3D Modeling of the Lateral Ventricles and Histological Characterization of Periventricular Tissue in Humans and Mouse
Summary
Using MRI scans (human), 3D imaging software, and immunohistological analysis, we document changes to the brain’s lateral ventricles. Longitudinal 3D mapping of lateral ventricle volume changes and characterization of periventricular cellular changes that occur in the human brain due to aging or disease are then modeled in mice.
Abstract
The ventricular system carries and circulates cerebral spinal fluid (CSF) and facilitates clearance of solutes and toxins from the brain. The functional units of the ventricles are ciliated epithelial cells termed ependymal cells, which line the ventricles and through ciliary action are capable of generating laminar flow of CSF at the ventricle surface. This monolayer of ependymal cells also provides barrier and filtration functions that promote exchange between brain interstitial fluids (ISF) and circulating CSF. Biochemical changes in the brain are thereby reflected in the composition of the CSF and destruction of the ependyma can disrupt the delicate balance of CSF and ISF exchange. In humans there is a strong correlation between lateral ventricle expansion and aging. Age-associated ventriculomegaly can occur even in the absence of dementia or obstruction of CSF flow. The exact cause and progression of ventriculomegaly is often unknown; however, enlarged ventricles can show regional and, often, extensive loss of ependymal cell coverage with ventricle surface astrogliosis and associated periventricular edema replacing the functional ependymal cell monolayer. Using MRI scans together with postmortem human brain tissue, we describe how to prepare, image and compile 3D renderings of lateral ventricle volumes, calculate lateral ventricle volumes, and characterize periventricular tissue through immunohistochemical analysis of en face lateral ventricle wall tissue preparations. Corresponding analyses of mouse brain tissue are also presented supporting the use of mouse models as a means to evaluate changes to the lateral ventricles and periventricular tissue found in human aging and disease. Together, these protocols allow investigations into the cause and effect of ventriculomegaly and highlight techniques to study ventricular system health and its important barrier and filtration functions within the brain.
Introduction
An ependymal cell monolayer lines the ventricular system of the brain providing bi-directional barrier and transport functions between the cerebral spinal fluid (CSF) and interstitial fluid (ISF) 1-3. These functions help to keep the brain toxicant-free and in physiological balance 2,3. In humans loss of portions of this lining through injury or disease does not appear to result in regenerative replacement as found in other epithelial linings; rather loss of ependymal cell coverage appears to result in periventricular astrogliosis with a meshwork of astrocytes covering regions denuded of ependymal cells at the ventricle surface. Serious repercussions to important CSF/ISF exchange and clearance mechanisms would be predicted to result from loss of this epithelial layer 1,2,4-7.
A common feature of human aging is enlarged lateral ventricles (ventriculomegaly) and associated periventricular edema as observed by MRI and fluid-attenuated inversion recovery MRI (MRI/FLAIR) 8-14. To investigate the relationship between ventriculomegaly and the cellular organization of the ventricle lining, postmortem human MRI sequences were matched with histological preparations of lateral ventricle periventricular tissue. In cases of ventriculomegaly, substantial areas of gliosis had replaced ependymal cell coverage along the lateral ventricle wall. When ventricle expansion was not detected by MRI-based volume analysis, the ependymal cell lining was intact and gliosis was not detected along the ventricle lining 6. This combinatorial approach represents the first comprehensive documentation detailing changes in cellular integrity of the lateral ventricle lining using wholemount preparations of portions or the entire lateral ventricle wall and 3D modeling of ventricle volumes 6. Several diseases (Alzheimer’s disease, schizophrenia) and injuries (traumatic brain injury) show ventriculomegaly as an early neuropathological feature. Denudation of areas of the ependymal cell lining thereby would be predicted to interfere with normal ependymal cell function and compromise the homeostatic balance between CSF/ISF fluid and solute exchange. Thus, a more thorough examination of changes to the ventricular system, its cellular composition, and the consequence to underlying or neighboring brain structures will ultimately begin to reveal more about the neuropathology associated with ventricle enlargement.
The lack of multimodal imaging data, and in particular longitudinal data sequences, together with limited access to corresponding histological tissue samples makes analysis of human brain pathologies difficult. Modeling phenotypes found in human aging or disease can often be achieved with mouse models and animal models become one of our best means to explore questions about human disease initiation and progression. Several studies in healthy young mice have described the cytoarchitecture of the lateral ventricle walls and the underlying stem cell niche 4,7-15. These studies have been extended to include 3D modeling and cellular analysis of the ventricle walls through aging 6,15. Neither periventricular gliosis nor ventriculomegaly are observed in aged mice, rather mice display a relatively robust subventicular zone (SVZ) stem cell niche subjacent to an intact ependymal cell lining 6,15. Thus, striking species-specific differences exist in both the general maintenance and integrity of the lateral ventricle lining during the process of aging 6,15. Therefore, to best use mice to interrogate conditions found in humans, differences between the two species need to be characterized and appropriately considered in any modeling paradigm. Here, we present procedures to evaluate longitudinal changes to the lateral ventricles and associated periventricular tissue in both humans and mouse. Our procedures include 3D rendering and volumetry of both mouse and human ventricles, and use of immunohistochemical analysis of whole mount preparations of periventricular tissue to characterize both cellular organization and structure. Together these procedures provide a means to characterize changes in the ventricular system and associated periventricular tissue.
