Summary
We describe a protocol for the light-catalyzed generation of hydrogen peroxide — a cofactor for oxidative transformations.
Abstract
Oxidoreductases belong to the most-applied industrial enzymes. Nevertheless, they need external electrons whose supply is often costly and challenging. Recycling of the electron donors NADH or NADPH requires the use of additional enzymes and sacrificial substrates. Interestingly, several oxidoreductases accept hydrogen peroxide as electron donor. While being inexpensive, this reagent often reduces the stability of enzymes. A solution to this problem is the in situ generation of the cofactor. The continuous supply of the cofactor at low concentration drives the reaction without impairing enzyme stability. This paper demonstrates a method for the light-catalyzed in situ generation of hydrogen peroxide with the example of the heme-dependent fatty acid decarboxylase OleTJE. The fatty acid decarboxylase OleTJE was discovered due to its unique ability to produce long-chain 1-alkenes from fatty acids, a hitherto unknown enzymatic reaction. 1-alkenes are widely used additives for plasticizers and lubricants. OleTJE has been shown to accept electrons from hydrogen peroxide for the oxidative decarboxylation. While addition of hydrogen peroxide damages the enzyme and results in low yields, in situ generation of the cofactor circumvents this problem. The photobiocatalytic system shows clear advantages regarding enzyme activity and yield, resulting in a simple and efficient system for fatty acid decarboxylation.
Introduction
The climate change and the foreseeable depletion of renewable resources poses a serious threat to our society. In this context, enzyme catalysis represents a still not fully exploited potential for the development of sustainable and 'greener' chemistry1. Oxidoreductases have the capacity to catalyze the introduction and modification of functional groups under mild reactions conditions and belong to the most important biocatalysts2. Most redox transformations require the supply of external of cofactors such as NAD(P)H. Methods for cofactor regeneration have been applied in industrial scale. However, they still result in high process costs, which limits their application mostly to high-value products. Interestingly, several peroxidases3,4 and P450 monooxygenases5 accept electrons from hydrogen peroxide via the so-called peroxide shunt. While H2O2 is a cheap co-reagent, it is reportedly harmful for many enzymes. A steady in situ formation of low concentrations of hydrogen peroxide is a viable approach to drive the reaction without impairing the operational stability of enzyme.
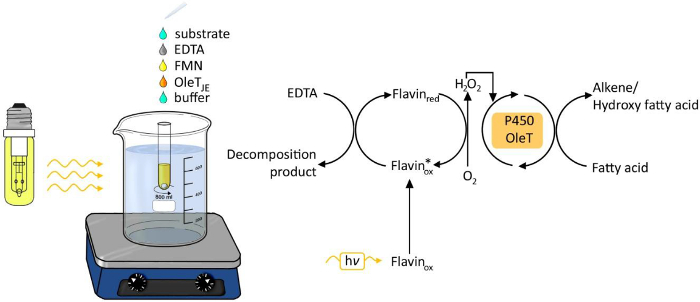
Figure 1. Experimental set-up of the photobiocatalytic decarboxylation of fatty acids by OleTJE. Please click here to view a larger version of this figure.
The use of light as energy source for chemical and biological processes has been receiving increasing attention in the last years6. Light-driven generation of hydrogen peroxide has emerged as an easy and robust method to supply hydrogen peroxide for redox transformations (Figure 1). A photocatalyst such as flavin adenine mononucleotide (FMN) allows the reduction of molecular oxygen to hydrogen peroxide, which then is used as cofactor for the enzymatic oxyfunctionalization reaction. Possible electron donors are ethylenediaminetetraacetic acid (EDTA), ascorbate or the inexpensive formiate. The method is generally applicable for H2O2-dependent enzymes, including peroxidases3,4 and P450 monooxygenases5.
We have recently investigated the application of a novel bacterial decarboxylase7 for the transformation of natural fats into olefins8. This would be a sustainable route for the synthesis of widely used platform chemicals from a bio-based source. The decarboxylase OleTJE from the gram-positive bacterium Jeotgalicoccus sp. catalyzes the oxidative decarboxylation of fatty acids and forms 1-alkenes as products. OleTJE is closely related to bacterial P450 monooxygenases and needs electrons from hydrogen peroxide for the reaction.
