contrast, in vitro transcribed mRNA is detectable on an RNase-free agarose gel in the presence of Phenol Red (Figure 3, lane 1). Given that there is no evidence for mRNA degradation on the gel, these results suggest that Texas Red dextran tends to trap mRNA. When used in an injection solution, Texas Red dextran might thus prevent the translation of the injected mRNA. Following this observation, the interactions of other dyes (FITC dextran, Rhodamine dextran and Oregon Green dextran) with mRNA were tested, both on RNase-free agarose gels and by injection into B. lanceolatum or zebrafish (Figure 3). The analyses indicate that FITC dextran does not trap mRNA on gel (Figure 3, lane 4) and leads to fluorescent protein expression in zebrafish (n = 1 experiment), but not in embryos of B. lanceolatum (n=1 experiment) (data not shown). Rhodamine dextran does not trap mRNA on gel (Figure 3, lane 5) and leads to fluorescent protein expression in zebrafish embryos (n = 1 experiment) (data not shown), but its activity could unfortunately not be tested in B. lanceolatum embryos. Finally, as the Oregon Green dextran migrates at the same level as the mRNA, mRNA trapping could not be assessed by agarose gel (Figure 3, lane 3). This latter dye prevented fluorescent protein expression in amphioxus (n = 1 experiment), but not zebrafish embryos (n=1 experiment) (data not shown). In sum, these data suggest that some dextrans can efficiently inhibit the translation of injected mRNA. Given that dose-response experiments were not carried out, these data are qualitative. Intriguingly, this inhibitory effect seems to be dependent on the animal species (zebrafish versus amphioxus), which highlights the need for further studies to understand the mechanisms underlying this effect. Furthermore, these results call for preparatory analyses, if dextrans are to be used as color tracers for injections.
The development of the microinjection technique for a given model opens the door for gene-specific manipulations. In amphioxus, for example, the first description of microinjections (in B. floridae) was followed by the successful knockdown of a gene, hox1, using morpholino oligonucleotide injections23,24. The number of B. lanceolatum embryos that can successfully be injected every day using the protocol presented here is certainly compatible with this type of study. Furthermore, with these methods and the novel constructs at hand, one can now also design and perform overexpression and knockdown studies by injection of mRNAs of interest (native or dominant-negative forms) or use other strategies for disrupting gene expression (for example microRNA- or shRNA-based strategies). So far, amphioxus injections can only be performed at the oocyte stage, simply because this is the only stage that can efficiently be immobilized. After injection at the oocyte stage, the molecule of interest is ubiquitously expressed in the embryo. Thus, if gene expression needs to be altered or monitored only in a subset of cells, DNA constructs need to be injected, as these are expressed mosaically in the developing amphioxus embryo15,25-27. DNA construct injections can also be used to test the activity of regulatory regions during amphioxus development in transient transgenic assays, with the aforementioned drawback of mosaic expression15,25-27.
Ultimately, the amphioxus microinjection technique also opens the door for stable transgenesis, including the targeted knock-out or knock-in of specific genetic loci. Systems for stable insertion of exogenous DNA already exist, for example with the fish transposon-based Tol2 system that seems to function in both insects and amniote vertebrates28. Furthermore, approaches based on modified zinc-finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs) or RNA-guided genome editing tools (CRISPR/Cas system) could be used to generate point mutations and knock-in modifications in specific regions of the genome29. These efforts will require the year-round breeding of amphioxus in captivity, which has already been set up for B. belcheri11,30 and B. japonicum30,31 and is currently being developed for A. lucayanum, B. floridae and the amphioxus species featured here, B. lanceolatum32.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the support by the “Animalerie Centrale de Gif-sur-Yvette” for animal husbandry. This work was supported by funds from ANR (ANR-09-BLAN-0262-02 and ANR-11-JSV2-002-01) to Michael Schubert, by the European Union FP6 grant “Embryomics” and by the ANR grant “ANR-10-BLAN-121801 Dev-Process” to Jean-François Nicolas and Nadine Peyriéras. João Emanuel Carvalho is financed by a FCT doctoral fellowship (SFRH/BD/86878/2012).
Requests for the vectors described here can be addressed directly to the authors.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Consumables | |||
| 35 mm Petri dishes | Falcon | 353001 | culture-treated |
| Filtration unit (Stericup 1L) | Fisher | W21719 | 0.22 micron filtration |
| Spin-X tubes | Costar | 8160 | 0.22 micron filtration tube |
| Needle storage jar for 1.2 mm diameter capillaries | WPI | E212 | |
| Pasteur Pipettes | 230 mm long | ||
| Aspiration tube | Dutscher | 75056 | |
| Capillaries for injection needles | Sutter | BF 120-94-10 | Borosilicate glass with filament, OD 1.20 mm, ID 0.94 mm, length 10 mm |
| Reagents | |||
| Low-melting agarose | SIGMA | A9414 | |
| Phenol Red | SIGMA | 114537 | |
| Glycerol | SIGMA | G2025 | |
| Poly-L-Lysine hydrobromide | SIGMA | P9155 | |
| H2O Dnase, Rnase-free | Gibco | 10977-035 | |
| mMessage mMachine SP6 Transcription kit | Ambion | AM1340 | mRNA synthesis kit |
| Phenol pH8 | SIGMA | P4557 | |
| 24:1 chloroform:isoamylic alcohol | SIGMA | C0549 | |
| 5:1 phenol pH4.7:chloroform | SIGMA | P1944 | |
| Reef Crystal salts (200 kg) | Europrix | Commercial salts | |
| Equipment | |||
| Fluorescent dissecting scope with 200x magnification | Leica | MZ16F | 25x oculars, DSR and GFP2 filters |
| Micromanipulator | Marzhauzer | M-33 | |
| Injector | Picospritzer | model II or III | |
| Needle puller | Sutter | P97 | heating-filament needle puller |
| Fine forceps | FINE SCIENCE TOOLS GMBH | 11252-30 | Dumont #5 |
References
- Weber, T., Köster, R. Genetic tools for multicolor imaging in zebrafish larvae. Methods. 62, 279-291 (2013).