Protocol
NOTE: Animal procedures were approved by the University of Connecticut IACUC and conform to NIH guidelines. Human tissue and data analysis and procedures were in compliance with and approved by the University of Connecticut IRB and conform to NIH guidelines.
1. Mouse: Analysis of Periventricular Cellular Integrity and 3D Modeling of the Lateral Ventricle
1.1) Preparation of Mouse Lateral Ventricle Wall Whole Mounts
- Prepare mouse lateral ventricle whole mounts for immunohistochemistry (IHC) as previously described 16,17.
1.2) Immunohistochemistry for Lateral Ventricle Analysis
- Fix and section mouse brain tissue as previously described 18,19.
- Briefly, flush mice transcardially with normal, room temperature saline, until organs have cleared (indicated by a pale coloration), followed by room temperature 4% paraformaldehyde (PFA) in 0.1 M phosphate buffered saline (PBS) until tissue has stiffened.
- Remove brain from the skull. Use iris dissection scissors to cut from base of skull along the sagittal suture.
- Expose skull and remove brain with forceps. Immerse the brain in 4% PFA fixative overnight at 4 °C. Then wash 3 times for 20 min with PBS.
- Prepare serial sections for immunohistochemistry and volume analysis.
- Using a microsurgical scalpel, make a small incision longitudinally along the cortex of the left hemisphere to allow the identification of the left hemisphere from the right hemisphere when floating sections are immunostained.
- Encase fixed brains in 3% agarose for added structural support during vibratome sectioning. Warm 3% agarose in distilled water (w/v) until completely dissolved using either a microwave or hot plate.
- With a razor blade, remove the caudal section of the brain at the cerebellum with a coronal cut, leaving a flat and even surface. Apply super glue to the tissue stage and position the brain on the newly cut surface with the olfactory bulbs facing upward.
- When liquid agarose solution has cooled to a temperature just warm to touch, apply evenly over the brain to completely encase the entire brain. Allow the agarose to solidify.
- Section agarose encased brains coronally on a vibratome into 50 µm sections, with sections placed serially into a 24-well plate containing PBS in order to preserve sequential order.
NOTE: Generally 12 wells are used for one brain, with collection of tissue beginning 5 sections prior to seeing the emergence of the lateral ventricles beginning with well A1, moving numerically through to B6 and cycling back to A1. This allows multiple distinguishable sections from the same brain to be processed within the same well. For lateral ventricle volume analysis, tissue collection ceased when the lateral ventricles and third ventricle merge, resulting in approximately 3 sections per well or 36 sections total. - Aspirate the PBS using a transfer pipette affixed with a 20-200 µl micropipette tip to ensure that floating sections are not accidently aspirated. Block and permeabilize floating sections in 10% serum, 0.1% Triton X-100 (TX) in PBS (PBS-TX) for 1 hr at room temperature. Use 300 µl of solution per well to cover free-floating tissue sections in a 24-well plate and perform all incubations of sections on a rocking plate.
- Aspirate the blocking solution, and incubate sections with primary antibodies in PBS-TX overnight (at least 12 hr) at 4 °C. For ventricle volume traces using coronal sections, use anti-S100β antibody to label ependymal cells.
- Aspirate primary antibody solution, and wash sections 3 times for 10 min each with PBS-TX.
- Incubate sections in dark for 1 hr at room temperature with fluorescent secondary antibodies, at concentrations of 1:1,000 in 10% serum, PBS-TX. Keep sections in the dark for all remaining incubation steps.
- Aspirate secondary antibody solution and incubate sections with 4’,6-diamidino-2-phenylindole in PBS (DAPI, 10 µg/ml) for 5 min to counterstain cell nuclei. Aspirate the DAPI solution and rinse sections 3 times for 10 min each with PBS.
- Mount sections serially onto slides using the cortical mark to preserve left versus right hemisphere orientation. Air dry sections in the dark and then coverslip by applying a thin layer of mounting media to the slide and gently place a glass coverslip on top. Allow coverslipped slides to dry overnight at room temperature.
1.3) Lateral Ventricle Segmentation for 3D Reconstructions
NOTE: Perform tracing of lateral ventricles using mapping software on an upright epifluorescence microscope with an automated stage and a digital CCD camera for fluorescence detection.
- Turn on the fluorescence microscope equipment and launch mapping software in fluorescence mode. Open ‘Display Options’ to load the proper toolbars and docking tool panels needed for tracing. Options>Display Options>Accessories, and select the ‘Main’ and ‘Marker’ toolbars. In the same menu, select ‘Camera Histogram’, ‘Multichannel Control’, ‘Camera Settings’, and ‘Image Acquisition’ under ‘Docking Tool Panels.’
- Observe beginnings of the ventricles based on strong S-100β fluorescence that labels ependymal cells that line the lateral ventricle. Identify the first piece of tissue containing the lateral ventricles.
- Create a new data file and designate a reference point for stage movement by clicking in the image window above the left lateral ventricle. Look for the small incision to orient left from right hemisphere. Keep the reference point in a consistent location relative to the lateral ventricles across tracing files to help when importing contours for serial reconstruction.
- Select appropriate preset contour type from the contour drop down menu, e.g., ‘Left Lateral Ventricle.’ Use a unique color for each of the left and right lateral ventricle tracings. If changes in the contour name and color are required, go to Options>Display Preferences>Contours, then modify the names of the contour colors or select ‘Add Contour Type’.
- With the ‘Left Lateral Ventricle’ contour selected, click along the apical surface of the ependymal lining to begin the trace contour. Trace the ventricle by continuing to click successively along the apical surface.
