Radiolabeling and Quantification of Cellular Levels of Phosphoinositides by High Performance Liquid Chromatography-coupled Flow Scintillation
Summary
Phosphoinositides are signaling lipids whose relative abundance rapidly changes in response to various stimuli. This article describes a method to measure the abundance of phosphoinositides by metabolically labeling cells with 3H-myo-inositol, followed by extraction and deacylation. Extracted glycero-inositides are then separated by high-performance liquid chromatography and quantified by flow scintillation.
Abstract
Phosphoinositides (PtdInsPs) are essential signaling lipids responsible for recruiting specific effectors and conferring organelles with molecular identity and function. Each of the seven PtdInsPs varies in their distribution and abundance, which are tightly regulated by specific kinases and phosphatases. The abundance of PtdInsPs can change abruptly in response to various signaling events or disturbance of the regulatory machinery. To understand how these events lead to changes in the amount of PtdInsPs and their resulting impact, it is important to quantify PtdInsP levels before and after a signaling event or between control and abnormal conditions. However, due to their low abundance and similarity, quantifying the relative amounts of each PtdInsP can be challenging. This article describes a method for quantifying PtdInsP levels by metabolically labeling cells with 3H-myo-inositol, which is incorporated into PtdInsPs. Phospholipids are then precipitated and deacylated. The resulting soluble 3H-glycero-inositides are further extracted, separated by high-performance liquid chromatography (HPLC), and detected by flow scintillation. The labeling and processing of yeast samples is described in detail, as well as the instrumental setup for the HPLC and flow scintillator. Despite losing structural information regarding acyl chain content, this method is sensitive and can be optimized to concurrently quantify all seven PtdInsPs in cells.
Introduction
Phosphoinositides (PtdInsPs) are important signaling phospholipids that help regulate a variety of cellular functions, including signal transduction, membrane trafficking and gene expression, which then modulate higher-order cell behavior such as cell division, organelle identity and metabolic activity1-3. There are seven species of PtdInsPs that are derived from the phosphorylation of the 3, 4, and/or 5 positions of the inositol head group of phosphatidylinositol (PtdIns), the parent phospholipid. Importantly, the seven PtdInsPs are unequally distributed and the local concentration of each PtdInsP species can increase or decrease at specific subcellular sites where they bind to a distinct set of protein effectors, which together permits each PtdInsP to control the identity and function of its host membrane3,4. In addition, the levels of each PtdInsP need to be tightly controlled since this can significantly impact the signal intensityproduced by a PtdInsP. The localization and levels of each PtdInsP depends on the targeting and activity of numerous lipid kinases, phosphatases and phospholipases that mediate the synthesis and turnover of each PtdInsP3,4. Hence, misregulation of the PtdInsP regulatory machinery can perturb cell function, leading to diseases such as cancer and degenerative diseases2,5,6. To fully understand the roles and functions of PtdInsPs and their regulatory machinery, both microscopy-based and biochemical-based techniques have been developed to track and quantify PtdInsPs.
In many cases, PtdInsPs bind to their protein effectors via a specific protein domain7-9. These protein modules often retain their proper fold and lipid recognition properties when expressed separately from the entire protein. This gave rise to PtdInsP probes by fusing a specific PtdInsP-binding protein domain to a fluorescent protein (FP) like green fluorescent protein (GFP) for the subcellular detection of PtdInsPs by microscopy. Indeed, many studies have used FP-fused PtdInsP-binding protein modules to identify the localization and dynamics of specific PtdInsP species by live-cell imaging1,10. For example, the Pleckstrin homology (PH) domain of phospholipase C δ1 (PLCδ1) fused to GFP specifically recognizes the phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] on the plasma membrane, whereas tandem copies of the FYVE domain of early endosome antigen 1 (EEA1) has been employed to track phosphatidylinositol-3-phosphate (PtdIns3P) on endosomes10-13. Overall, microscopy-based techniques are great to visualize PtdInsP localization and dynamics, but there are several caveats including that PtdInsP-binding domains may also interact with additional factors other than the target PtdInsP species and that they cannot detect changes below cytosolic fluorescence of the FP-probes.
Biochemical techniques including thin-layer chromatography, mass spectrometry and radioisotope labeling can also be used to characterize and quantify the levels of each PtdInsP14-16. These methods require the isolation of lipids for the detection of cellular levels of PtdInsPs. Mass spectrometry can be used to characterize phospholipids from lipid extracts and is invaluable for determining the acyl chain composition of PtdInsPs14,17. However, mass spectrometry is mostly semi-quantitative and it remains difficult to resolve and concurrently quantify PtdInsP species of the same molecular weight14,17. In comparison, radioisotope labeling of PtdInsPs followed by high performance liquid chromatography (HPLC)-coupled flow scintillation is useful for the separation and concurrent quantification of all seven species of PtdInsPs18. The use of HPLC with a strong anion exchange (SAX) column achieves separation based on molecular weight, charge and shape, thus fractionating deacylated PtdInsPs (Gro-InsPs) even of the same molecular weight and charge. Coupling the HPLC eluent to a flow scintillator then generates radioactive-based signal peaks for each Gro-InsPs species relative to the original parent glycerol-inositol (Gro-Ins)18. This ultimately corresponds to relative levels of PtdInsPs in cells.
