Studying the Protein Quality Control System of D. discoideum Using Temperature-controlled Live Cell Imaging
Summary
The social amoebae Dictyostelium discoideum has recently been established as a system to study protein misfolding and proteostasis. Here, we describe a new imaging-based methodology to study temperature-induced protein aggregation and the cellular stress response in D. discoideum.
Abstract
The complex lifestyle of the social amoebae Dictyostelium discoideum makes it a valuable model for the study of various biological processes. Recently, we showed that D. discoideum is remarkably resilient to protein aggregation and can be used to gain insights into the cellular protein quality control system. However, the use of D. discoideum as a model system poses several challenges to microscopy-based experimental approaches, such as the high motility of the cells and their susceptibility to photo-toxicity. The latter proves to be especially challenging when studying protein homeostasis, as the phototoxic effects can induce a cellular stress response and thus alter to behavior of the protein quality control system.
Temperature increase is a commonly used way to induce cellular stress. Here, we describe a temperature-controllable imaging protocol, which allows observing temperature-induced perturbations in D. discoideum. Moreover, when applied at normal growth temperature, this imaging protocol can also noticeably reduce photo-toxicity, thus allowing imaging with higher intensities. This can be particularly useful when imaging proteins with very low expression levels. Moreover, the high mobility of the cells often requires the acquisition of multiple fields of view to follow individual cells, and the number of fields needs to be balanced against the desired time interval and exposure time.
Introduction
Dictyostelium discoideum are solitary soil living amoebae that feed on bacteria and other microorganisms, which are taken up by phagocytosis. It has a unique and remarkable life cycle that has been a major area of research since its discovery1. The early interest in multicellular development2 and the molecular basis of chemotaxis3 was soon complemented by studies focusing on cell motility, cell polarity, innate immunity. In addition, D. discoideum was introduced as a model system for biomedical research4,5.
Recently, we established D. discoideum as a new system to study the protein quality control (PQC) system6,7. Its proteome is enriched in aggregation-prone prion-like proteins, which poses a challenge to protein quality control8. To investigate whether D. discoideum has developed special molecular mechanisms to control its highly aggregation-prone proteome, we studied the behavior of aggregation-prone marker proteins both under normal growth conditions and during stress. Stress conditions, such as heat stress, can be used to increase the rate of protein misfolding9. Therefore, we sought a system where we could induce temperature changes and simultaneously follow the behavior of marker proteins. For this purpose, we combined live-cell imaging with Peltier-controlled heating using a thermal stage insert (cooling chamber). This method allowed us to maintain a constant and uniform temperature as well as to induce a rapid, yet precise temperature change.
Live-cell imaging is used to study a variety of biological processes in D. discoideum. However, this approach faces two major limitations. First, the cells display a high motility and tend to migrate out of the field of view, thus tracking of individual cells often requires imaging of a large area. The cell mobility can be reduced by Agar overlay10, however, these conditions are not suitable for long term imaging due to a decline in viability. Second, D. discoideum cells show a particularly high sensitivity to photo-toxicity, which results in cell rounding and mitotic arrest11. Previous protocols addressed this issue by addition of ascorbate as a radical scavenger and reduction of exposure times12. The latter can be critical if the protein of interest is expressed at low levels and shows a weak fluorescence signal. The authors also suggest to provide a constant temperature of 21 °C either by imaging in an air-conditioned room or by using temperature-controlled incubation boxes, which cover the objective and microscope stage12.
Here, we describe a method with improved temperature control by using a cooling chamber set at 23 °C. During imaging our set-up significantly enhances the resistance to photo-toxicity. It allows the usage of higher exposure times and higher excitation light intensities. This is of particular importance during time-lapse imaging, as the time intervals need to be carefully balanced against the number of positions imaged and the exposure time used. The possibility to increase the numbers of imaged positions also allows the coverage of a wider imaging area and facilitates tracking of individual cells over a longer period of time.
Protocol
1. Cell Preparation
- Growing cells
- Grow D. discoideum cells in AX medium at 23 °C under light in tissue culture plates. Avoid keeping cells at high densities above 75% confluency.