- For irregular areas requiring more finesse, click and hold the left mouse button in order to trace free hand. Press Ctrl+Z, or go to Options>Undo to undo the previous contour point. Move the tracing window using the arrow keys or by making a contour point off screen and allowing the window to automatically center itself. To finish the ventricle tracing right click and select ‘Close Contour’.
- Select a new contour color (contour type) for the right lateral ventricle using the drop down menu. Trace the right ventricle as in Step 1.3.4 for the left.
- If changes in the contour name and color are required, right click on the contour and select Change Contour Type to select the appropriate color.
- Add markers along the midline to assist in aligning serial contours for 3D reconstruction. To add markers, select the appropriate marker (use the ‘X’) from the Markers toolbar on the left and click to drop them on the tracing screen. Import markers along with the contours for each tissue section trace.
- To save the tissue traces, select File>Save Data File As and create a new folder and file name. Number the tracings [slide #]-[tissue #] for easy identification of traces from serial sections. For example, the third tissue section on the second slide has the file name ‘2-3’ within the directory for each brain.
- Repeat Steps 1.3.4 to 1.3.9 for each serial section of brain tissue to trace the ventricles through the entire brain, excluding regions of ventricle wall adhesion.
- If the ventricle has a region of adhesion, sub-segment the anterior region from those dorsal and ventral to the adhesion site. Create two new contour types for each ventricle (e.g., ‘Dorsal Left Ventricle’ and ‘Ventral Left Ventricle’). On the first tissue slice with adhesion, trace the lateral ventricle with both the original lateral ventricle contour and the new dorsal and ventral contours to ensure a complete ventricle reconstruction can be created.
- In sections posterior to a ventricle wall adhesion, create a set of new contour colors for the posterior portion of each ventricle (e.g., ‘Posterior Left Ventricle’). Double-trace this first re-joined section with both the current adhesion and new posterior contour types.
1.4) Lateral Ventricle 3D Reconstruction
- Align and compile serial contour tracings of the mouse lateral ventricles to generate 3D reconstructions and volumetric data.
- Open 3D reconstruction program. Click File>Open and select the contour file of the first traced serial section. Import the contour tracings of the left and right ventricle as well as any added markers.
- To select contours, click the ‘Select Objects’ button (cursor icon) and select the two ventricles and markers by holding ‘Ctrl’. To establish the Z position of the contours, right click and select ‘Modify Z Position’. Set the first tissue slice to 0 µm.
- To save the reconstruction, select File>Save Data File As. Save after every file import, so if a mistake is made recovery is as easy as re-loading the previous file.
- To add the next tissue slice to the reconstruction, click File>Append to Display. Select the .DAT file to be appended, and check the ‘Merge’ box.
- Follow Step 1.4.3 to again adjust the Z position of the newly imported trace file. Select only the newly added left and right contours in the main window to avoid moving the other traces. Set each successive trace file to 50 µm from the Z position of the previous contour for 50 µm sections. (First contour: z = 0, second contour: z = 50, third contour: z = 100, etc.)
- To adjust the X, Y position of selected contours and markers, click the ‘Enable Translation with Mouse’ button and drag the contours to align with the previous tissue contours. Keep both the left and right contours of the current trace selected to allow synchronous movement.
- To rotate the traces clockwise or counter clockwise to line up the midline markers, select the ‘Z-axis Rotation’ button. To flip contours across the Y-axis (if the left contour is on the right) select both imported contours and markers, right click, select ‘Set Rotation Angle’, and enter ‘180’ into the ‘Y rotation’.
- Ensure that the ventricles and midline markers are best aligned before saving the reconstruction file, as in Step 1.4.4.
- Repeat Steps 1.4.5 to 1.4.9 for each subsequent tissue slice until all have been imported and properly aligned.
- To view volume data of the serial reconstruction, click Analysis>‘Markers and Region Analysis’ and select ‘3D Contour Summary’. Select all contours in the left hand panel and then click the ‘3D Visualization’ button to display the final 3D reconstruction.
2. Human: Analysis of Periventricular Cellular Integrity and 3D Modeling of the Lateral Ventricle
2.1) Human MRI Data Analysis
NOTE: Protocols are listed to create 3D image reconstructions and volumetric quantification of the lateral ventricles and assess volumetric changes over time using longitudinal overlay analysis. It is important to note that consistency in MR data collection (e.g., machine and magnet strength, section thickness, orientation and resolution) and post-acquisition processing are extremely important criteria for inclusion of data sets 20.
- Ventricular Segmentation
NOTE: ITK/Snap is used to segment the ventricles from high-resolution T1-weighted MR images. Alternatively, other freeware software such as Freesurfer can be used.- Open ITK/Snap, File > Open Grayscale Image. Select desired file and open as a .nii file (NITFI file).
- Scroll through each anatomical plane, noting ventricular abnormalities (stenosed regions, large or small temporal horns, etc.). In the Main Toolbox click ‘Snake ROI Tool’. Click ‘Segment 3D’. Select the ‘Intensity Region’ option.
- Click ‘Preprocess Image’. Select ‘Above’, to include regions below a certain intensity threshold in order to highlight ROI in white. Record maximum intensity using sliding bar. Set the threshold to 15% of maximum intensity (record this value). Set smoothness parameter to 10. Click ‘OK’, then ‘Next’.