Unfortunately, addition of H2O2 to a solution of substrate and enzyme resulted in low conversions and a poor reproducibility of the results, presumably due to a harmful effect of hydrogen peroxide on the stability of OleTJE. Generation of a fusion protein with the NADPH-reductase RhFred made an NADPH-dependent decarboxylation possible.9 Nevertheless, the high price of NADPH and the current limited possibilities for a cost-efficient regeneration prompted us to investigate cheaper electron donors. Inspired by the similarity of OleTJE with P450 monooxygenases, we used the light-catalyzed generation of H2O2. We were pleased to obtain high conversions (up to >95%) using cell-free extracts or purified enzyme solutions.
With the example of fatty acid decarboxylation, we present a general protocol for light-driven enzymatic redox transformations using FMN as photocatalyst and hydrogen peroxide as cofactor. The presented methods include the production of the enzyme in recombinant cell of E. coli, purification of the enzyme, the application for the synthesis of 1-alkenes and the analysis of the reaction products.
Protocol
Caution: Please consult all relevant material safety data sheets (MSDS) before use. Several of the chemicals used in these syntheses are acutely toxic and carcinogenic. All steps involving harmful organic solvents, particularly the extraction of the reaction products and the derivatization of fatty acids must be carried out under a fume hood using personal protective equipment (safety glasses, gloves, lab coat, full-length pants, closed-toe shoes). All operations involving genetically modified organisms in this work require installations that are approved for handling of genetically modified organisms of the safety level S1.
1. Expression of the Recombinant Decarboxylase OleTJE in E. coli
- Preparation of the production strain
- Prepare a synthetic gene of OleTJE from Jeotgalicoccus and cloned into the expression vector pASK-IBA37plus as described.8
NOTE: To avoid degradation of fatty acids by enzymes from the bacterial metabolism, E. coli strain JW5020 from the Keio collection with a dysfunctional pathway for fatty acid degradation was used.
- Prepare a synthetic gene of OleTJE from Jeotgalicoccus and cloned into the expression vector pASK-IBA37plus as described.8
- Cultivation and induction of enzyme expression
- Prepare a pre-culture by inoculating the E. coli JW5020 OleT mutant from an LB-plate (containing 100 µg ml-1 ampicillin) in two 3 ml LB-medium in test tubes. Pipette ampicillin (100 µg ml-1) from a stock solution into the medium for selection and incubate at 37 °C for 15 hr in a shaking incubator at 180 rpm.
- Add 2 ml of the pre-cultures to two 200 ml LB-medium, containing ampicillin (100 µg ml-1), in 1 L shake flasks. Incubation proceeds at 37 °C and 180 rpm.
- Monitor the change in OD600nm after the inoculation. When the OD600 reaches a value between 0.6 and 0.8 induce the expression by adding tetracycline (0.2 µg ml-1). Incubate the cultures for 15 hr at 20 °C and 180 rpm.
NOTE: Since OleTJE contains a heme cofactor, addition of δ-Amino levulinic acid (0.5 mM) prior to induction is needed to provide the culture with a precursor for cofactor synthesis.
- Cell lysis
- Carry out the following steps on ice. Transfer the cultures by decanting or pipetting into centrifuge tubes and use a scale to ensure equal distribution. Centrifuge the cultures for 20 min at 12,000 x g at 4 °C.
- Carefully discard the supernatant and resuspend the pellets in 50 ml buffer (Tris 50 mM, NaCl 200 mM, pH 7.5) by pipetting and transfer the suspension in a conical centrifuge tube.
- After centrifugation for 15 min at 4,000 x g at 4 °C discard the supernatant, resuspend each pellet in 3 ml buffer by pipetting.
- Perform cell lysis by sonicating the solutions on ice (three cycles x 30 sec) leaving a 1 min pause in between the cycles. Centrifuge the solutions for 20 min at 15,000 x g at 4 °C to remove cell debris and transfer the supernatant into a conical centrifugation tube without disturbing the pellet by pipetting. Discard the pellet.
NOTE: One solvent fraction will be used directly for biocatalysis after small molecules are removed. The second fraction will be used for purification of the His-tagged OleTJE.
