Optimization of Synthetic Proteins: Identification of Interpositional Dependencies Indicating Structurally and/or Functionally Linked Residues
Summary
Synthetic protein sequences based on consensus motifs typically ignore co-evolving residues, that imply interpositional dependencies (IPDs). IPDs can be essential to activity, and designs that disregard them may result in suboptimal results. This protocol uses StickWRLD to identify IPDs and help inform rational protein design, resulting in more efficient results.
Abstract
Protein alignments are commonly used to evaluate the similarity of protein residues, and the derived consensus sequence used for identifying functional units (e.g., domains). Traditional consensus-building models fail to account for interpositional dependencies – functionally required covariation of residues that tend to appear simultaneously throughout evolution and across the phylogentic tree. These relationships can reveal important clues about the processes of protein folding, thermostability, and the formation of functional sites, which in turn can be used to inform the engineering of synthetic proteins. Unfortunately, these relationships essentially form sub-motifs which cannot be predicted by simple “majority rule” or even HMM-based consensus models, and the result can be a biologically invalid “consensus” which is not only never seen in nature but is less viable than any extant protein. We have developed a visual analytics tool, StickWRLD, which creates an interactive 3D representation of a protein alignment and clearly displays covarying residues. The user has the ability to pan and zoom, as well as dynamically change the statistical threshold underlying the identification of covariants. StickWRLD has previously been successfully used to identify functionally-required covarying residues in proteins such as Adenylate Kinase and in DNA sequences such as endonuclease target sites.
Introduction
Protein alignments have long been used to evaluate the similarity of residues in a protein family. Frequently the most interesting features of a protein (e.g., catalytic or other binding sites) are the result of protein folding bringing distal regions of the linear sequence into contact, and as a result these apparently unrelated regions in the alignment tend to evolve and change in a coordinated fashion. In other cases, the function of a protein can be dependent on its electrostatic signature, and mutations that affect the electronic dipole are compensated for by changes to distant charged residues. Allosteric effects can also induce long-range sequential and spatial dependencies between residue identities. Regardless of their origin, these functionally-required covariations of residues – inter-positional dependencies (IPDs) – may not be obvious with visual examination of the alignment (Figure 1). Identification of IPDs – as well as of which specific residues within those positions tend to covary as a unit – can reveal important clues about the processes of protein folding and the formation of functional sites. This information can then be used to optimize synthetic (engineered) proteins in terms of thermostability and activity. It has long been known that not all point mutations toward consensus provide improved stability or activity. More recently, proteins designed to take advantage of known IPDs in their sequence have been shown to result in greater activity than the same protein designed strictly from consensus1,2 (manuscript in preparation), similar to the idea of stabilizing point mutations3.
Unfortunately, traditional consensus-building models (e.g., majority rule) only capture IPDs by accident. Consensus and Position Specific Scoring Matrix methods are ignorant of IPDs and only 'correctly' include them in models, when the dependent residues are also the most popular residues for those positions in the family. Markov Chain models can capture IPDs when they are sequentially proximal, but their typical implementation ignores everything except immediate sequential neighbors, and even at their best, Hidden Markov Model calculations (see Figure 2) become intractable when dependencies are separated in the sequence by more than a dozen or so positions4. Since these IPDs essentially form “sub-motifs” which cannot be predicted by simple “majority rule” or even HMM-based consensus models5,6 the result can be a biologically invalid “consensus” which is not only never seen in nature but is less viable than any extant protein. Systems based on Markov Random Fields, such as GREMLIN7, attempt to overcome these problems. Additionally while sophisticated biological/biochemical techniques such as noncontiguous recombination3,8 can be used to identify essential protein elements by region, they require considerable time and bench work for single-base-pair precision to be achieved.
StickWRLD9 is a Python based program that creates an interactive 3D representation of a protein alignment that makes IPDs clear and easy to understand. Each position in the alignment is represented as a column in the display, where each column is comprised of a stack of spheres, one for each of the 20 amino acids that could be present in that position within the alignment. The sphere size is dependent on the frequency of occurrence of the amino acid, such that the user can immediately glean the consensus residue or the relative distribution of amino acids within that position by simply looking at the size of the spheres. The columns representing each position are wrapped around a cylinder. This gives every sphere representing a possible amino acid at each position in the alignment, a clear 'line of sight' to every other amino acid possibility at every other position. Prior to visualization, StickWRLD calculates the the correlation strength between all possible combinations of residues to identify the IPDs9. To represent IPDs, lines are drawn between residues which are coevolving at a higher, or lower than would be expected if the residues present in the positions were independent(IPDs).