NOTE: For an optimal response to heat stress, cells need to be in the exponential growth phase and should not have reached stationary phase. - The day before imaging, split cells into low fluorescence medium (LFM) to 1 x 104 cells/ml.
NOTE: This step is necessary to reduce background fluorescence caused by the AX medium. The incubation time can be reduced to a minimum of 2 hr. However, vesicles taken up in AX medium can still generate some background signal after 2 hr.
- Grow D. discoideum cells in AX medium at 23 °C under light in tissue culture plates. Avoid keeping cells at high densities above 75% confluency.
- Preparing cells for microscopy
- Before imaging, harvest cells from the tissue plate in LFM and transfer an adequate number of cells into glass bottom dishes.
NOTE: Usually, 1 x 105 cells/ml is sufficient; however, if the experimental setup requires long imaging periods (longer than 8 hr), cell numbers need to be carefully adjusted, as D. discoideum cells will transition to the developmental cycle and start to stream and aggregate if cell numbers are high while nutrients are low. - Allow the cells to settle for 20 min. Remove non-settled cells by carefully replacing the used medium with fresh medium.
- Before imaging, harvest cells from the tissue plate in LFM and transfer an adequate number of cells into glass bottom dishes.
2. Imaging
NOTE: The protocol can be used on wide-field systems as well as in confocal microscopy setups. The usage of a temperature-controlled incubation box covering the objectives and the microscope stage is helpful, yet not essential for temperature control. The temperature of the incubation box is set to the highest temperature used in the experiment, e.g., to 40 °C if heat stress is performed. Otherwise, the temperature is set to 25 °C. Temperature changes should be made in advance to allow the system to adapt, as changes in temperature during imaging can affect the focus.
NOTE: During imaging, the temperature is controlled using a cooling chamber. The default temperature for imaging non-stressed cells is 23 °C. For heat stress conditions, the temperature is increased accordingly to the desired level. The piezo element in the cooling chamber can respond very quickly to the temperature shift. However, the focus of the images will change slightly (it can deviate to up to 20 µm depending on the temperature shift). Thus manual refocusing is required within the first minutes of image acquisition, unless the used microscope is equipped with a hardware auto-focus.
- Image acquisition under non-stressed conditions
- Image cells under non-stressed conditions at 23 °C, acquire a minimum of 6 fields of view (FOVs).
NOTE: Non-stress condition can be visualized either by recording a short time-lapse (5-30 min) or acquisition of representative stills. For each condition or cell line acquire a minimum of 6 fields of view (FOVs).
NOTE: In case individual cells are of interest, position the respective cells in the middle of the FOV and record a 3 x 3 tile series with the FOV containing the cells of interest in the middle. D. discoideum cells are motile and might migrate out of the FOV. However, the 3 x 3 tile series increases the area potentially explored by the cell of interest and it remains traceable for time-lapse periods of ~6 hr. - Acquire z-stacks of maximum 7 µm in 1-0.25 µm steps to capture the whole volume of the cell.
- Acquire a reference bright field image to assess the total number of cells.
- Image cells under non-stressed conditions at 23 °C, acquire a minimum of 6 fields of view (FOVs).
- Image acquisition for heat stress
- Adjust the temperature of the cooling chamber to 30 °C according to the manufacturer's instructions.
- After the temperature display has reached the desired value, refocus manually in the bright field channel and check all recorded FOVs before starting the time-lapse.
- Start a time-lapse for 4 hr of heat stress with a time interval of 5-10 min between time points. Check for correct focusing during the acquisition of the first two time points.
- Image acquisition during recovery
- After heat stress, reduce the temperature back to 23 °C, wait for the temperature display to adjust and refocus manually.
- Record time-lapse movies in the recovery phase for 8-10 hr in 10-20 min time intervals. Check for correct focusing during the acquisition of the first time point.
3. Image Analysis
NOTE: To quantify the number of cells harboring cytosolic foci, assess the total number of cells and the number of cells with cytosolic foci in each time frame. Due to the difficulty of segmenting cells from bright field images, the quantification needs to be done manually. In the following, data analysis is described for the free image processing package ((http://fiji.sc/Fiji). It requires the plugin “Cell Counter” (http://fiji.sc/Cell_Counter).