- Add 10-12 small (size 2) ‘bubbles’ to each ventricle: two in the anterior frontal horn, seven evenly-spaced along the superior surface of the lateral ventricle body, one in the occipital horn and one in the temporal horn of the lateral ventricle. ‘Select Parameters’ and set curvature force to 0.4. Click the Play Symbol to begin active contour evolution.
- Click ‘Update Mesh’ to visualize surface; check ‘Auto-Update’. Stop segmentation when all areas of the lateral ventricle are included in the region of interest (ROI). Avoid inclusion of the third ventricle. Click ‘Finish’.
- Manually edit image if necessary to remove undesired regions by the following methods (typically need to erase third ventricle leakage). Manually use the ‘3D Scalpel’ tool to remove excess regions or manually use the ‘Paintbrush’ tool with ‘Clear Label’ as the active drawing label to erase any inclusion beyond ROI.
- Save segmentation results as a .nii image (‘NIFTI’ file format).
- To obtain total ventricle volume, select ‘Segmentation>Volume’ and Statistics; obtain the total volume of the ventricle using set color label.
- Longitudinal Representation Of Lateral Ventricles From MRIScans
NOTE: Using Mango software, longitudinal ROIs are overlaid to qualitatively and quantitatively demonstrate changes in ventricle volume within subjects over time.- Open Mango. Click File>Load ROI of the first ROI time point (baseline) as GREEN. File>Load ROI of the second ROI time point as RED. Save longitudinal overlay by selecting File>Save as; assign each image as ‘file name_[first time point age]-[second time point age].nii’.
- Open ITK-SNAP. Reopen the combined ROI as ‘greyscale image’, by selecting Segmentation>Load From Image>combined ROI file. To obtain longitudinal volume data run the following: Segmentation>Volume and Statistics>Label 1 (red) = expansion volume; Label 2 (green) = stenosis volume; Label 3 (blue) = base volume.
2.2) Human Periventricular Tissue Preparation and Analysis
- Safety instructions for working with human tissue
- Obtain appropriate safety training (e.g., Blood-borne Pathogens course).
- Observe standard (universal) safety precautions. Wear gloves. Change gloves before touching any common equipment or surfaces that are not disposable. Label any items routinely handled when working with human tissue with ‘human tissue use’ designations.
- Follow decontamination practices for all items contacting human tissue. Bleach all contaminated liquids for a minimum of 24 hr prior to disposal, treat contaminated surfaces with a bleach soaked towel (e.g., bench top) or by placing object directly in bleach (e.g. forceps).
- Prepare contingency plan for contact with human tissue directly on skin, which may include soaking a paper towel in bleach and holding on the affected area (5-10 min).
- Use appropriate waste disposal for all items coming in contact with human tissue.
- Lateral Ventricle Whole Mount Preparation
- Beginning with an intact hemisphere (fixed in 10% formalin and rinsed thoroughly with 0.1 M PBS), slice 1.5 cm thick coronal sections through entire hemisphere using large knife.
NOTE: All fixation protocols require prior antibody verification. - Label sections front to back starting with first section containing the lateral ventricle. Use an alpha-numeric labeling system, labeling the slices with letters (anterior to posterior) and subsection with numbers (top down). Avoid disrupting ependymal surface.
- Using microsurgical scalpel, dissect ventricle wall 1 cm deep, maintaining one continuous section for entire lateral wall. Subdivide wall as needed to create appropriately sized sections for staining well. Notch section on opposite side of ventricle wall to identify superior/inferior orientation.
- Perform antigen retrieval by incubating tissue sections in 10 mM sodium citrate buffer (pH 6.0) at 100 °C for 10-20 min using a double boiler (optimal incubation time is determined empirically). Rinse 3 times in PBS/0.1%Triton X-100 (PBS-TX).
- Block in 10% horse serum using PBS/ PBS-TX for 1 hr at room temperature.
- Incubate with primary antibodies for 48 hr at 4 °C. To visualize ventricle wall lining, immunostain whole mounts with the following antibody combinations: mouse anti-β-catenin (1:250); goat anti-GFAP (1:250); rabbit anti-AQP4 (1:400).
- Perform all subsequent steps in the dark. Rinse tissue 3 times in PBS, incubate with secondary antibodies (1:500) for 24 hr at 4 °C or 2 hr at room temperature, and rinse another 3 times in PBS. Incubate tissue in DAPI for 7 min at room temperature followed with another 3 rinses in PBS.
- Prepare tissue for final dissection by covering a wax bottom dish with parafilm. Fill dish with PBS, place tissue in dish using fine forceps, avoiding making contact with ependymal wall.
- Under the dissecting microscope, firmly secure tissue on its side using pins. With a 22.5° microsurgical stab knife, create uniform thin sections (approximately 300 µm thick) of lateral ventricle wall. For large sections or sections with difficult curvature, cut into smaller cubes before cutting into final sections for mounting and note location.
- Carefully place dissected section ependyma side up onto slide using fine forceps, taking note of regional location and orientation on the slide. Cover tissue with thin layer of aquapolymount, taking care to avoid bubbles. Place coverslip over tissue, add additional aquapolymount as necessary to fill any gaps under coverslip.
- Place mounted sections in dark and dry for 2-3 days at room temperature. Store in slidebox at 4 °C.
- Beginning with an intact hemisphere (fixed in 10% formalin and rinsed thoroughly with 0.1 M PBS), slice 1.5 cm thick coronal sections through entire hemisphere using large knife.