Not only does this visualization show which sequence positions interact evolutionarily, but as the IPD edge lines are drawn between the amino acid spheres in each column, the user can quickly determine which specific amino acids tend to be coevolving at each position. The user has the ability to rotate and explore the visualized IPD structure, as well as dynamically change the statistical thresholds controlling the display of correlations, making StickWRLD a powerful discovery tool for IPDs.
Applications such as GREMLIN7 similarly display complex relational information between residues – but these relationships are computed via more traditional Markov models, which are not designed to determine any conditional relationships. As such, these are capable of being displayed as 2D projections. By contrast, StickWRLD can compute and display multi-node conditional dependencies, which may be obfuscated if rendered as a 2D graph (a phenomenon known as edge occlusion).
StickWRLD’s 3D view also has several other advantages. By allowing users to manipulate the visual – panning, rotating, and zooming – features that may be obfuscated or unintuitive in a 2D representation can be more easily seen in the 3D cylinder of StickWRLD. StickWRLD is essentially a visual analytics tool, harnessing the power of the human brain’s pattern recognition ability to see patterns and trends, and the ability to explore the data from various perspectives lends itself to this.
Protocol
1. Software Download & Installation
- Use a computer has an Intel i5 or better processor with at least 4 Gb of RAM, and is running Mac OS X or GNU/Linux (e.g., Ubuntu) OS. In addition, Python 2.7.610 and the wxPython 2.811, SciPy12, and PyOpenGL13 python libraries are required – download and install each from their respective repositories.
- Download StickWRLD as a zip archive containing all of the relevant Python scripts. Download the “fasta2stick.sh” script for converting standard FASTA DNA/protein sequence alignments to StickWRLD format.
- Extract the archive and put the resulting StickWRLD folder on your Desktop. Place the “fasta2stick.sh” script on the desktop as well.
2. Prepare the Alignment
- Create an alignment of the protein sequences using any standard alignment software (e.g., ClustalX14). Save the alignment on the desktop in FASTA format.
- Open the terminal application on the Mac or GNU/Linux computer and navigate to the desktop (the location of the “fasta2stick.sh” shell script) by typing cd ~/Desktop and pressing return. Execute the “fasta2stick.sh” script by typing ./fasta2stick.sh in the terminal. If the script does not execute, ensure that it is executable – in the terminal type chmod +x fasta2stick.sh to make the script executable.
- Follow the onscreen instructions provided by the script to specify the input file name (the file created in 1.2 above) and the desired output name. Save the output file (which is now in the correct format for StickWRLD) on the desktop.
3. Launching StickWRLD
- Navigate into the StickWRLD executables folder using the terminal application of the Mac or GNU/Linux computer. For example, if the StickWRLD folder is on the desktop, type cd ~/Desktop/StickWRLD/exec in the terminal.
- Launch StickWRLD by typing python-32 stickwrld_demo.py in the terminal.
- Verify that the StickWRLD Data Loader panel is visible on the screen (Figure 3).
4. Loading the Data
- Load the converted protein sequence alignment by pressing the “Load Protein…” button.
- Select the file created in step 3 above and press “Open”. StickWRLD will open several new windows, including “StickWRLD Control” (Figure 4) and “StickWRLD – OpenGL” (Figure 5).
- Select the “StickWRLD – OpenGL” window. Choose “Reset View” from the “OpenGL” menu to display the default StickWRLD visualization in a “top-down” view through the cylinder representing the data in the resizable OpenGL windows..
5. View Options
- Select the boxes for “Column Labels” and “Ball Labels” in the “StickWRLD Control” pane (Figure 4) to display values for columns and balls.
- Deselect the box for “Column Edges” in the “StickWRLD Control” pane to hide the column edge lines.
- Set the “Column Thickness” to 0.1 in the “StickWRLD Control” pane to draw a thin line through the columns, making it easier to navigate the 3D view. Press return to accept the change.
- Reset the view in the “StickWRLD – OpenGL” window as in step 5.3 above, then press the “full screen” button to maximize the view.
6. Navigation
- Rotate the 3D StickWRLD display by holding down the left mouse button while moving the mouse in any direction.
- Zoom the 3D StickWRLD display by holding down the right mouse button while moving the mouse up or down.
7. Finding Interpositional Dependencies (IPDs)
- Browse the view by panning and zooming as described in step 6. Coevolving residues exceeding the threshold requirements of both p and residual are connected via edge lines as seen in Figure 6. If there are too many or too few edges connecting residues, change the Residual threshold (on the “StickWRLD Control” pane) to show fewer, or more, edges.