- Removal of small molecules of cell-extracts
- Before using the crude extract in biocatalysis remove small molecules which might interfere with hydrogen peroxide formation or electron transfer by pipetting 3 ml of cell-free extract into a centrifuge filter unit with a 10 kDa membrane and centrifuge at 4,000 x g at 4 °C. Resuspend the remaining protein in 3 ml Tris-HCl-buffer.
- Purification of OleT
- Apply 3 ml of the second cell extract to His Pur Ni-NTA spin columns, which have been equilibrated before with equilibration buffer (Tris 20 mM, NaCl 300 mM, imidazole 10 mM).
- Seal the columns with the bottom plugs and upper screw-caps and shake them lightly until the resin is distributed equally with the cell-free extract. Incubate the loaded columns for 30 min in an overhead shaker at 4 °C.
- Wash the columns with 1 ml of washing buffer (Tris 20 mM, NaCl 300 mM, imidazole 20 mM, pH 7.4) by centrifuging at 700 x g for 2 min. Repeat this step twice.
- Place the columns into a fresh conical centrifugation tube and add 1 ml of elution buffer (Tris 20 mM, NaCl 300 mM, imidazole 250 mM, pH 7.4) OleTJE . Centrifuge at 700 x g for 2 min and repeat this step two times.
NOTE: Imidazole will bind to nickel and thus remove OleTJE from the column. - Transfer the elution to a centrifuge filter unit (10 kDa membrane) and centrifuge at 4,000 x g at 4 °C to remove imidazole.
- Check the purity of the enzyme by an SDS-Page (15%).9 Mix 3 µl of the elution with a SDS-buffer (final concentration 1x) and incubate the solution at 95 °C for 5 min for denaturation. Subsequently apply the total amount on a SDS-Page. Run the gel at 35 mA using a commercial protein standard. Detect the protein at 50 kDa.
- To determine the protein concentration use a commercial available BSA-kit. Pipette 50 µl of diluted protein samples (1:100, 1:200, 1:500) and BSA standard (0, 20, 30, 40,50, 60, 80, 100 mg ml-1) in a 96-well plate. Add 200 µl of Bradford reagent, measure the absorbance at 595 nm after 15 min with a fluorimeter and calculate the concentrations using the standard curve.
2. Light-catalyzed Biotransformation
- Biocatalytic reactions without hydrogen peroxide addition
- Prepare a 10 ml stock solution of 10 mM stearic acid (Mr 284.5 g mol-1) by adding 10%(v/v) tergitol and 0.0284 g stearic acid to distilled water. Heat the solution in a heating chamber to 60 °C until the fatty acid is completely dissolved.
- Biocatalysis and sampling
- Prepare two 2 ml reaction mixtures containing 50 mM EDTA, 0.01 mM FMN, 0.5 mM stearic acid in Tris-HCl-buffer. Add a magnetic stirring bar for sufficient oxygen supply and place the glass tubes in a water bath, heated to 25 °C.
- Add 200 µg ml-1 of purified enzyme or 10% (v/v) cell-free extract to each of the reaction mixtures and illuminate them with a clear glass light bulb (120 W) in 15 cm distance of the reaction tubes.
- Take 200 µl samples at specific time points (0, 10, 30, 60 and 120 min and over night) stopping the reaction with 20 µl 37% HCl. Add 5 µl myristic acid (10 mM stock solution) as internal standard.
- Analysis of reaction products
- For extraction add 500 µl ethyl acetate to the samples twice, invert the tubes and centrifuge for 1 min at 13,000 x g.
- Take off 400 µl of the supernatant and let the solvent completely evaporate.
- Derivatize the carboxylic acids into trimethylsilylcarboxylic acid by adding 200 µl N-methyl-N-(trimethylsilyl)trifluoro acetamide (MSTFA). Incubate the solution at 60 °C for 30 min in order to convert hydroxyl groups to trimethylsilyl ethers.