Radiolabeling of PtdInsPs and HPLC-coupled flow scintillation is a useful tool to investigate the regulation and function of phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2], a PtdInsP that only constitutes ~0.1-0.3% of PtdIns16,19,20. Synthesis of PtdIns(3,5)P2 is performed by the PtdIns3P 5-kinase called Fab1 in yeast and PIKfyve in mammals21. This reaction is counteracted by a PtdIns(3,5)P2 5-phosphatase called Fig4 in yeast or mFig4/Sac3 in mammals22-24. Interestingly, both PIKfyve/Fab1 and mFig4/Fig4/Sac3 exist in a single complex and are regulated by the scaffolding adaptor protein, Vac14 in yeast or mVac14/ArPIKfyve in mammals25,26. In VAC14-deleted yeast cells, Fab1 does not function efficiently leading to a 90% decrease in PtdIns(3,5)P2 levels27,28. On the other hand, Atg18 is a PtdIns(3,5)P2 effector protein that may control vacuolar fission 29. Atg18 is also a negative regulator of PtdIns(3,5)P2 since the deletion of its gene, ATG18, causes a 10 to 20-fold increase in PtdIns(3,5)P2 levels29,30. Overall, changes in the levels of PtdIns(3,5)P2 severely impacts the function of the yeast vacuole and the mammalian lysosomes, consequently affecting processes such as membrane trafficking, phagosome maturation, autophagy and ion transport 6,19,21,31.
This article describes the process of radioisotope labeling of PtdInsPs with 3H-myo-inositol in yeast to detect the relative levels of PtdIns(3,5)P2 in wild-type, vac14Δ and atg18Δ yeast strains. Using this as an example, the resolving capabilities of HPLC for the separation of individual PtdInsPs as well as the sensitivity of flow scintillation for detection of trace amount of 3H-myo-inositol is shown. We also elaborate on how one might optimize the methodology for labeling and separating 3H-labelled PtdInsPs from mammalian cells, whose samples tend to be more complex since these cells possess all seven PtdInsP species.
Protocol
Note: The text below describes in detail a method to measure PtdInsPs in yeast. It provides the experimental details for labeling yeast cells with 3H-myo-inositol, extracting and deacylating lipids and an HPLC-elution protocol to fractionate and quantify deacylated PtdInsPs. Please note that labeling, deacylation, resolution and quantification of PtdInsPs in mammalian cells require optimization and longer HPLC-elution profiles. These details can be found elsewhere, though we discuss some of aspects in the Discussion. Overall, the methodology to extract lipids, deacylate and extract the water-soluble Gro-InsPs from yeast is given and is illustrated in Figure 1.
1. Protocol for Labeling and Analysing PtdInsPs in Yeast
- Cell culture and radiolabeling
- Grow a 20 ml liquid culture of the yeast strain SEY6210 at 30 °C with constant shaking in complete synthetic media to mid-log phase or an optical density at 600 nm (OD600) of approximately 0.6-0.7.
Note: Do not use YPD media as this may affect 3H-myo-inositol incorporation. This culture size should provide sufficient material for 2-3 HPLC runs. - Pellet a total of 10-14 OD of the yeast cells (note that 1 ml culture with an OD600 = 1 contains ~1×107 cells) by centrifugation at 800 x g for 5 min in a 12 ml round-bottom tube. Resuspend with 2 ml of inositol-free media (IFM) (see Table 1 for IFM composition).
- Repeat centrifugation and aspirate the media. Resuspend the pellet with 440 µl of IFM and incubate at RT for 15 min.
- Add 60 µl of 3H-myo-inositol (1 µl = 1 µCi) to each sample and incubate at the growing temperature of 30 °C for 1 to 3 hr with constant shaking.
Caution: 3H-myo-inositol is a radioactive material that should be handled after appropriate training. In addition, all consumables that make contact with 3H-containing material should be disposed appropriately and designated as radioactive waste.
Note: The growth temperature can be changed as required as long as all strains to be compared are grown at the same temperature. - Transfer the cell suspension (500 µl) to a microcentrifuge tube containing 500 µl of 9% perchloric acid and 200 µl of acid washed glass beads. Mix by inversion and incubate on ice for 5 min.
- Vortex the sample at top speed for 10 min and transfer the lysates to a new microcentrifuge tube using a gel-loading tip to avoid the glass beads.
- Pellet the sample at 12,000 x g for 10 min at 4 °C and aspirate the supernatant.
- Resuspend the pellet by bath sonication in 1 ml of ice-cold 100 mM EDTA. Pellet again as before.
- Grow a 20 ml liquid culture of the yeast strain SEY6210 at 30 °C with constant shaking in complete synthetic media to mid-log phase or an optical density at 600 nm (OD600) of approximately 0.6-0.7.
- Deacylation and extraction
- Freshly prepare 5.0 ml of the deacylation reagent by combining 2.3 ml of methanol, 1.3 ml of 40% methylamine, 0.55 ml of 1-butanol and 0.80 ml of water. Invert to mix.
- Aspirate the EDTA and sonicate the pellet in 50 µl of water.
- Add 500 µl of the deacylation reagent and sonicate to mix. Incubate at RT for 20 min.
- Heat lipids for 50 min at 53 °C in a heat block. Dry the samples completely by vacuum centrifugation over 3 hr or O/N.
- Ensure that a chemical trap is installed between the vacuum-centrifuge and the vacuum pump to prevent vapours from entering the air.
- Resuspend the pellet with 300 µl of water by bath sonication and incubate at RT for 20 min. Dry the samples by vacuum centrifugation for 3 hr or O/N. Repeat this step once more.
- Sonicate the pellet with 450 µl of water. This is the aqueous phase during extraction.
- Freshly prepare 10 ml of the extraction reagent by adding 8.0 ml of 1-butanol, 1.6 ml of ethyl ether and 0.40 ml of ethyl formate. Invert to mix.
- Add 300 µl of the extraction reagent to the aqueous phase. Vortex the mixture at top speed for 5 min and separate the layers by centrifugation at top speed (18,000 x g) for 2 min.
- Collect the bottom aqueous layer into a new microfuge tube while avoiding the top organic layer, the interface, and the pellet. Repeat extraction of the aqueous phase twice more with fresh extraction reagent.
- Completely dry the collected aqueous layer by vacuum centrifugation. Disperse the pellet in 50 µl of water by bath sonication.
- Add 2 µl of each sample to 4 ml of scintillation fluid in a 6 ml polyethylene scintillation vial. Determine the number of counts (in CPM) in each sample by liquid scintillation using an open window.
- Store the samples at -20 °C until ready for HPLC.