- Increase the residual threshold on the StickWRLD Control Pane until no IPD edge lines are shown and slowly ramp down until relationships appear. Continue increasing the residual until you have a sufficient number of relationships to examine.
- Identify relationships that involve either residues of known interest (e.g., within a motif or binding/functional site) or residues that are distal to one another within the alignment (suggesting that they are proximal in the folded protein)
8. Selecting and Saving Findings
- Using command + left click on any edges of interest. The StickWRLD Control pane will indicate the columns and connect specific residues, e.g., “(124|G) (136|H)” (Figure 7). Solid lines represent positive associations; dashed lines represent negative associations.
- Press the “Output Edges” button on the “StickWRLD Control” panel to save a plain-text formatted file (edge_residual.csv) of all of the visible edges, including the joined residues and their actual residual values, in the /StickWRLD/exec/ directory.
Representative Results
StickWRLD has been used previously to detect interpositional dependencies (IPDs) between residues in both DNA3 and protein15-17 alignments. These co-evolving residues, while often distal from one another in the sequence alignment, are often proximal to one another in the folded protein. StickWRLD allows rapid discovery of residue-specific co-occurrence at such sites, e.g., an alanine at position “x” is strongly correlated to a threonine at position “y”. Such correlations can be indicative of provable structural relationships, and typically are sites that, by necessity, co-evolve. StickWRLD is able to detect these relationships even when more “traditional” approaches using HMMs to describe motifs fail. For example, analysis of the PFAM alignment of the ADK lid domain using StickWRLD reveals a strong positive correlation between cysteines (C) at positions 4 and 8 and a coordinated pair of C at positions 35 and 38. At the same time, StickWRLD showed a similar strong positive relationship between histidine (H) and serine (S) at 4 and 8, with a strong negative relationships between these and the C quartet at 4, 8, 35, and 38, and a strong positive relationship with aspartic acid (D) and threonine (T) at positions 35 and 38 respectively. Additional IPDs exist between the H,S,D,T motif and a T and G at position **** 10 and 29 in b subtilis **** highlighting the conditional nature of these IPDs – the tetracysteine motif does not 'care' about the identities at these two positions, while the hydrophilic H,S,D,T triad requires specific residues in these positions almost absolutely. These two completely different position-dependent residue motifs can fulfill the same role the ADK lid. As can be seen in Figure 6, a large cluster of IPDs, including a 3-node association between G (glycine) at position 132, Y (tyrosine) at position 135, and a P (proline) at position 141, is visible in the foreground (Figure 6A). In Figure 6B, the view has been skewed to position the user slightly above the cylinder, revealing an IPD between an H (histidine) at position 136 and an M (methionine) at position 29, 107 residues distant. A PFAM HMM-derived motif of the same domain (Figure 2), meanwhile, not only does not detect these as specifically co-occuring motif variants, but also defines the overall groupings in a biologically unsupported scheme16.

Figure 1. “Subway Map” representation of the B. subtilis Adenosine Kinase (ADK) Lid domain structure. Arrows indicate IPDs identified in the PFAM alignment of ADK Lid domain by StickWRLD. StickWRLD is able to correctly identify IPDs within a cluster of residues which are in close proximity in the folded protein. Of particular interest are the T and G pair at positions 9 and 29, which only form an IPD when the tetrad of residues at 4, 7, 24, and 27 is not C,C,C,C). Residue numbers displayed represents B. subtilis position and not PFAM alignment positions. Please click here to view a larger version of this figure.

Figure 2. Skylign18 Hidden Markov Model (HMM) Sequence Logo for the ADK lid domain. While HMMs are powerful tools for determining probabilities at each position as well as the contribution of each site to the overall model, the positional independence of HMMs makes them unsuitable for detecting IPDs. This model does not suggest any of the dependencies seen in the StickWRLD representations (Figure 6). Please click here to view a larger version of this figure.
Figure 3. The StickWRLD Data Loader. Users can choose from existing demo data or load their own data in the form of DNA or Protein sequence alignments.

Figure 4. The StickWRLD Control window. The Control pane allows the user to change various view properties as well as regulate the thresholds controlling the display of edge lines indicating relationships between residues (IPDs). Circled in red are the defaults that typically need to be adjusted for best viewing of any dataset. The Residual value sets the threshold of (observed-expected) for which connector/association lines are drawn. The controls for Column and Ball labels control whether or not the column position and residue values (e.g., “A” for arginine) are displayed. The Column Edge Line control toggles on and off the display of edge lines connecting columns – for dense data sets this is better turned off. The Column Thickness controls whether or not the column itself is displayed – setting this to a very small value (e.g., 0.1) will draw a line through the spheres in the column, making it easy to distinguish the columns from one another. Please click here to view a larger version of this figure.