- Immunohistochemistry and Histological Analysis
- Using confocal microscopy to view fluorescent immunohistochemistry, set the imaging focal plane at the ventricle surface, as determined by the use of β-catenin (marker for apical adherens junction proteins of ependymal cells). Delineate regions of ependymal cell coverage and surface astrogliosis (GFAP staining at ventricle surface).
- To document and montage the entire ventricle surface, use overlapping images to cover entire surface. To create montage of serial images, open Adobe Photoshop. Use File>Automate>Photomerge>Interactive layout to select images to overlay, making manual adjustments if necessary.
- Create new layer to trace montage to generate cartoon representation of intact ependyma cell coverage or ventricle surface gliosis. Repeat for each slide or ROI. Compile all sections to reconstruct map of ventricle wall and link to corresponding region on MRI reconstruction.
Representative Results
Contour tracing of the mouse lateral ventricles based on immunostained 50 µm coronal sections and 3D reconstructions (Figure 3) allows volume data to be collected in different experimental paradigms using mouse as a model system for disease or injury. Critical to this procedure is the exclusion of regions where the lateral ventricle walls adhere to each other. By subsegmenting regions of the ventricles and designating a different color for each region (Figure 3C), contiguous sections can be complied and regional and total volume can be calculated from compiled subsegments.
Similar studies can be performed using MRI scans of the brain together with semi-automated segmentation (ITK-SNAP) of the lateral ventricles for 3D renderings. For direct pairing of ventricle volumes and periventricular tissue analysis, pre- or post-mortem MRI scans are segmented and aligned to create 3D reconstructions (Figure 4). If longitudinal MRI scans are available, 3D reconstructions can be aligned and overlaid for analysis of expansion over time (Figure 5). Longitudinal analysis provides information about regions of particular interest for future immunohistochemical analysis. Corresponding tissue is analyzed immunohistochemically to reveal regions of astrogliosis versus intact ependymal cell monolayer at the ventricle surface (Figure 6A). Tissue images are captured and then montaged to cover large areas of ventricle surface (Figure 6B). Compiled montages are converted to cartoon representations and then mapped onto an MRI-based 2D model to show regional alterations in periventricular cellular integrity (Figure 6C). Whole mount preparation of the ventricle wall allows for panoramic view of large expanses of the ventricle surface, or as we have demonstrated the entire lateral ventricle surface 6. Increased levels of Aquaporin-4 expression in areas of surface gliosis suggesting edema can be used to assess ventricle lining integrity 6.
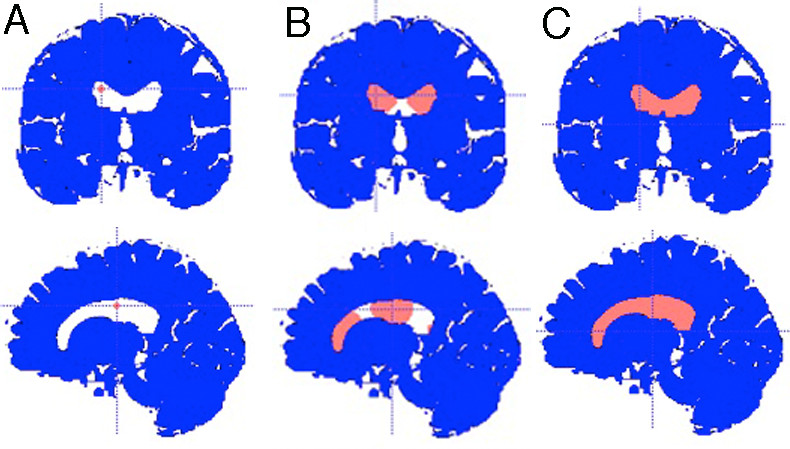
Figure 1: Performing lateral ventricle segmentation using the ITK/Snap Snake ROI Tool (A) ‘Bubbles’ are added to lateral ventricles. (B) ‘Bubbles’ expand during active contour evolution. (C) Segmentation is complete when entire lateral ventricle is filled. (Care is taken to avoid the 3rd ventricle.)
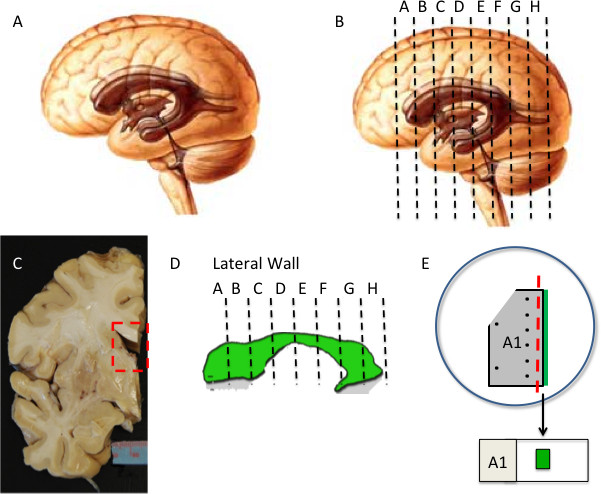
Figure 2: Human lateral ventricle wall dissection (A-D) Intact hemisphere is cut into 1.5 cm thick coronal slabs. Isolated 2D representation of ventricle is shown in (D). (E) A section of ventricle wall is dissected out and processed for IHC. Subdivision of tissue may be required depending on size and curvature. To maintain orientation, tissue is notched at top on opposite side of ventricle surface (green). Pins (black dots) are used to secure tissue and guide final dissection (red dashed line). Final whole mount preparation is mounted on slide with ventricle surface facing up (green).