- Separation of 3H-glycero-inositides by HPLC
- Prepare buffer A by filtering 1 L of ultrapure water with a bottle top 0.22 µm vacuum filter, followed by degassing.
- Prepare buffer B by making a 1 L solution of 1 M ammonium phosphate dibasic (APS, MW 132.06) in water and adjust the pH to 3.8 with phosphoric acid. Filter the ammonium phosphate with a bottle top 0.22 µm vacuum filter, followed by degassing.
- Assemble the injection vial by outfitting a 2 ml vial with a spring-loaded 250 µl small volume insert. Load 10 million CPM and add water for a total volume of 55 µl. Prepare a blank vial with 55 µl of water.
- Cap the vials with screw caps outfitted with a PTFE/silicone septa (red side facing the inside of the vial). Place each vial in the auto-sampling tray of the HPLC, starting with the blank vial.
- Use an HPLC and associated software that controls buffer flow, degasses buffers and controls sample injection. Use an online flow scintillator and its associated software to regulate the scintillant flow rate and to monitor the 3H-decay signal.
- Use a SAX liquid chromatography column with dimensions of 250 mm x 4.6 mm and containing 5 µm silica resin in the HPLC. Outfit the column with a column guard to prevent injection of contaminants.
- Install a 3H-compatible 500 µl flow cell in the flow scintillator.
NOTE: Fractionation can be done with an Agilent 1200 infinity series HPLC system controlled by Chemstation software or with other commercially available HPLC systems. Flow scintillation of the HPLC eluent can be done with a β-RAM flow scintillator controlled by the Laura software or another commercially available flow scintillator or compatible software.
- Initialize the quaternary pump at a flow rate of 1.0 ml/min with 100% buffer A.
- Set up an equilibration profile on the HPLC to control the gradient of Buffers A and B (call this "Protocol A equilibration"): 1% Buffer B for 5 min, 1-100% B for 5 min, 100% B for 5 min, 100-1% B for 5 min, 1% B for 20 min. Set the pressure limit at 400 bar, 1.0 ml/min flow rate, and 30 min run time.
- Set up an elution profile (Figure 2) on the HPLC to control the gradient of Buffers A and B (call this "Protocol A"): 1% B for 5 min, 1-20% B for 40 min, 20-100% B for 10 min, 100% B for 5 min, 100-1% B for 20 min, 1% B for 10 min. Set the pressure limit at 400 bar, 1.0 ml/min flow rate, and 90 min run time.
- Set up a detection protocol on the flow scintillator (call this "Protocol A detection") to run for 60 min, with a scintillation fluid flow rate of 2.5 ml/min and an 8.57 sec dwell time.
- Program an automated injection sequence on the HPLC to begin with the water blank on "Protocol A equilibration", followed by each radiolabelled sample on "Protocol A". Initialize the sequence when ready.
- While the run is still on the equilibration protocol, create a batch on the flow scintillator to measure all of the samples with "Protocol A detection." The injection of each new sample will initialize "Protocol A" on the HPLC while triggering the flow scintillator to begin "Protocol A detection."
- At the completion of all runs, flush the HPLC, the column and flow scintillator with 100% Buffer A for 30 min at 1.0 ml/min flow rate.
- Data analysis
- Use Laura software or any other software that can quantify the chromatography spectra. The steps described in this section are depicted in Figure 3.
- Open the files by clicking "File", "Open", and select the raw data file for each sample.
- In the "chromatograms" tab, zoom in to stretch each of the lesser peaks (about 1000 counts [y-axis]) while retaining the time resolution. Zoom into each individual peak if necessary.
- Highlight each of the peaks for analysis using the "Add ROI" tool. Identify peaks by time of elution: Parental Gro-Ins at 10 min, Gro-Ins3P at 18 min, Gro-Ins4P at 20 min, Gro-Ins(3,5)P2 at 29 min and Gro-Ins(4,5)P2 at 32 min.
- In the "regions tables" tab, record the "area (counts)" of each peak. Note the "start (mm:ss)" time and "end (mm:ss)" time of each peak.
- For background subtraction, highlight a region adjacent to the peak spanning the same amount of time. Subtract the number of counts from the corresponding peak.
- Normalize the area of each peak against parental Gro-Ins (expressed as "% of total PtdIns"). Then, normalize each of the peaks in each experimental condition against the control condition (expressed as "n-fold increase compared to control").
NOTE: Normalization of each peak can also be done against total counts but since the parental Gro-Ins is much more abundant than all other PtdInsPs the results tend to be similar. - Export the data for the chromatographs by pressing "File", "Save as…", and choose the "CSV (comma separated)" file format. Plot the data on a spreadsheet and present as necessary.
Representative Results
Using this method, yeast PtdInsPs were metabolically labelled with 3H-myo-inositol. After labeling, the phospholipids were precipitated with perchloric acid, followed by phospholipid deacylation and extraction of the water-soluble Gro-InsPs (Figure 1). At this stage, it is important to quantify the total radioactive signal associated with the extracted Gro-InsPs by liquid scintillation to ensure sufficient signal-to-noise ratio for the very low-abundance PtdInsPs like PtdIns(3,5)P2; a total of 5-10 million CPMs should be injected. Since yeast cells exhibit only four PtdInsPs, resolution can be achieved with a 1 hr elution method as described (Figure 2).
Here, wild-type, atg18Δ and vac14Δ yeast mutants were labelled and processed to analyse changes in PtdIns(3,5)P2, the least abundant of the PtdInsPs in yeast16,19,27. As observed in Figure 4A-C, the elution profile for all three strains produced 3H-associated signal peaks. The parental Gro-Ins peak elutes first (8-9 min; shown in Figure 3, but not in Figure 4 since Gro-Ins overshadows the signal of the phosphorylated species), followed by Gro-Ins3P (~18 min), Gro-Ins4P (~20 min), Gro-Ins(3,5)P2 (~29 min) and Gro-Ins(4,5)P2 (~32:00 min), which respectively corresponds to the acylated phospholipids PtdIns, PtdIns3P, PtdIns4P, PtdIns(3,5)P2 and PtdIns(4,5)P2. There are a couple of additional minor peaks at 4 min and one that tends to immediately follow the parent Gro-Ins (10 min); these likely represent inositol and a non-glycerated inositide phosphates (Figure 3). The elution pattern is consistent and characteristic, though the exact time may vary when employing a different HPLC system and/or a new column.