- Assessing total number of cells
- Load the bright field image stack (XYT) by clicking "File" and "Open".
- Open the "Cell Counter" plugin ("Plugins" -"Cell Counter"). Load the image stack by clicking "Initialize".
- Choose the appropriate type of counters by ticking the appropriate box.
NOTE: use type 1 as counter for total number of cells and type 2 as counter for cells with cytosolic foci. Count all cells. - Save the markers by clicking "Save Markers".
- Assessing the number of cells with cytosolic foci
- Load the fluorescence image hyperstack (XZYT) ("File" – "Open") and make a maximum intensity projection using the "Z-Projector" (Image/Stacks).
- Open the "Cell Counter" plugin. Load the resulting image stack (XYT) by clicking "Initialize".
- Load the markers by clicking "Load Markers".
CAUTION: The file names of the reference image and the fluorescence stack must be identical. If necessary, rename accordingly before initializing the image stack. - Choose the appropriate marker type (see 3.1.3) and count cells with cytosolic foci. The existing markers can be used as guidance for the position of individual cells.
- Retrieve the number of counts per slice by clicking on "Results" and save the counts elsewhere for further calculations.
Representative Results
The described imaging protocol can be used to analyze the behavior of aggregation-prone prion-like proteins in the amoeba D. discoideum during heat stress. Figure 1 shows the distribution of GFP-tagged polyglutamine protein Q103 in cells under normal growth conditions (Figure 1A), after 2 hours of heat stress at 30 °C (Figure 1B) and after 6 hours of stress recovery and growth at normal growth conditions (Figure 1C). Upon temperature increase from 23 °C to 30 °C, the Q103-GFP marker coalesces from a diffuse distribution to punctate structures. Upon stress release and growth at normal temperature, these structures dissolve. The redistribution of Q103-GFP and formation of heat stress-induced cytosolic foci can be quantified using image analysis. Figure 2A shows the analysis of individual FOVs using Fiji and the plugin "Cell counter". Figure 2B shows the quantification of the heat stress response. This shows that the cytosolic Q103-GFP foci increase in number with continuing heat stress. During heat stress, the protein quality control (PQC) system is overwhelmed, thus aggregation-prone proteins such as the prion-like protein Q103 cannot be maintained in a soluble state and increasingly aggregate in cytosolic foci. The cells also show a characteristic rounding, as depicted in Figure 1B. The reduction of cytosolic foci after stress removal suggests that the aggregated proteins are dissolved.
The imaging quality is sensitive to the initial cell density. Figure 3 shows an example of cells after stress recovery, comparing appropriate initial cell density (Figure 3A) and high initial cell density (Figure 3B). If the initial cell density was too high, cells started to form streams as depicted in Figure 3b. This which imposes difficulties on the subsequent quantification as the cells have moved out of the focal plane.
The described method can be used to monitor temperature-induced changes as described above. It can also be used to reduce photo-toxicity when imaging at physiological growth temperature. Figure 4 shows a comparison of cell imaged at 23 °C in a cooling chamber (Figure 4A) and cells imaged without temperature control in an air conditioned room, set to 25 °C (Figure 4B). Upon constant exposure to the excitation light, cells round up when not imaged under temperature-controlled conditions. This change in cell shape is indicating stress.
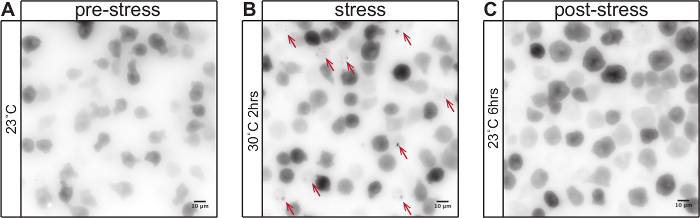
Figure 1: Aggregation-prone prion-like proteins change their cellular distribution during heat stress. GFP-tagged polyglutamine-rich huntingtin exon 1 (Q103) is constitutively expressed in AX2-214 D. discoideum cells (GFP signal depicted in inverted gray scale lookup table). (A) Under normal growth conditions at 23 °C, Q103-GFP is diffusely distributed in the cytoplasm. (B) After 2 hr of heat stress at 30 °C, Q103-GFP coalesces into cytosolic foci (red arrows). The cells show a characteristic shape change and round up. (C) Upon stress release and growth at 23 °C, the cytosolic foci are dissolved. After 6 hours, no cytosolic foci can be observed. Scale bar = 10 µm. Please click here to view a larger version of this figure.