Figure 3: Tracing and 3D reconstruction of mouse lateral ventricles (A Coronal mouse brain section including lateral ventricles, outlined by S100β-immunoreactive ependymal cells. (*, Lateral Ventricles; bracket, region of adhesion; scale bar, 500 µm). (B) Mouse lateral ventricles are traced as ‘contours’ and arranged as sub-segmented sections, excluding regions of intraventricular adhesion from interfering with volumetric analysis. (C)3D reconstruction of lateral ventricle contours. Yellow, contour of whole brain volume spanning the region of interest containing the lateral ventricles.
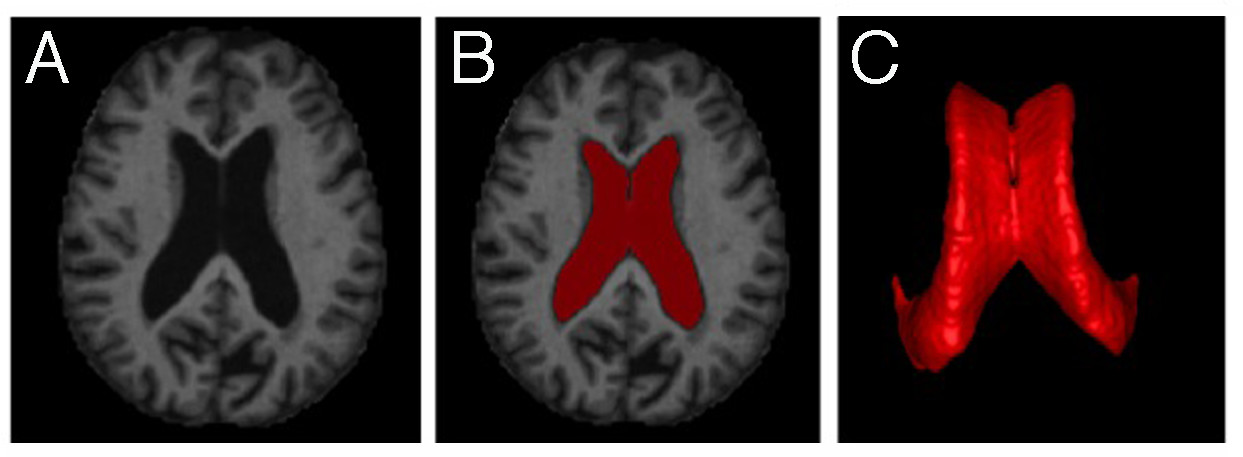
Figure 4: MRI-based lateral ventricle segmentation (A) MR Images are assembled. (B) Lateral ventricle is defined as ROI (red). (C) 3D image reconstruction allows for volumetric quantification and qualitative visualization of lateral ventricle.
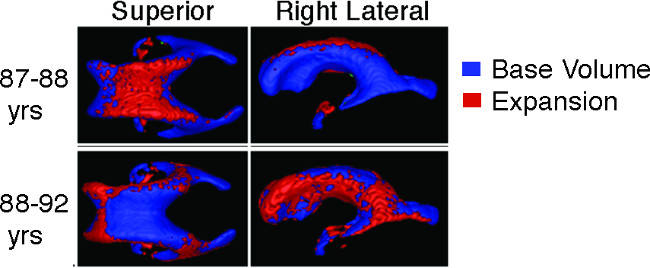
Figure 5: Assessment of longitudinal ventricle expansion Longitudinal MRIs from multiple points can be aligned and overlaid to visualize and quantify ventricle volume expansion within subjects over time.
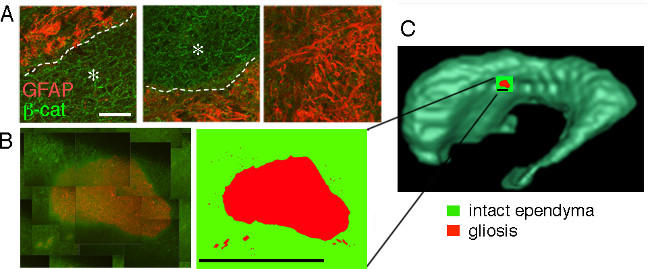
Figure 6: Immunohistochemical evaluation and regional mapping of human lateral ventricle surface (A) Areas of intact ependyma cells, outlined by β-catenin staining, show a cobblestone appearance (asterisk) and are demarcated by a dotted line from regions of astrogliosis on ventricle surface (GFAP+ staining). (B) Serial confocal images are overlaid to generate a regional montage and traced using Adobe Photoshop for a cartoon representation of cellular organization at the ventricle surface. (C) Cartoon of image montage is mapped to ventricle surface on corresponding MRI based 3D ventricle reconstruction. (Scale bar (A), 40 µm; Scale bar (B), 1 mm)
Discussion
We present tools and protocols that can be used to evaluate the integrity of the brain’s ventricular system in mice and in humans. These tools, however, can also be applied to other brain structures or organ systems that undergo changes due to injury, disease, or during the process of aging 14,21,22. The strategies presented take advantage of software that allows the alignment of cross-sectional and longitudinal MRI sequences to generate 3D volume representations of specific regions or structures of interest. Longitudinal MRI sequences allow compilation of 3D volume changes that occur over time and can be extended to include total brain volume, for lateral ventricle to total brain volume ratios, and/or other brain structures (e.g., subthalamic nucleus 23 or corpus callosum white matter tracts using diffusion-weighted tensor imaging). Together, a comprehensive analysis of brain structural changes can be performed. Ultimately, to evaluate age-related and disease-related changes to brain structures a collection of multimodal imaging techniques are needed to provide the most accurate picture of brain health status 24. Documentation and compilation of critical structural data, together with subject-subject variability and the range of variability across subjects, could ideally be used to generate ultra-high field MRI atlases to best guide clinical diagnosis of tissue in health or disease.