The peak associated with PtdIns(3,5)P2 (Figure 4, red arrow) undergoes the most dramatic change between wild-type, atg18Δ and vac14Δ yeast strains. Relative to wild-type cells (A), there is a very large Gro-Ins(3,5)P2-associated signal in atg18Δ cells and a smaller peak in vac14Δ yeast cells. In order to quantify the total radioactive signal associated with each peak, the counts were integrated under the area of the peak (Figure 3). In addition, since absolute radioactive signal varies between samples, the parental Gro-Ins species was used as an internal control by normalizing against it (one can also choose to normalize against total counts). By doing this, the data suggest that PtdIns(3,5)P2 constitutes only 0.07% of PtdIns in wild type yeast cells, while PtdIns(3,5)P2 corresponds to 1.2% of the parental PtdIns in atg18Δ cells, or a dramatic 17-fold increase in PtdIns(3,5)P2 relative to wild-type cells (Figure 4A, B and D). In comparison, PtdIns(3,5)P2 levels was only 0.01% of the PtdIns signal in VAC14-deleted yeast cells, an 85% loss in PtdIns(3,5)P2 (Figure 4C and D). Overall, these measurements are in agreement with published results showing that PtdIns(3,5)P2 is about 0.1% of PtdIns in wild-type cells, 90% reduced in vac14D, and 10 to 20-times higher in atg18D cells relative to wild-type cells27,29.
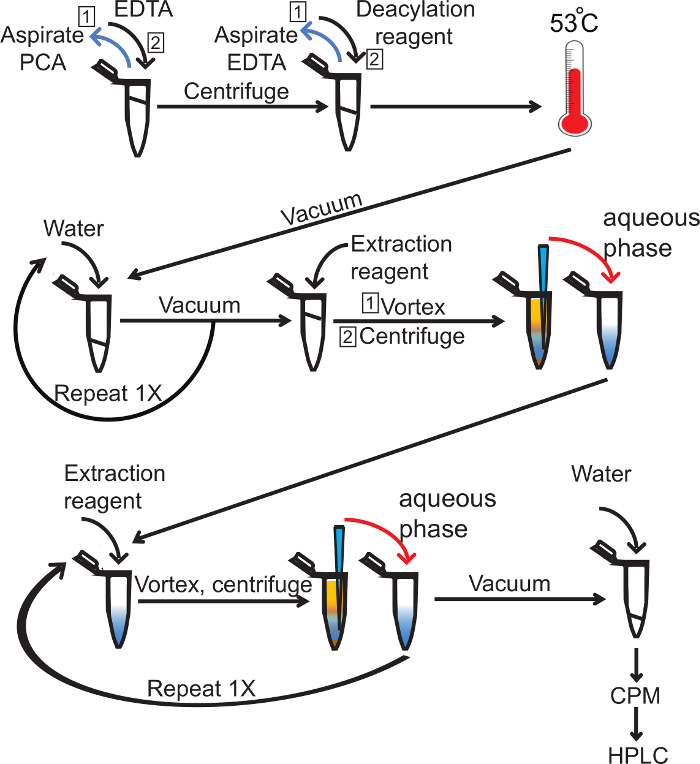
Figure 1. Deacylation and extraction of 3H-Gro-InsP. Radiolabeled lipid precipitate in perchloric acid is washed with aqueous solution of EDTA. This is then aspirated and deacylation reagent added and incubated at 53 °C. The deacylation reaction mixture is then vacuum dried and washed with water twice, followed by the addition of extraction reagent. This is then vortexed, centrifuged and the aqueous phase is collected. The extraction step is repeated three times total to remove organic and insoluble contaminants. The water-soluble fraction (blue), which includes 3H-Gro-InsP, is then vacuum dried and rehydrated with 60 µl of water. The amount of radioactivity is determined by liquid scintillation (CPM) and equal counts across samples are loaded into the HPLC. Please click here to view a larger version of this figure.
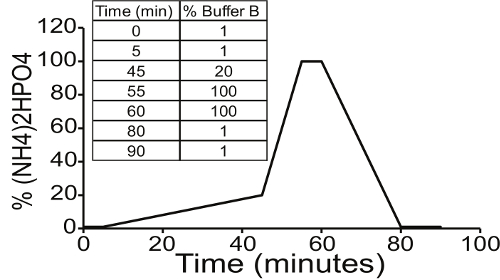
Figure 2. Gradient of ammonium phosphate buffer for elution of 3H-Gro-InsP. Resolution of Gro-InsP by strong anion exchange chromatography depends on the application of ammonium phosphate dibasic, pH 3.8 (Buffer B). The choice of gradient depends on the mixture of Gro-InsP species available for separation. Protocol A is used for yeast samples, which possess only four PtdInsPs. Resolution of each of the four peaks is sufficient with a rapid increase in (NH4)2HPO4 concentration. Please click here to view a larger version of this figure.