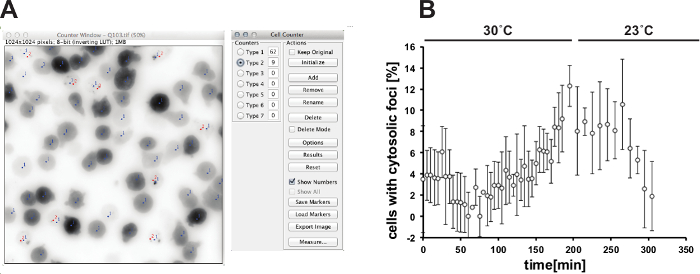
Figure 2: Quantification of heat-induced cytosolic foci formation. (A) Analysis of cells using the free image-processing package Fiji and the plugin "Cell Counter". The total amount of cells (type 1, blue counter markers) is determined in the bright field image. The markers are loaded in the GFP-channel and the number of cells with cytosolic foci (type 2, red counter markers) is determined. (B) Quantification of foci after 3 hr of heat stress at 30 °C and 3 hours of stress recovery at 23 °C (n = 4 fields of view, error bar = SD). Please click here to view a larger version of this figure.
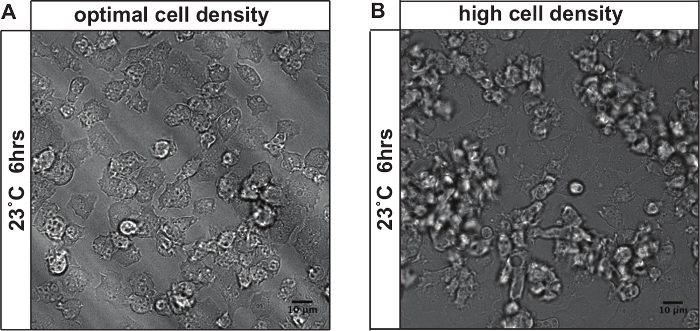
Figure 3: Effect of cell density on long-term time-lapse imaging. (A) Cells imaged at initial cell densities <105 cells/ml. After 3 hr of heat stress at 30 °C and 6 hr of stress recovery at 23 °C, cells still remain in the vegetative cycle and divide. Only few cells exit the focal plane. (B) Cells imaged at initial cell densities >106 cells/ml. After 9 hr of imaging (settings as in A), cells have entered the developmental cycle and started streaming and aggregating. Cells in the aggregates cannot be clearly distinguished. A large portion of cells exits the focal plane and cannot be analyzed. Scale bar = 10 µm. Please click here to view a larger version of this figure.
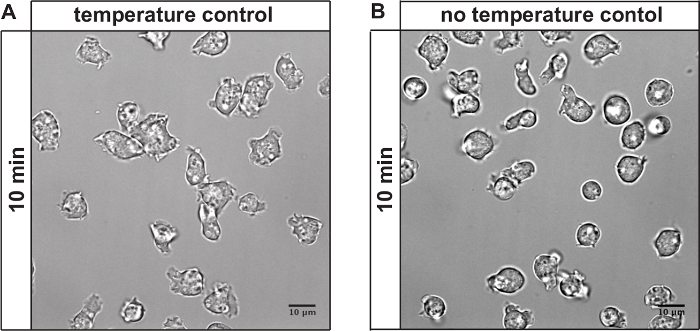
Figure 4: Effect of temperature control on phototoxic effects. (A) Cells imaged for 10 min with temperature control at 23 °C: 6 fields of view, z-stack (spacing 0.5 µM, sample thickness 8 µM), time lapse (total time 10 min, lapse 1 min), GFP channel (0.07 msec exposure time, 10% laser intensity), RFP channel (0.1 msec exposure time, 10% laser intensity). Cells show no signs of the characteristic morphological changes associated with phototoxic stress. (B) Cells imaged for 10 min without temperature control (same conditions as in A). Cells exhibit signs of phototoxic induced rounding, indicating stress. Scale bar = 10 µm. The cells were imaged on a system based on a microscope with a 100x/1.4 numerical aperture (N.A.) objective. The images were collected with a CCD camera as 1,024 x 1,024 pixel files using 1 x 1 binning. Please click here to view a larger version of this figure.