- Determination of conversion by GC/FID
NOTE: Determine the formation of 1-heptadecene (RT: 8.34 min), α- and β-hydroxy stearic acid (RT: 12.05 and 12.1 min) and decrease of the substrate (RT: 11.15 min) using GC/FID.- Set the temperature profile described below Set the injection temperature to 340 °C. Hold the initial oven temperature is at 90 °C for 3.5 min and subsequently increase with 45 °C min-1. At 220 °C, hold the temperature for 2 min. After a repeated increase of 45 °C min-1 , hold the temperature at 280 °C for 2 min and then increase with 60 °C min-1.
NOTE: Maximum temperature of 330 °C is held for 1.44 min. - Inject 4 µl of the sample, with a split ratio of 5 to get rapid volatilization and homogenous mixing with the carrier gas helium.
NOTE: In dependence on the increasing temperature the substrate and products will enter the gas phase. The substances are detected by the GC/FID detector after passing the column at specific retention times and are displayed as peaks on the screen. Observe the peak formation on the monitor. - Calculate the concentration of formed product by the following formula:
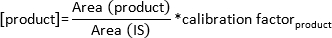
NOTE: The calibration factor has been determined specifically for this column for each substrate and product by calculating the standard curve from known amounts of the derivatized substances and their corresponding peak area.
- Set the temperature profile described below Set the injection temperature to 340 °C. Hold the initial oven temperature is at 90 °C for 3.5 min and subsequently increase with 45 °C min-1. At 220 °C, hold the temperature for 2 min. After a repeated increase of 45 °C min-1 , hold the temperature at 280 °C for 2 min and then increase with 60 °C min-1.
- Identify olefin and β-hydroxy acid peaks by GC/MS.
- Identify olefin and β-hydroxy acid peaks by GC/MS. Set the temperature profile. Set the injection temperature to 250 °C. Hold the oven temperature at 100 °C for 5 min and subsequently increase with 20 °C min-1.
NOTE: Maximum temperature is held for 7.5 min. - Set the mass spectrometer detector temperature to 250 °C and scan at 50 to 500 m/z in electron impact mode.
NOTE: Electron impact ionization removes a single electron resulting in the formation of a radical cation which will be passed forward for mass analysis. Due to the high intramolar energy covalent bonds within the molecule break creating fragments of lower m/z. These fragments form a fingerprint specific for a substance. - Inject 1 µl of the sample. Detect 1-Heptadecene after 11.4 min retention time by the corresponding fingerprint (Figure 2).
- Identify olefin and β-hydroxy acid peaks by GC/MS. Set the temperature profile. Set the injection temperature to 250 °C. Hold the oven temperature at 100 °C for 5 min and subsequently increase with 20 °C min-1.
Representative Results
While the addition of hydrogen peroxide to the reaction mixture resulted in low to moderate conversions (<10%), in situ generation of hydrogen peroxide increased the conversion up to 80% conversion. Analysis by GC/MS shows the formation of olefins from fatty acids (Figure 2).
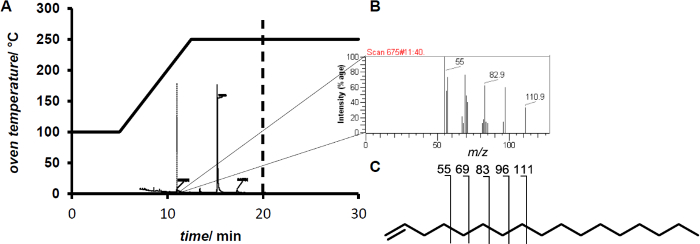
Figure 2. (A) Temperature profile for GC/MS measurements. (B) Fingerprint of the 1-heptadecene eluting after 11.4 min. (C) Characteristic secondary CnH2n-1 fragment ions of terminal olefins exemplary shown for 1-heptadecene. Please click here to view a larger version of this figure.
The light-catalyzed reaction achieved high conversions with cell-free extracts of E. coli. A purified enzyme solution showed a clear correlation between enzyme concentration and conversion. Increasing the concentration of the light-harvesting molecule FMN lead to higher conversions. However, increasing the concentration above 10 mM did not further accelerate the reactions, indicating that the amount of hydrogen peroxide is sufficiently available and no longer the limiting factor.