Several critical steps are important to note in order to decrease acquisition and processing variability between and within subject data sets. Ideally to obtain matched data sequences, the same MRI scanner and sequence type should be used throughout the study. Postmortem brain tissue is often of varying quality; critical factors such as the cause of death, the postmortem interval, quality of the perfusion, and length of time in fixative all may affect staining efficacy and the need for an antigen retrieval protocol. In addition, the size of the human brain requires multiple slice sections with each slice requiring further manipulation to obtain tissue containing the regions of interest. Therefore, it is critical that the location and orientation of each section is clearly marked and recorded. Ultimately, the primary problem with postmortem tissue analysis is precisely that – it is postmortem – and therefore provides only a snapshot view of the tissue at the end of life. Improved multimodal imaging techniques is required for real-time analysis of changes to brain structures and morpho-functional information with cellular resolution.
Mouse studies allow interrogation of the human condition variable by variable and do not present many of the difficulties found when working with human tissue. Mouse studies used to model human disease or injury through analysis of causal structural dynamics may offer improvements for disease-model validation. However, species-specific differences should be noted. In our analysis of the lateral ventricles, it is important to note that mice retain an active stem cell niche along the lateral wall of the lateral ventricles and stem cell-mediated regenerative repair of the ventricle lining occurs in cases of modest ependymal cell loss, as found in aged mice or when limited ependymal cell denudation occurs through exposure of the ependyma to neuraminidase 19. In contrast, humans do not maintain a robust stem cell niche along the lateral ventricle walls 25-27 and little, if any, regeneration is likely. In contrast to aged mice, aged humans show extensive astrogliosis at the ventricle surface associated with ventricle expansion6. This level of denudation and ‘scarring’ of the ventricle lining can be modeled in mouse using neuraminidase to denude the ventricle lining of ependymal cells 6,28. In addition, it is important to recognize that vivarium-maintained mice do not experience infections, disease or trauma, presenting a much different medical history from most human subjects. Furthermore, mice typically do not show age-associated neurodegenerative disease, ventriculomegaly or periventricular gliosis. Therefore, to observe these phenotypes they have to be modeled. In the end, no animal model can fully recapitulate the ontogeny, pathophysiology, and symptomatic complexity of human disease and these limitations need to be acknowledged and properly communicated.
In summary, we present techniques and protocols to assess lateral ventricle volumes in mouse and human. Ventricles can be rendered in 3D and longitudinal analysis allows compilation of spatiotemporal volume changes. In addition, using paired brain tissue we demonstrate how cellular features can be linked to changes in ventricle volumes. Recent results demonstrating increased levels of Aquaporin-4 expression in areas of periventricular gliosis suggest edema along the ventricle surface. Therefore future investigations pairing FLAIR-MRI with tissue histology will improve our understanding of CSF and interstitial fluid exchange, and provide critical information about ventricle system health and how its deterioration affects various brain functions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
An NINDS Grant NS05033 (JCC) supported this work. The University of Connecticut RAC, SURF and OUR programs provided additional support.
Materials
| Name of the Materal/Equipment | Company | Catalog Number | Comments/Description |
| Phosphate buffered saline (PBS) | Life Technologies | 21600-069 | |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | 19210 | Use at 4% in PBS, 4 °C |
| Normal Horse Serum | Life Technologies | 16050 | 10% in PBS-TX (v/v) |
| Normal Goat Serum | Life Technologies | 16210 | 10% in PBS-TX (v/v) |
| Triton X-100 (TX) | Sigma-Aldrich | T8787 | 0.1% in PBS (v/v) |
| Vibratome | Leica | VT1000S | |
| Fluorescence Microscope | Zeiss | Imager.M2 | |
| Camera | Hamamatsu | ORCA R2 | |
| Microscope Stage Controller | Ludl Electronic Products | MAC 6000 | |
| Stereology software | MBF Bioscience | Stereo Investigator 11 | |
| Stereology software | ImageJ/NIH | NIH freeware | |
| 3D Reconstruction software | MBF Bioscience | Neurolucida Explorer | |
| Confocal Microscope | Leica | TCS SP2 | |
| MRI Software | |||
| Freesurfer | https://surfer.nmr.mgh.harvard.edu/fswiki/DownloadAndInstall | Segmentation and Volume | |
| ITK-Snap | http://www.itksnap.org/pmwiki/pmwiki.php | Segmentation and Volume | |
| Multi-image Analysis GUI (Mango) | http://ric.uthscsa.edu/mango/ | Longitudinal overlay | |
| Whole Mount Equipment | |||
| 22.5° microsurgical straight stab knife | Fisher Scientific | NC9854830 | |
| parafilm | |||
| wax bottom dissecting dish | |||
| pins | |||
| fine forceps | |||
| aquapolymount | |||
| Dissecting Microscope | Leica | MZ95 | |
| Whole Mount Antibodies | |||
| mouse anti-b-catenin | BD Bioschiences, San Jose, CA, USA | 1:250 | |
| goat anti-GFAP | Santa Cruz Biotechnology | 1:250 | |
| rabbit anti-AQP4 (aquaporin-4) | Sigma-Aldrich | 1:400 | |
| Coronal Antibodies | |||
| Anti-S100β antibody | Sigma-Aldrich | 1:500 | |
| 4’,6-diamidino-2-phenylindole (DAPI) | Life Technologies | D-1306 | 10 µg/mL in PBS |
References
- Del Bigio, M. R. Ependymal cells: biology and pathology. Acta Neuropathol. 119, 55-73 (2010).