Discussion
The protocol described here can be used to study the behavior of a particular protein of interest in response to heat-induced stress. Temperature increase to 30 °C has been reported to trigger heat stress response and under these conditions the viability of D. discoideum is markedly reduced.
Modifications
The protocol can be modified to compare the behavior of different proteins under the same stress conditions. For this, cells expressing different proteins with the same fluorescent tag are transferred into multi-well dishes, such as four chamber dishes (section 1.3.1). The protocol can also be applied on cells co-expressing proteins with different markers, such as GFP or RFP. This can be for example used to monitor the behavior of different components of the protein quality control (PQC) system. Cells expressing GFP-tagged aggregation marker and different RFP-tagged PCQ components can be observed using multi-well dished for imaging. This ensures the same stress conditions (speed of temperature increase/decrease, duration of the temperature increase/decrease) and allows for comparative studies.
Moreover, the protocol can be used to study the influence of the PQC system on the heat stress response. The activity of the components can be modulated by changing expression levels using genetic tools such as knock-out or overexpression or by using commercially available specific inhibitors6. The proteasome can be inhibited by adding MG132 (100 µM) or lactacystin (10 µM) to the growth medium. The chaperone Hsp90 can be inhibited using geldanamycin (6 µM) or radicicol (10 µM). The chaperone Hsp70 can be inhibited with VER-155088, although we could not confirm the efficacy of the inhibition in our experimental settings so far. Inhibitors should be added one day before imaging and the cells are incubated for 14 hr.
Critical Steps within the Protocol
The critical step for the assessment of heat-induced perturbation is the state of the cells prior to the imaging experiment. Studies in yeast showed that cells acquire resistance to a variety of environmental stresses during stationary phase13. We also observed minimal responsiveness to applied heat stress if D. discoideum cells had reached stationary phase prior to imaging. Therefore, maintaining a constant cell number <5 x 105 cells/ml is critical.
Moreover, high cell numbers can induce transition from the vegetative cycle of D. discoideum to the developmental cycle, thus triggering starvation pathways, which might lead to a different response to heat stress. If high cell numbers are reached in the recovery phase after heat stress, streaming and aggregating cells can interfere with data analysis as the cells move out of focus (see Figure 4).
Limitation of the Technique
The loss of focus is the bottleneck of the protocol. During temperature changes, the focal plane can shift drastically, which requires manual refocusing in a time period immediately after changing the temperature. The jump in the focus can sometimes be too extreme to be overcome by software autofocus, especially if the time between the time points is high. However, if the used microscope is equipped with a hardware autofocus, this bottleneck can be circumvented.
Significance of the Technique with Respect to Existing Methods
In comparison with existing setups such as the use of air-conditioned rooms, temperature-controlled incubation boxes covering the objectives and the microscope stage or temperature-controlled stages and objective collars, the use of a cooling chamber provides a more precise control of the ambient temperature. The Peltier-element in the cooling chamber can maintain a constant and uniform temperature throughout the experimental setup. In classical setups, local temperature differences might lead to different observation outcomes. Moreover, it can respond quickly and rapidly to induced changes in the temperature, while classical setups adapt very slowly to induced temperature changes, especially lowering the temperature. The Peltier-element in the cooling chamber can also cover a wider temperature range (15-40 °C) than classical setups, with which reaching temperatures such as 15 °C or 40 °C is very difficult.
Dictyostelium discoideum is particularly sensitive to photo-toxicity. Previous studies used ascorbate as scavenger to reduce photo-toxicity. However, long imaging periods require additional supplementation. Moreover, the use of ascorbate is limited to studies where the mechanism of interest is not affected by the antioxidant supplement. We propose that temperature-controlled imaging can be used as an alternative approach to minimize photo-toxicity and could be combined with addition of ascorbate to further decrease toxicity.