In addition to the decarboxylation of fatty acids, OleTJE also catalyzes a hydroxylation in β-position. In the conversion of stearic acid, the decarboxylation is about three times faster than the hydroxylation. In a typical experiment using a solution of 0.5 mM stearic acid and 10 µM FMN, 99% of the substrate were converted to a mixture of 1-heptadecene and 2-hydroxystearic acid with a ratio of 3.3:1 (Figure 3)8. An investigation of the substrate spectrum of the light-driven biocatalysis showed that for fatty acids with longer acyl chains, OleTJE preferentially catalyzes the decarboxylation, while the relative amount of hydroxyl-fatty acids in the product mixture increased with shorter chain length. Fatty acids shorter than myristic acid (C14) did not undergo decarboxylation.
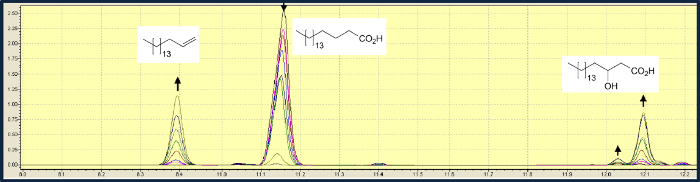
Figure 3. Gas chromatographic analysis of OleTJE-mediated decarboxylation of stearic acid (11.15 min) into 1-heptadecene (8.4 min), α-hydroxy stearic acid (12.04 min) β-hydroxy stearic acid (12.1 min). The change of the peak areas from samples taken over a time-course of 2 hr is shown. Please click here to view a larger version of this figure.
Surprisingly, the unsaturated acids oleic acid (C18:1d9 cis) and linoleic acid (C18:2d9d12) were not accepted as substrates, indicating that the twisted configuration of cis double bonds cannot be accommodated in a productive binding mode in the enzyme. Addition of oleic acid (10%) also prevented the conversion of stearic acid. Interestingly, stearic acid (C18:1d9 trans) was converted by OleTJE, yet with slightly lower activity then stearic acid.
Discussion
The light-driven generation of hydrogen peroxide can be applied for a range redox transformations, including peroxygenases3, chloroperoxidases10 and P450 monooxygenases5. It is an easy and practicable approach. In the long term, the use of visible light opens up the perspective to utilize sunlight for chemical transformations, which is a sustainable alternative for energy-rich reactions.
The method is applicable with purified enzyme or with cell-free extract. While the latter requires less cost and work, it should be noted that small molecules in the crude extract may interfere with the light catalyzed conversion. A practicable approach is to remove these small components with a micromembrane (i.e., by centrifugation in a Centrifugal filter unit or by dialysis). The concentration of the light-harvesting molecule FMN determines the concentration of the hydrogen peroxide. Depending on the affinity of the oxidoreductase, this concentration is decisive for the enzymatic activity. Another important factor is the concentration of the sacrificial electron donor EDTA. The most important parameter, however, is the operational stability and activity of the enzyme.
The olefinization of fatty acids is an elegant reaction for the conversion of bio-based fatty acids into olefins that belong to the major commodities for the chemical industry. The light-driven biocatalytic decarboxylation can be carried out at room temperature and at neutral pH, which offers clear advantages in terms of sustainability.
Our results show that in situ generation of hydrogen peroxide is a strategy to supply the cofactor without impairing enzyme stability, leading to a high conversion. Present methods for cofactor regeneration use agricultural products or petrol-based chemicals. Light-driven reactions are emerging as renewable alternative. Future research will be dedicated to methods for the substitution of the sacrificial reagent EDTA by cheaper molecules and to reduce the amount of the light-harvesting molecule FMN.
Disclosures
The authors have nothing to disclose.
Acknowledgements
R.K. and F.H. are grateful for the EU-commision for financial support within the Marie-Sklodowska ITN Biocascades (Nr. 634200).