Figure 5. Initial view of the StickWRLD OpenGL window with the Adenylate Kinase lid domain protein data set loaded. The initial perspective looks “down” through the cylinder comprised of the sequence alignment positions. The user can rotate the cylinder using left-mouse-click-drag, and zoom in/out using right-mouse-click-drag. The initial view is quite dense because the default display shows even small rates of co-evolution. For many proteins, at this setting, distinct modules can be detected, but even in densely co-evolving proteins the display can be rapidly and interactively simplified to find the most important IPDs using the StickWRLD interface. Please click here to view a larger version of this figure.

Figure 6. Closeup view of a StickWRLD visualization of the Adenylate Kinase lid domain protein. Here we have changed the default Residual to 0.2. This increases the threshold for display of inter-residue edges, showing fewer edges. The edges that remain indicate strongly associated IPDs. In addition the view has been rotated and zoomed to allow for easier viewing of the edges. (A) A large cluster of IPDs is visible in the foreground, including a 3-node association between G (glycine) at position 132, Y (tyrosine) at position 135, and a P (proline) at position 141. (B) The view has been skewed to position the user slightly above the cylinder, revealing an IPD between an H (histidine) at position 136 and an M (methionine) at position 29, 107 residues distant. Please click here to view a larger version of this figure.
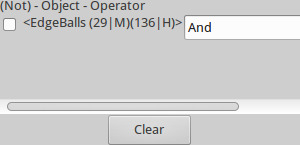
Figure 7. StickWRLD Control window lower-right information view. CTRL+Left clicking on an object (e.g., sphere or edge) in the OpenGL window displays the information for the object in the lower right of the StickWLRD Control window. Here we see the information for an IPD edge between a methionine at position 29 and a histidine at position 136.
Discussion
StickWRLD has been successfully used to identify such IPDs in the Adenylate Kinase Lid domain16, as well as associated DNA bases in rho-dependent terminators9, and a novel splice-site specificity in Archaeal tRNA intron endonuclease6 target sites. These IPDs were not detectable via a direct examination of the alignments.
StickWRLD displays each position of an alignment as a column of 20 “spheres”, where each sphere represents one of the 20 amino acid residues and the size of the sphere indicates the frequency of occurrence of that particular residue within that column (Figure 4). Columns are arranged in a cylinder, with edge lines connecting residues in different columns (indicating an IPD). These edge lines are only drawn if the corresponding residues are covarying at a frequency surpassing both the p-value (significance) and residual (expected – observed) thresholds.
Detection of co-occurring interdependent residues, or IPDs, in distal regions of a DNA or protein sequence alignment is difficult using standard sequence alignment tools6. While such tools generate a consensus, or motif, sequence, this consensus is in many cases a simple majority-rule averaging and does not convey covariation relationships that may form one or more sub-motifs – groups of residues that tend to co-evolve. Even HMM models, which are capable of detecting neighboring dependencies, cannot accurately model sequence motifs with distal IPDs5. The result is that the calculated consensus may in fact be a “synthetic” sequence not found in nature – and engineered proteins based on such computational consensus may not, in fact, be optimal. In fact, the Pfam HMM for ADK would suggest that a chimeric protein containing half of the tetracysteine motif, and half of the H,S,D,T motif, is functionally just as acceptable as any actually existing ADK. This is not the case, as such chimeras (and many other blendings of these motifs) Are catalytically dead4,19.
When looking for correlations, it is critical that the residual threshold be adjusted to allow for the discovery of relevant correlations by setting the threshold above the level at which any edges are seen and then gradually ramping the threshold back down. This ensures that only the most significant edges are considered initially.
An alternate approach is to start with the residual threshold set extremely low. This results in the display of all significant edges. From here the residual threshold can slowly be increased, allowing edges to drop out until patterns emerge. While this approach is less useful when looking for the inclusion of specific nodes (e.g., application of domain knowledge), it allows for the discovery of unexpected relationships using StickWRLD as a visual analytical tool to discover emerging patterns in the data visualization.
StickWRLD is limited primarily by the available memory of the system on which it is run as well as the resolution of the display device. While there is no theoretical limit to the number of data points StickWRLD can examine, and sequences up to 20,000 positions have been tested, in practice StickWRLD performs best with sequences up to around 1,000 positions.