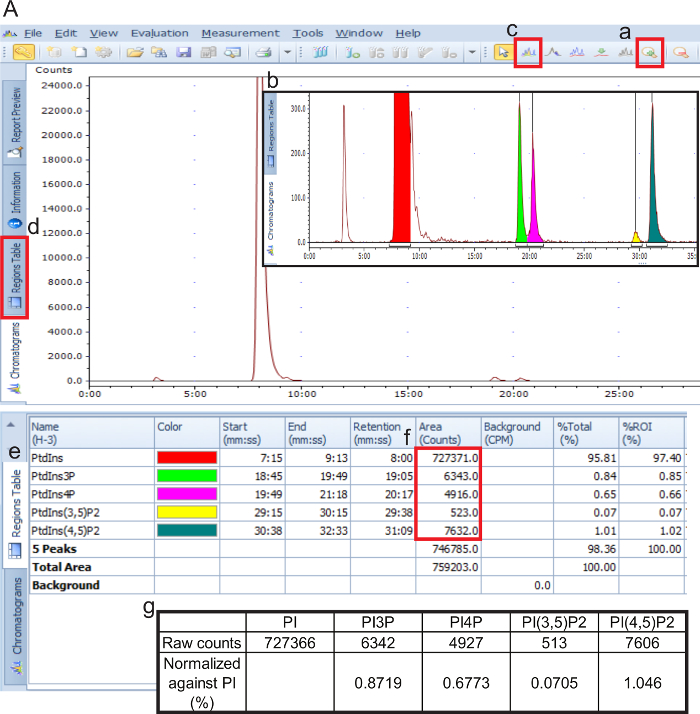
Figure 3. Guided instruction for data analysis of PtdInsP abundance. Data files are loaded on the Laura software and evaluated visually using the chromatograph (open by default). Using the "zoom in" tool (a), magnify the peaks (b). Select distinct peaks on the chromatograph using the "add ROI" tool (c). In the "region table" tab (d), all resulting information from the highlighted regions of interest is displayed as a single table (e). Export the raw counts of each peak (f) and normalize each PtdInsP species against the parental phosphatidylinositol (g) or against total counts. Please click here to view a larger version of this figure.
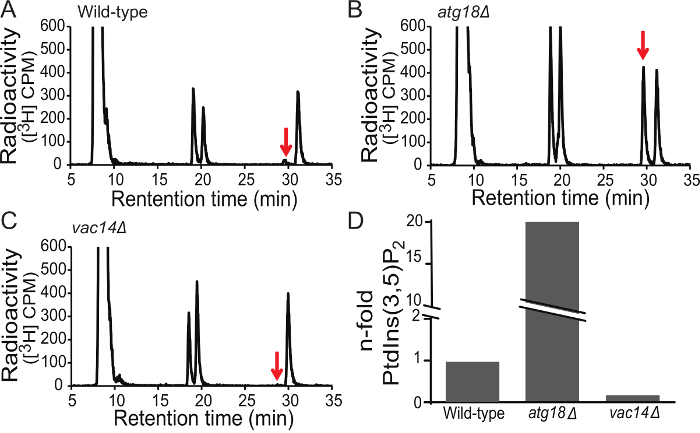
Figure 4. PtdIns(3,5)P2 levels in wild-type and mutant yeast cells. Representative chromatographs are shown for the flow scintillation of the extracted 3H-Gro-InsPs from wild-type (A), atg18Δ (B), and vac14Δ (C) yeast strains using Protocol A. The raw counts from the regions of interest corresponding to Gro-Ins(3,5)P2 (arrow) are extracted from the chromatograph and background subtracted. The resulting values are compared to wild-type and expressed as fold-change in Gro-Ins(3,5)P2 levels (D). The levels and changes in Gro-Ins(3,5)P2 are interpreted to show levels and changes in the original PtdIns(3,5)P2. Please click here to view a larger version of this figure.
| Inositol-free media (IFM) |
| 27.8 mM (NH4)2SO4 |
| 2% Glucose |
| 0.01 mM KH2PO4 |
| 1X Amino acids |
| 1X LPSM |
| 1X Trace elements |
| 5X Vitamins |
| 10X Low phosphate and sulfate media (LPSM) |
| 9 mM CaCl2 |
| 17 mM NaCl |
| 67 mM KCl |
| 63 mM MgCl2 |
| 1,000X Trace elements |
| 8 mM Boric acid |
| 250 µM CuSO4 |
| 602 µM KI |
| 1.23 mM FeCl3 |
| 2.65 mM MnSO4 |
| 971 µM Na-molybdate |
| 2.48 mM ZnSO4 |
| 500X Vitamins |
| 4 µM Biotin |
| 839 µM Ca-pantothenate |
| 2.27 µM Folic acid |
| 1.62 mM Niacin |
| 729 µM p-aminobenzoic acid |
| 973 µM Pryidoxine-HCl |
| 266 µM Riboflavin |
| 593 µM Thiamine-HCl |
Table 1. Composition of the yeast inositol-free media. Yeast cells are radiolabelled with IFM as the base media to be supplemented with 3H-myo-inositol. Prepare a 100 ml solution of IFM using the solutions indicated, filter sterilize using a bottle top 0.22 µm vacuum filter and store at RT. To prepare low phosphate and sulfate media (LPSM), generate a 100 ml solution using the indicated salts, filter sterilize using a bottle top 0.22 µm vacuum filter and store at RT. Dissolve the indicated salts to prepare a 1 L 1,000x solution of trace elements. Mix thoroughly before use and/or store aliquots at -20 °C. Dissolve the indicated vitamins to prepare a 1 L 500x vitamin solution. Mix thoroughly before use and store aliquots at -20 °C.
Discussion
This article details the experimental protocol required to quantify cellular levels of PtdnsPs by HPLC-coupled flow scintillation from yeast. The methodology enables the metabolic labeling of PtdInsPs with 3H-myo-inositol, followed by lipid processing and extraction of water-soluble 3H-Gro-InsPs, HPLC fractionation and analysis. Using this method, the relative levels of PtdInsPs in cells under various conditions can be quantified, as is shown for PtdIns(3,5)P2 in wild-type, vac14D and atg18D yeast cells (Figure 4).