Materials
| Chemicals | |||
| Ampicillin | Sigma Aldrich | 69-52-3 | |
| Bradford reagent | Roth | K015.1 | |
| BSA | Sigma Aldrich | 90604-29-8 | |
| DMSO | Sigma Aldrich | 67-68-5 | |
| Ethyl acetate | Fisher Chemical | 141-78-6 | |
| Ethylenediaminetetraacetic acid (EDTA) | Roth | 8043.1 | |
| Riboflavin 5-monophosphate sodium salt hydrate | Sigma Aldrich | 130-40-5 | |
| Hydrochlorid acid 37% | Sigma Aldrich | 7647-01-0 | |
| Hydrogen peroxide 30% | Sigma Aldrich | 7722-84-1 | |
| δ-Amino levulinic acid | Sigma Aldrich | 5451-09-2 | |
| N-Methyl-N-(Trimethylsilyl)trifluoro acetamide (MSTFA) | Sigma Aldrich | 24589-78-4 | |
| Myristic acid >99% | Sigma Aldrich | 208-875-2 | |
| Imidazole | Sigma Aldrich | 288-32-4 | |
| Sodium chloride | Fisher Chemical | 7647-14-5 | |
| Stearic acid >99% | Sigma Aldrich | 57-11-4 | |
| Tetracycline | Sigma Aldrich | 60-54-8 | |
| Tergitol | Sigma Aldrich | MFCD01779855 | |
| Tris(hydroxymethyl)-aminomethan | Sigma Aldrich | 77-86-1 | |
| Name | Company | Catalog Number | Comments |
| Device | |||
| Incubator shaker | G-25CK | New Brunswick Scientific | |
| Ecotron | Infors HT | ||
| Centrifugation | Labofuge 400R | Heraeus | |
| RC 5B Plus | Sorvall | ||
| Fresco 17 | Thermo Scientific | ||
| Centrifugation rotors | SS34 | Sorvall | |
| SLA | Sorvall | ||
| Clean bench | Envirco | Ceag Schirp Reinraum technik | |
| Column GC-FID | CP-Sil 5CB (30 m x 0.25 mmx 0.25 µm) | Agilent Technologies | |
| Column GC-MS | FactorFour Capillary Coloumn (VF-5 ms + 5 m EZ Guard) | Varian | |
| GC-FID | GC-2010 plus | Shimadzu | |
| GC-MS | IST-40 | Varian | |
| Magnetic stirrer | RCT classic | IKA | |
| pH meter | SevenEasy | Mettler toledo | |
| Sonicator | Branson Sonifier 250 | Branson | |
| Spectral photometer | FLUOstar Omega | BMG Labtech | |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| Affinity chromatography column | His Pur Ni-NTA spin column | Thermo Scientific | |
| Centricon | Vivaspin turbo 15 | VWR International | |
| Microtiter plates | 96 Well Multiply®PCR Plates | Sarstedt |
References
- Kourist, R., Domìnguez de Marìa, P., Miyamoto, K. Biocatalytic strategies for the asymmetric synthesis of profens – recent trends and developments. Green Chem. , 2607-2618 (2011).
- Holtmann, D., Fraaije, M. W., Arends, I. W., Opperman, D. J., Hollmann, F. The taming of oxygen: biocatalytic oxyfunctionalisations. Chemical Comm. 50, 13180-13200 (2014).
- Churakova, E., et al. Specific photobiocatalytic oxyfunctionalization reactions. Ang. Chem. In. Ed. 123, 10904-10907 (2011).
- Hollmann, F., Arends, I., Buehler, K. Biocatalytic Redox Reactions for Organic Synthesis: Nonconventional Regeneration Methods. ChemCatChem. 2, 762-782 (2010).
- Girhard, M., Kunigk, E., Tihovsky, S., Shumyantseva, V. V., Urlacher, V. B. Light-driven biocatalysis with cytochrome P450 peroxygenases. Biotechnol. Appl. Biochem. 60, 111-118 (2013).
- Bartsch, M., et al. Photosynthetic production of enantioselective biocatalysts. Microb. Cell. Fact. 14, 53 (2015).
- Rude, M. A., et al. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718-1727 (2011).
- Zachos, I., et al. Photobiocatalytic decarboxylation for olefin synthesis. Chem. Comm. 51, 1918-1921 (2015).
- Liu, Y., et al. Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol. Biofuels. 7, 28 (2014).
- Perez, D. I., Grau, M. M., Arends, I. W., Hollmann, F. Visible light-driven and chloroperoxidase-catalyzed oxygenation reactions. Chem. Comm. 40 (44), 6848-6850 (2009).