- Weil, T. T., Parton, R. M., Davis, I. Making the message clear: visualizing mRNA localization. Trends. Cell. Biol. 20, 380-390 (2010).
- Zhang, Y., Yu, L. C. Microinjection as a tool of mechanical delivery. Curr. Opin. Biotechnol. 19, 506-510 (2008).
- Stepicheva, N. A., Song, J. L. High throughput microinjections of sea urchin zygotes. J. Vis. Exp. , e50841 (2014).
- Schubert, M., Escriva, H., Xavier-Neto, J., Laudet, V. Amphioxus and tunicates as evolutionary model systems. Trends Ecol. Evol. 21, 269-277 (2006).
- Bertrand, S., Escriva, H. Evolutionary crossroads in developmental biology: amphioxus. Development. 138, 4819-4830 (2011).
- Holland, L. Z. Evolution of new characters after whole genome duplications: insights from amphioxus. Semin. Cell Dev. Biol. 24, 101-109 (2013).
- Lemaire, P. Evolutionary crossroads in developmental biology: the tunicates. Development. 138, 2143-2152 (2011).
- Yu, J. K., Holland, L. Z. Cephalochordates (amphioxus or lancelets): a model for understanding the evolution of chordate characters. Cold Spring Harbor Protocols. 2009, (2009).
- Fuentes, M., et al. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J. Exp. Zool. B. 308, 484-493 (2007).
- Li, G., Shu, Z., Wang, Y. Year-round reproduction and induced spawning of Chinese amphioxus, Branchiostoma belcheri, in laboratory. PLoS One. 8, e75461 (2013).
- Theodosiou, M., et al. Amphioxus spawning behavior in an artificial seawater facility. J. Exp. Zool. B. 316, 263-275 (2011).
- Holland, L. Z., Yu, J. K. Cephalochordate (amphioxus) embryos: procurement, culture, and basic methods. Methods Cell Biol. 74, 195-215 (2004).
- Holland, L. Z., Onai, T. Analyses of gene function in amphioxus embryos by microinjection of mRNAs and morpholino oligonucleotides. Methods Mol. Biol. 770, 423-438 (2011).
- Liu, X., Li, G., Feng, J., Yang, X., Wang, Y. Q. An efficient microinjection method for unfertilized eggs of Asian amphioxus Branchiostoma belcheri. Dev. Genes Evol. 223, 269-278 (2013).
- Harvey, K. J., Lukovic, D., Ucker, D. S. Membrane-targeted green fluorescent protein reliably and uniquely marks cells through apoptotic death. Cytometry. 43, 273-278 (2001).
- Maruyama, J., Nakajima, H., Kitamoto, K. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol. Biochem. 65, 1504-1510 (2001).
- Rupp, R. A., Snider, L., Weintraub, H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8, 1311-1323 (1994).
- Turner, D. L., Weintraub, H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434-1447 (1994).
- Nakagawa, S., Niimura, Y., Gojobori, T., Tanaka, H., Miura, K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 36, 861-871 (2008).
- Nakamura, Y., Gojobori, T., Ikemura, T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28, 292 (2000).
- Deheyn, D. D., et al. Endogenous green fluorescent protein (GFP) in amphioxus. Biol. Bull. 213, 95-100 (2007).
- Schubert, M., et al. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 132, 61-73 (2005).
- Schubert, M., Holland, N. D., Laudet, V., Holland, L. Z. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev. Biol. 296, 190-202 (2006).
- Yu, J. K., Holland, N. D., Holland, L. Z. Tissue-specific expression of FoxD reporter constructs in amphioxus embryos. Dev. Biol. 274, 452-461 (2004).
- Beaster-Jones, L., Schubert, M., Holland, L. Z. Cis-regulation of the amphioxus engrailed gene: insights into evolution of a muscle-specific enhancer. Mech. Dev. 124, 532-542 (2007).
- Holland, L. Z., et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100-1111 (2008).
- Urasaki, A., Mito, T., Noji, S., Ueda, R., Kawakami, K. Transposition of the vertebrate Tol2 transposable element in Drosophila melanogaster. Gene. 425, 64-68 (2008).
- Li, G., et al. Mutagenesis at specific genomic loci of amphioxus Branchiostoma belcheri using TALEN method. J. Genet. Genomics. 41, 215-219 (2014).
- Zhang, Q. J., et al. Continuous culture of two lancelets and production of the second filial generations in the laboratory. J. Exp. Zool. B. 308, 464-472 (2007).
- Yasui, K., Igawa, T., Kaji, T., Henmi, Y. Stable aquaculture of the Japanese lancelet Branchiostoma japonicum for 7 years. J. Exp. Zool. B. 320, 538-547 (2013).
- Benito-Gutiérrez, E., Weber, H., Bryant, D. V., Arendt, D. Methods for generating year-round access to amphioxus in the laboratory. PLoS One. 8, e71599 (1371).