There are a number of critical steps and modifications that can be done to optimize results and/or troubleshoot. One of the most important parameters to achieve reliable quantification of PtdInsPs is the total radioactive signal level. Typically, injecting 3-5 million counts into the HPLC is often sufficient to reliably quantitate changes in the higher abundance PtdInsP species such as PtdIns(4,5)P2. However, injection of up to 10 million counts may be necessary for quantification of lower abundance species such as PtdIns(3,5)P2. There are various parameters that might impact the level of radioactive signal achieved including the total number of cells used to prepare the sample, the labeling time and temperature, the concentration of 3H-myo-inositol, the turnover rate of PtdInsPs and the endogenous synthesis of inositol, which will compete with the radiolabeled inositol. For example, radiolabeling yeast cells for at least one doubling time allows for metabolic incorporation of 3H-myo-inositol into PtdInsPs, but extension of the labeling time for yeast mutants that grow slower or have slower metabolic activity may be necessary. In addition, the protocol described here uses a diauxic shift of the yeast culture, where cells grown to mid-log phase are then concentrated for labeling over one doubling time. However, one can reduce the diauxic shift by labeling cells during early and mid-log phase and for longer-periods of time as previously done32. In doing this, these authors observed higher levels of PtdIns(3)P, PtdIns(4)P and PtdIns(4,5)P2 than cells undergoing diauxic shift32.
Similarly, the labeling period of mammalian cells may require optimization since some mammalian cells like the RAW 264.7 macrophage line doubles every 12 hr, whereas others may divide much slower, or as in the case of neurons, not at all20,35,39. In addition, one can increase the concentration of 3H-myo-inositol to achieve an ideal radioactive signal level. However, please note that some 3H-myo-inositol is packaged in 90% ethanol;adding excessive 3H-myo-inositol will increase the ethanol content in the media and may have an unintended effect on the cells. Finally, prolonged radiolabeling can also starve the cells of inositol. This is particularly evident in mammalian cell culture where complete DMEM contains 40 µM myo-inositol, compared to 0.5 µM 3H-myo-inositol when labeled with 10 µCi/ml. Thus, if necessary, simply increasing the number of mammalian cells during labeling is recommended.
The cellular lysis and lipid processing steps are also critical for obtaining an optimal yield, as well as maintaining the health of the HPLC column. Lysis with perchloric acid allows for precipitation of lipids and other macromolecules, a method first employed for tissue culture and later used for yeast28,33. Compared to the extraction of total lipids using the traditional methanol/HCl/chloroform method, use of perchloric acid enhances the recovery of PtdInsPs, including PtdIns(3,5)P2, with less variation between experiments28. Subsequent application of methylamine breaks ester bonds between the glycerol backbone and the two hydrophobic acyl chains, leaving the water soluble Gro-InsP to be phase-separated from free fatty acids and intact lipids28,34. Particular care must be taken to avoid carrying forward unwanted organic solvents and insoluble material during the extraction step. This may clog the system, cause pressure spikes and/or chemically damage the column. In addition the column must be conditioned at the beginning of each day by exposing the column to a high concentration of ammonium phosphate (buffer B) before usage; thus, a short "Equilibration" protocol is employed before running experimental samples. Failure to do this conditioning step results in a delayed elution profile of the Gro-InsPs peaks in the first experimental sample that is run.
There are several key points related to the analysis and quantification to consider as well. First, the quantification itself should be an integration of signal and area of each peak, not just the peak height. Moreover, an internal control is usually necessary because the absolute signal integration for each peak can vary significantly between samples and experiments without reflecting a change in PtdInsP abundance in a cell. The parental Gro-Ins peak is typically employed as the internal control by normalizing each Gro-InsP peak against it. The Gro-Ins peak typically holds nearly 90% of the radioactive signal and does not tend to change between conditions. Alternatively, one can choose to normalize against total counts, especially if one suspects that the parental PtdIns suffers a significant change in the experimental conditions. Finally, for low abundance PtdInsPs, background subtraction is recommended by defining a nearby non-peak area with the same "time scale" as the peak of interest (Figure 3D).
One final consideration relates to the analysis of mammalian samples, which possess seven PtdInsP species, including three mono-phosphorylated and three bis-phosphorylated species. Unfortunately, it is difficult to fully resolve PtdIns4P from PtdIns5P, and PtdIns(3,5)P2 from PtdIns(3,4)P2 in mammalian samples, resulting in conjoined double-peaks. There are several methods previously published that increase the resolution of these peaks. Typically, this involves modifying the length of elution, the starting concentration of ammonium phosphate buffer, the rate and steps by which the ammonium phosphate buffer concentration changes during the elution profile and the pH of the ammonium phosphate buffer33,35-38. Perhaps, the most successful method developed to separate PtdIns(4)P and PtdIns(5)P was recently described by Sarkes and Rameh39, which employed two-tandem SAX columns and an ammonium phosphate buffer at pH 6.0.
As with most methods, using HPLC-coupled flow scintillation to quantify PtdInsPs in cells has advantages and disadvantages compared to other available methods. For instance, online flow scintillation coupled to HPLC offers live-detection of Gro-InsPs but limits the scintillation counting time, reducing the sensitivity of this method. Alternatively, the HPLC eluent can be fraction-collected with an auto-sampler instead of feeding into a flow scintillator. The manual fractions can then be read by an off-line liquid scintillation counter for longer periods of time, thus offering higher sensitivity – nevertheless, this approach is more time consuming and labor intensive and reduces resolution depending on the number of fractions collected38,40. Additionally, analysis of whole cells unambiguously measures the levels of a specific PtdInsP, but it will not inform about changes to the subcellular distribution of that PtdInsP, though one could couple flow scintillation to sub-cellular fractionation methods as previously done39. In comparison, FP-based PtdInsP-binding domains are useful to investigate the location and dynamics of a PtdInsP species in a cell, but may also bind to factors other than the target PtdInsP. Also, the method described here simplifies the analysis of PtdInsP levels by eliminating the acyl chains, which greatly increases the molecular diversity of PtdInsPs. But by the same token, this is also a key disadvantage, since deacylation of lipids leads to a loss of structural information regarding the acyl chain, an underappreciated aspect of PtdInsP signaling41,42. If this is of great concern, then liquid chromatography coupled to mass spectrometry (LC-MS) should be employed14,17. However, it is currently difficult to concurrently analyse all PtdInsPs species by LC-MS43. Thus, a combination of methods is the best approach in order to investigate the location, dynamics, molecular diversity and levels of all seven PtdInsPs species.