Phototoxic effects can be minimized by providing a constant and uniform temperature of 23 °C using a cooling chamber. Cells imaged using temperature control show fewer signs of photo-toxicity, such cell rounding, for a longer time period. This also allows imaging with higher intensities, smaller time intervals or more fields of view (FOVs).
General Application
Heat stress at higher temperatures has been shown to trigger a different heat stress response14. Therefore, increasing the temperature to 34 °C or 37 °C might induce a different response. In addition to the applied temperature stress, the duration of a particular stress situation can be modified to study the immediate response or long-term adaptation to heat stress.
In general, the described protocol can be expanded to a wide set of applications. Because the precise temperature control considerably reduces photo-toxic effects, the protocol can be used in settings which require high exposure times and/or higher excitation light intensities, e.g., for visualizing proteins with a low expression level, or in combination with short time intervals between imaging, e.g., during time-lapse imaging. It can also be advantageous for imaging objects spanning the whole cell, e.g., microtubules, as these settings require a high number of optical z-sections.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors have no Acknowledgements.
Materials
| AX Medium | ForMedium | AXM0102 | |
| LoFlo medium | ForMedium | LF1001 | |
| MG132 | Sigma | C2211 | |
| Lactacystin | Sigma | L6785 | |
| Geldanamycin | Santa Cruz | sc-200617 | |
| Radicicol | Santa Cruz | sc-200620 | |
| MatTek disch 35 mm | MatTek corporation | P35G-1.5-14-C | glass bottom imaging dish |
| CellViell cell culture dish | Greiner | 627870 | 4-compartments glass bottom imaging dish |
| Thermal Stage Heater/Cooler Insert | Warner Istruments | TB-3/CCD | |
| Bipolar temperature controller | Warner Istruments | CL-100 | |
| Liquid cooling System | Warner Instruments | LCS-1 |
References
- Bonner, J. T. Evidence for the Formation of Cell Aggregates by Chemotaxis in the Development of the Slime Mold Dictyostelium Discoideum. Journal of Experimental Zoology. 106 (1), 1-26 (1947).
- Loomis, W. F. Cell signaling during development of Dictyostelium. 발생학. 391 (1), 1-16 (2014).
- Nichols, J. M. E., Veltman, D., Kay, R. R. Chemotaxis of a model organism: progress with Dictyostelium. Current Opinion in Cell Biology. 36, 7-12 (2015).
- Williams, J. G. Dictyostelium finds new roles to model. 유전학. 185 (3), 717-726 (2010).
- Müller-Taubenberger, A., Kortholt, A., Eichinger, L. Simple system – substantial share: The use of Dictyostelium in cell biology and molecular medicine. European Journal of Cell Biology. 92 (2), 45-53 (2013).
- Malinovska, L., Palm, S., Gibson, K., Verbavatz, J. M., Alberti, S. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc Natl Acad Sci U S A. 112 (20), E2620-E2629 (2015).
- Malinovska, L., Alberti, S. Protein misfolding in Dictyostelium: Using a freak of nature to gain insight into a universal problem. Prion. 9 (5), 339-346 (2015).
- Schneider, K., Bertolotti, A. Surviving protein quality control catastrophes – from cells to organisms. Journal of Cell Science. 128 (21), 3861-3869 (2015).
- Dobson, C. M. Protein folding and misfolding. Nature. 426 (6968), 884-890 (2003).
- Fukui, Y., Yumura, S., Yumura, T. K. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods in cell biology. 28, 347-356 (1987).
- Graf, R., Rietdorf, J., Zimmermann, T. Live cell spinning disk microscopy. Microscopy Techniques. 95, 57-75 (2005).
- Samereier, M., Meyer, I., Koonce, M. P., Graef, R. Live Cell-Imaging Techniques for Analyses of Microtubules in Dictyostelium. Methods in cell biology. 97, 341-357 (2010).
- Herman, P. K. Stationary phase in yeast. Current opinion in microbiology. 5 (6), 602-607 (2002).
- Loomis, W. F., Wheeler, S., Wheeler, S. Heat shock response of Dictyostelium. 발생학. 79 (2), 399-408 (1980).