- Johanson, C., et al. The distributional nexus of choroid plexus to cerebrospinal fluid, ependyma and brain: toxicologic/pathologic phenomena, periventricular destabilization, and lesion spread. Toxicol Pathol. 39, 186-212 (2011).
- Roales-Bujan, R., et al. Astrocytes acquire morphological and functional characteristics of ependymal cells following disruption of ependyma in hydrocephalus. Acta Neuropathologica. 124, 531-546 (2012).
- Cserr, H. F. Physiology of the choroid plexus. Physiol Rev. 51, 273-311 (1971).
- Iliff, J. J., et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Science Translational Medicine. 4, 147ra111 (2012).
- Shook, B. A., et al. Ventriculomegaly associated with ependymal gliosis and declines in barrier integrity in the aging human and mouse brain. Aging Cell. , (2013).
- Xie, L., et al. Sleep drives metabolite clearance from the adult brain. Science. 342, 373-377 (2013).
- Fazekas, F., et al. Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology. 43, 1683-1689 (1993).
- Meier-Ruge, W., Ulrich, J., Bruhlmann, M., Meier, E. Age-related white matter atrophy in the human brain. Ann N Y Acad Sci. 673, 260-269 (1992).
- Resnick, S. M., Pham, D. L., Kraut, M. A., Zonderman, A. B., Davatzikos, C. Longitudinal magnetic resonance imaging studies of older adults: a shrinking brain. The Journal Of Neuroscience : The Official Journal Of The Society For Neuroscience. 23, 3295-3301 (2003).
- Sener, R. N. Callosal changes in obstructive hydrocephalus: observations with FLAIR imaging, and diffusion MRI. Comput Med Imaging Graph. 26, 333-337 (2002).
- Sze, G., et al. Foci of MRI signal (pseudo lesions) anterior to the frontal horns: histologic correlations of a normal finding. AJR Am J Roentgenol. 147, 331-337 (1986).
- Tisell, M., et al. Shunt surgery in patients with hydrocephalus and white matter changes. Journal of Neurosurgery. 114, 1432-1438 (2011).
- Valdes Hernandez Mdel, C., et al. Automatic segmentation of brain white matter and white matter lesions in normal aging: comparison of five multispectral techniques. Magn Reson Imaging. 30, 222-229 (2012).
- Shook, B. A., Manz, D. H., Peters, J. J., Kang, S., Conover, J. C. Spatiotemporal changes to the subventricular zone stem cell pool through aging. The Journal of Neuroscience : The Official Journal Of The Society For Neuroscience. 32, 6947-6956 (2012).
- Mirzadeh, Z., Merkle, F. T., Soriano-Navarro, M., Garcia-Verdugo, J. M., Alvarez-Buylla, A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 3, 265-278 (2008).
- Mirzadeh, Z., Doetsch, F., Sawamoto, K., Wichterle, H., Alvarez-Buylla, A. The subventricular zone en-face: wholemount staining and ependymal flow. J Vis Exp. , (2010).
- Luo, J., Daniels, S. B., Lennington, J. B., Notti, R. Q., Conover, J. C. The aging neurogenic subventricular zone. Aging Cell. 5, 139-152 (2006).
- Luo, J., Shook, B. A., Daniels, S. B., Conover, J. C. Subventricular zone-mediated ependyma repair in the adult mammalian brain. J Neurosci. 28, 3804-3813 (2008).
- Marcus, D. S., Fotenos, A. F., Csernansky, J. G., Morris, J. C., Buckner, R. L. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. J Cogn Neurosci. 22, 2677-2684 (2010).
- Giorgio, A., De Stefano, N. Clinical use of brain volumetry. J Magn Reson Imaging. 37, 1-14 (2013).
- Caspers, S., et al. Studying variability in human brain aging in a population-based German cohort-rationale and design of 1000BRAINS. Front Aging Neurosci. 6, 149 (2014).
- Keuken, M. C., et al. Ultra-high 7T MRI of structural age-related changes of the subthalamic nucleus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 33, 4896-4900 (2013).
- Marti-Bonmati, L., Sopena, R., Bartumeus, P., Sopena, P. Multimodality imaging techniques. Contrast Media Mol Imaging. 5, 180-189 (2010).
- Bergmann, O., et al. The age of olfactory bulb neurons in humans. Neuron. 74, 634-639 (2012).
- Sanai, N., et al. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 478, 382-386 (2011).
- Wang, C., et al. Identification and characterization of neuroblasts in the subventricular zone and rostral migratory stream of the adult human brain. Cell Res. 21, 1534-1550 (2011).
- Carmen Gomez-Roldan, D. e. l., M, , et al. Neuroblast proliferation on the surface of the adult rat striatal wall after focal ependymal loss by intracerebroventricular injection of neuraminidase. The Journal of Comparative Neurology. 507, 1571-1587 (2008).