Disclosures
The authors have nothing to disclose.
Acknowledgements
C.Y.H. was supported by an Ontario Graduate Scholarship from the Government of Ontario. This article was made possible by funding held by R.J.B. from the Natural Sciences and Engineering Research Council, the Canada Research Chairs Program and Ryerson University.
Materials
| 1-Butanol | Biobasic | BC1800 | Reagent grade |
| Ammonium phosphate dibasic | Bioshop | APD001 | ACS grade |
| Ammonium sulfate | Biobasic | ADB0060 | Ultra Pure grade |
| Autosampler | Agilent | G1329B | Agilent 1260 infinity series |
| Biotin | Sigma | B4501 | |
| Boric acid | Biobasic | BB0044 | Molecular biology grade |
| Calcium Chloride | Biobasic | CT1330 | Ahydrous, industrial grade |
| Calcium pantothenate | Sigma | C8731 | |
| Copper(II) sulfate | Sigma | 451657 | Anhydrous |
| D-Glucose | Biobasic | GB0219 | Anhydrous, biotech grade |
| Dulbecco's modification of Eagle's Medium | Life | 11995-065 | With 4.5 g/L glucose, 110 mg/L pyruate, L-glutamine |
| Dulbecco's modification of Eagle's Medium | MP biomedicals | 0916429 | With 4.5 g/L glucose, without L-gluatmine, without inositol |
| EDTA | Biobasic | EB0107 | Acid free, ultra pure grade |
| Ethyl ether | Caledon labs | 1/10/4700 | Anhydrous, reagent grade |
| Ethyl formate | Sigma | 112682 | Reagent grade |
| Fetal bovine serum | Wisent | 080-450 | US origin, premium quality, heat inactivated |
| Fetal Bovine Serum, Dialyzed | Life | 26400044 | US origin |
| FlowLogic U | LabLogic Systems Ltd | SG-BXX-05 | Scintillation fluid for flow scintillation |
| Folic acid | Biobasic | FB0466 | USP grade |
| HEPES buffer solution | Life | 15630080 | 1 M solution |
| Inositol, Myo-[2-3H(N)] | Perkin Elmer | NET114005MC | 9:1 ethanol to water |
| Insulin-Transferrin-Selenium-Ethanolamine | Life | 51500056 | 100x solution |
| Iron(III) chloride | Sigma | 157740 | Reagent grade |
| Laura – Chromatography data collection and analysis software | LabLogic Systems Ltd | Version 4.2.1.18 | Flow scintillator software |
| L-glutamine | Sigma | G7513 | 200 mM, solution, sterile-filtered, BioXtra, suitable for cell culture |
| Magnesium Chloride | Sigma | M8266 | Anhydrous |
| Manganese sulfate | Biobasic | MB0334 | Monohydrate, ACS grade |
| Methanol | Caledon labs | 6701-7-40 | HPLC Grade |
| Methylamine solution | Sigma | 426466 | 40% (v/v) |
| Monopotassium phosphate | Biobasic | PB0445 | Anhydrous, ACS grade |
| Nicotinic acid | Biobasic | NB0660 | Reagent grade |
| OpenLAB CSD ChemStation | Agilent | Rev. C.01.03 | HPLC software |
| p-aminobenzoic acid (PABA) | Bioshop | PAB001.100 | Free acid |
| Penicillin-Streptomycin | Sigma | P4333 | 100X, liquid, stabilized, sterile-filtered, cell culture tested |
| Perchloric acid | Sigma | 244252 | ACS reagent, 70% |
| PhenoSpher SAX column | Phenomenex | 00G-315-E0 | 5µm, 80Å, 250 x 4.6 mm |
| Phosphoric acid | Caledon labs | 1/29/8425 | Reagent grade |
| Potassium Chloride | Biobasic | PB0440 | ACS grade |
| Potassium iodide | Biobasic | PB0443 | ACS grade |
| Pyridoxine hydrochloride | Sigma | P9755 | |
| Quaternary pump | Agilent | G1311C | Agilent 1260 infinity series |
| Riboflavin | Bioshop | RIB333.100 | USP grade |
| Sodium Chloride | Biobasic | DB0483 | Biotech grade |
| Sodium molybdate | Sigma | 243655 | |
| Thermostatted Column Compartment | Agilent | G1316A | Agilent 1260 infinity series |
| Thiamine hydrochloride | Sigma | T4625 | Reagent grade; make solution of 0.02% (w/v), forms a suspension. mix and freeze aliquots |
| Ultima Gold | Perkin Elmer | 6013321 | Scintillation coctail for liquid scintillation counting |
| Zinc sulfate | Biobasic | ZB2906 | Heptahydrate, reagent grade |
| β-RAM 4 | IN/US systems | Model 4 | Flow scintillator – 500 µL flow cell; alternative Radiomatic Flow Scintillator Analyser by Perkin Elmer |
References
- Di Paolo, G., De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443 (7112), 651-657 (2006).
- Bridges, D., Saltiel, A. R. Phosphoinositides and Disease. Curr. Top. Microbiol. Immuno. 362, 61-85 (2012).
- Botelho, R. J. Changing phosphoinositides “on the fly”: How trafficking vesicles avoid an identity crisis. BioEssays. 31 (10), 1127-1136 (2009).
- Simonsen, A., Wurmser, A. E., Emr, S. D., Stenmark, H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell. Biol. 13 (4), 485-492 (2001).
- Katso, R., Okkenhaug, K., Ahmadi, K., Timms, J., Waterfield, M. D. Cellular Function of Phosphoinositide 3-Kinases: Implications for Development, Immunity, Homeostasis, and Cancer. Annu. Rev. Cell Dev. Biol. 17, 615-675 (2001).