The primary advantage of StickWRLD lies in its ability to identify groups of residues that covary with one another. This is a significant advantage over the traditional approach of the statistical consensus sequence, which is a simple statistical averaging and does not take coevolution into account. While in some cases covarying residues may simply be an artifact of phylogeny, even these residues have withstood the “test of selection”, and as such are unlikely to detract from the functionality of any protein engineered to include them.
While using StickWRLD to identify IPDs in a canonical DNA or protein sequence consensus/motif prior to engineering synthetic variants will reduce the potential for error and support rapid optimization of function, it should be noted that StickWRLD can be used as a generalized correlation identification tool and is not limited exclusively to protein data. StickWRLD can be used to visually discover the co-occurance of any variables in any properly encoded data set.
Disclosures
The authors have nothing to disclose.
Acknowledgements
StickWRLD was made possible in part through funding provided to Dr. Ray by the Research Institute at Nationwide Children’s Hospital, and by NSF grant DBI-1262457.
Materials
| Mac or Ubuntu OS computer | Various | NA | Any Mac or linux (e.g. Ubuntu) computer capable of running python & associated shell scripts |
| Python programming language | python.org | NA | Python version 2.7.6 or greater recommended |
| wxPython library | wxpython.org | NA | Latest version recommended |
| SciPy library | scipy.org | NA | Latest version recommended |
| PyOpenGL library | pyopengl.sourceforge.net | NA | Latest version recommended |
| StickWRLD Python scripts | NCH BCCM | NA | Available from http://www.stickwrld.org |
| fasta2stick.sh file converter | NCH BCCM | NA | Available from http://www.stickwrld.org |
| Protein and/or DNA sequence data | NA | NA | Samples available at http://www.stickwrld.org |
References
- Ray, W. C. Addressing the unmet need for visualizing conditional random fields in biological data. BMC. 15, 202 (2014).
- Sullivan, B. J., Durani, V., Magliery, T. J. Triosephosphate isomerase by consensus design: dramatic differences in physical properties and activity of related variants. Journal of molecular biology. 413, 195-208 (2011).
- Smith, M. A., Bedbrook, C. N., Wu, T., Arnold, F. H. Hypocrea jecorina cellobiohydrolase I stabilizing mutations identified using noncontiguous recombination. ACS synthetic biology. 2, 690-696 (2013).
- Ray, W. C. Understanding the sequence requirements of protein families: insights from the BioVis 2013 contests. BMC proceedings. 8, S1 (2014).
- Eddy, S. R. What is a hidden Markov model?. Nature biotechnology. 22, 1315-1316 (2004).
- Ray, W. C., Ozer, H. G., Armbruster, D. W., Daniels, C. J. Beyond identity – when classical homology searching fails, why, and what you can do about it. Proceedings of the 4th Ohio Collaborative Conference on Bioinformatics. , 51-56 (2009).
- Ovchinnikov, S., Kamisetty, H., Baker, D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife. 3, e02030 (2014).
- Trudeau, D. L., Lee, T. M., Arnold, F. H. Engineered thermostable fungal cellulases exhibit efficient synergistic cellulose hydrolysis at elevated temperatures. Biotechnology and bioengineering. 111, 2390-2397 (2014).
- Ray, W. C. MAVL and StickWRLD: visually exploring relationships in nucleic acid sequence alignments. Nucleic acids research. 32, W59-W63 (2004).
- . Python Language Reference v.2.7.6 Available from: https://www.python.org/download/releases/2.7.6/ (2014)
- . . PyOpenGL The Python OpenGL Binding. , (2014).
- Larkin, M. A. Clustal W and Clustal X version 2.0. Bioinformatics. 23, 2947-2948 (2007).
- Ozer, H. G., Ray, W. C. MAVL/StickWRLD: analyzing structural constraints using interpositional dependencies in biomolecular sequence alignments. Nucleic acids research. 34, W133-W136 (2006).
- Ray, W. C. MAVL/StickWRLD for protein: visualizing protein sequence families to detect non-consensus features. Nucleic acids research. 33, W315-W319 (2005).
- Ray, W. C. A Visual Analytics approach to identifying protein structural constraints. IEEE. , 249-250 (2010).
- Wheeler, T. J., Clements, J., Finn, R. D. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC bioinformatics. 15, 7 (2014).
- Perrier, V., Burlacu-Miron, S., Bourgeois, S., Surewicz, W. K., Gilles, A. M. Genetically engineered zinc-chelating adenylate kinase from Escherichia coli with enhanced thermal stability. The Journal of biological chemistry. 273, 19097-19101 (1998).