- Lenk, G. M., Meisler, M. H. Mouse models of PI(3,5)P2 deficiency with impaired lysosome function. Methods. Enzymo. 534, (2014).
- Lemmon, M. A. Phosphoinositide recognition domains. Traffic. 4 (4), 201-213 (2003).
- Cullen, P. J., Cozier, G. E., Banting, G., Mellor, H. Modular phosphoinositide-binding domains-their role in signalling and membrane trafficking. Curr. Biol. 11 (21), R882-R893 (2001).
- Balla, T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell. Sci. 118 (Pt 10), 2093-2104 (2005).
- Burd, C. G., Emr, S. D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 2 (1), 157-162 (1998).
- Lemmon, M. A., Ferguson, K. M., O’Brien, R., Sigler, P. B., Schlessinger, J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 92 (23), 10472-10476 (1995).
- Scott, C. C., et al. Phosphatidylinositol-4, 5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J. Cell. Biol. 169 (1), 139-149 (2005).
- Gillooly, D. J., et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19 (17), 4577-4588 (2000).
- Haag, M., Schmidt, A., Sachsenheimer, T., Brügger, B. Quantification of Signaling Lipids by Nano-Electrospray Ionization Tandem Mass Spectrometry (Nano-ESI MS/MS). Metabolites. 2 (1), 57-76 (2012).
- Furutani, M., Itoh, T., Ijuin, T., Tsujita, K., Takenawa, T. Thin layer chromatography-blotting, a novel method for the detection of phosphoinositides. J. Biochem. 139 (4), 663-670 (2006).
- Cooke, F. T., et al. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in. 8 (22), 1219-1222 (1998).
- Milne, S. B., Ivanova, P. T., DeCamp, D., Hsueh, R. C., Brown, H. A. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J. Lipid. Res. 46 (8), 1796-1802 (2005).
- Rusten, T. E., Stenmark, H. Analyzing phosphoinositides and their interacting proteins. Nat. methods. 3 (4), 251-258 (2006).
- Ho, C. Y., Alghamdi, T. A., Botelho, R. J. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 13 (1), 1-8 (2012).
- Samie, M., et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cel. 26 (5), 511-524 (2013).
- Shaw, J. D., Hama, H., Sohrabi, F., DeWald, D. B., Wendland, B. PtdIns(3,5)P2 is required for delivery of endocytic cargo into the multivesicular body. Traffic. 4 (7), 479-490 (2003).
- Botelho, R. J., Efe, J. A., Emr, S. D. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol. Biol. Cell. 19, 4273-4286 (2008).
- Rudge, S. A., Anderson, D. M., Emr, S. D. Vacuole size control: Regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell. 15, 24-36 (2004).
- Sbrissa, D., et al. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport: Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 282 (33), 23878-23891 (2007).
- Alghamdi, T. A., et al. Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J. Biol. Chem. 288 (13), 9363-9372 (2013).
- Ikonomov, O. C., Sbrissa, D., Fenner, H., Shisheva, A. PIKfyve-ArPIKfyve-Sac3 core complex: Contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J. Biol. Chem. 284 (51), 35794-35806 (2009).
- Dove, S. K., et al. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12 (11), 885-893 (2002).
- Bonangelino, C. J., et al. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell. Biol. 156 (6), 1015-1028 (2002).
- Dove, S. K., et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23 (9), 1922-1933 (2004).
- Efe, J. A., Botelho, R. J., Emr, S. D. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol. Biol. Cell. 18 (1), 4232-4244 (2007).
- Odorizzi, G., Babst, M., Emr, S. D. Fab1p Ptdlns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95 (6), 847-858 (1998).
- Duex, J. E., Nau, J. J., Kauffman, E. J., Weisman, L. S. Phosphoinositide 5-phosphatase Fig4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryotic Cell. 5 (4), 723-731 (2006).
- Whiteford, C. C., Best, C., Kazlauskas, A., Ulug, E. T. D-3 phosphoinositide metabolism in cells treated with platelet-derived growth factor. Biochem J. 319 (3), 851-860 (1996).
- Murthy, P. P., et al. Evidence of two isomers of phosphatidylinositol in plant tissue. Plant physiology. 98 (4), 1498-1501 (1992).
- Zolov, S. N., et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. 109 (43), 17472-17477 (2012).
- McEwen, R. K., et al. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombe and murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. J. Biol. Chem. 274 (48), 33905-33912 (1999).
- Tolias, K. F., et al. Type I phosphatidylinositol-4-phosphate 5-kinases synthesize the novel lipids phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5- phosphate. J. Biol. Chem. 273 (29), 18040-18046 (1998).
- Ikonomov, O. C., et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/- embryos but normality of PIKfyve+/- mice. J. Biol. Chem. 286 (15), 13404-13413 (2011).
- Sarkes, D., Rameh, L. E. A Novel HPLC-Based Approach Makes Possible the Spatial Characterization of Cellular PtdIns5P and Other Phosphoinositides. Biochem J. 428 (3), 375-384 (2011).
- Sbrissa, D., Ikonomov, O. C., Filios, C., Delvecchio, K., Shisheva, A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am. J. Physiol. Cel. Physiol. 303 (4), 436-446 (2012).
- D’Souza, K., Epand, R. M. Enrichment of phosphatidylinositols with specific acyl chains. Biochim. Biophys. Acta – Biomembr. 1838 (6), 1501-1508 (2014).
- Shulga, Y. V., Anderson, R. A., Topham, M. K., Epand, R. M. Phosphatidylinositol-4-phosphate 5-kinase isoforms exhibit acyl chain selectivity for both substrate and lipid activator. J. Biol. Chem. 287 (43), 35953-35963 (2012).
- Pettitt, T. R., Dove, S. K., Lubben, A., Calaminus, S. D. J., Wakelam, M. J. O. Analysis of intact phosphoinositides in biological samples. J. Lipid. Res. 47 (7), 1588-1596 (2006).

