Analysis of the Ambient Particulate Matter-induced Chromosomal Aberrations Using an In Vitro System
Summary
This protocol describes techniques for the quantification and characterization of chromosomal aberrations in vitro in RAW264.7 mouse macrophages after treatment with ambient air particulate matter.
Abstract
Exposure to particulate matter (PM) is a major world health concern, which may damage various cellular components, including the nuclear genetic material. To assess the impact of PM on nuclear genetic integrity, structural chromosomal aberrations are scored in the metaphase spreads of mouse RAW264.7 macrophage cells. PM is collected from ambient air with a high volume total suspended particles sampler. The collected material is solubilized and filtered to retain the water-soluble, fine portion. The particles are characterized for chemical composition by nuclear magnetic resonance (NMR) spectroscopy. Different concentrations of particle suspension are added onto an in vitro culture of RAW264.7 mouse macrophages for a total exposure time of 72 hr, along with untreated control cells. At the end of exposure, the culture is treated with colcemid to arrest cells in metaphase. Cells are then harvested, treated with hypotonic solution, fixed in acetomethanol, dropped onto glass slides and finally stained with Giemsa solution. Slides are examined to assess the structural chromosomal aberrations (CAs) in metaphase spreads at 1,000X magnification using a bright-field microscope. 50 to 100 metaphase spread are scored for each treatment group. This technique is adapted for the detection of structural chromosomal aberrations (CAs), such as chromatid-type breaks, chromatid-type exchanges, acentric fragments, dicentric and ring chromosomes, double minutes, endoreduplication, and Robertsonian translocations in vitro after exposure to PM. It is a powerful method to associate a well-established cytogenetic endpoint to epigenetic alterations.
Introduction
It has been estimated that exposure to particulate matter (PM) causes over 3 million excess deaths annually, primarily from cardiopulmonary disease and lung cancer1. Indeed, PM was recognized as carcinogenic to humans by the International Agency for Research on Cancer (Group 1), as an increased risk of lung cancer with increasing levels of exposure to PM has been shown2. Interestingly, almost all the cancer cells harbor numerical and/or structural chromosomal abnormalities. Particulate organic carbon is a variant, complex, and heterogeneous mixture, whose composition and size distribution depends on emissions as well as physical and chemical transformations. It accounts from 35-55% of urban PM2.5 mass (PM 2.5 µm in size and smaller) and more than 60% of rural and continental background PM2.5 mass3, 4. The water soluble fraction accounts for 30-90% of organic aerosol. A large number of organic compounds have been identified, including aliphatic hydrocarbons, polycyclic aromatic hydrocarbons, and their oxygenated and nitrated derivatives, aliphatic aldehydes and alcohols, free fatty acids and their salts, di-carboxylic acids, multifunctional compounds, proteins, and humic-like macromolecules (HULIS) using chromatographic methods coupled with mass spectrometric techniques5-8. These compounds represent less than 10-20% of particulate organic carbon, thus most of organic carbon is unknown9.
Experimental evidence suggests that cytotoxicity, oxidative stress, and inflammation are involved in the development of PM-associated pathological states. It has been recently shown, however, that exposure to PM also results in a number of epigenetic alterations, including alterations in DNA methylation of repetitive elements, in both experimental in vitro systems and in human subjects10-12. Of particular interest are the effects of PM on satellite DNA — major and minor satellites — which are found in the heterochromatic region around the centromere of chromosomes. It was shown that these effects may be persistent by nature, as they can be detected for at least 72 hr after exposure12. Alterations in DNA methylation, particularly around centromeres, may lead to accumulation of satellite DNA mRNA transcripts, compromise chromosomal integrity during cell division and, subsequently, result in the development of a variety of pathological states13.
Adaptation of cytogenetic approach for the analysis of CAs as an end-point of epigenetic alterations caused by PM, thus, is of high importance. Here, we report the approach for the ambient PM collection and preparation, in vitro exposure and analysis of CAs using the murine macrophage RAW264.7 model system. Macrophages comprise the first line of defense against the inhaled foreign objects and, therefore, this cell line serves as an established and the most frequently used model in particle toxicology11, 12, 14, 15.
Protocol
1. Particle Collection and Preparation
- Calibration of the high volume total suspended particles sampler
- Disconnect the sampler motor from the mass flow controller and connect the motor to a stable AC power source (Figure 1).
NOTE: A filter is not used during this procedure. - Mount the calibrator orifice and top loading adapter plate to the sampler. Tighten the top loading adapter hold-down nuts securely to ensure that no air leaks are present. Allow the sampler motor to warm up to its normal operating temperature (approximately 10-15 min).
- Conduct a leak test by covering the holes on top of the orifice and pressure tap on the orifice with hands. Listen for a high-pitched squealing sound made by escaping air. If this sound is heard, re-tighten the top loading adapter hold-down nuts as a leak is present. If the sound is lower, the leak is near one of the other gaskets in the system.
- Connect one side of a water manometer to the pressure tap on the side of the orifice with a rubber vacuum tube. Leave the opposite side of the manometer open to the atmosphere. Hold a manometer vertically to ensure accurate readings. Tapping the backside of the continuous flow recorder will help to center the pen and provide accurate readings.
- Repeat step 1.1.2 using five flow rates representing operating flow rates from 30 to 60 ft/min (approximately every 5-6 ft3/min). Adjust the knob on the variable orifice to five different positions and take five different readings.
- Record the ambient air temperature, ambient barometric pressure, sampler serial number, orifice serial number, orifice slope and intercept with date last certified, the date, site location, and operator's initials on the calibration sheet.
- Disconnect the sampler motor from the mass flow controller and connect the motor to a stable AC power source (Figure 1).
- Total suspended particles sample collection and gravimetric analysis
- Wrap an 8" x 10" quartz fiber filter in two layers of aluminum foil. Place the wrapped filter in the furnace and set the temperature at 550 °C. Bake the filter for at least 4 hr. Let the furnace cool and retrieve the filter. Place the filter in the desiccator with CaCO3.
- Weigh the filter in a microbalance with precision of 10 µg. Repeat the measurement three times during a day. Individual measurements should be within 25 µg. If this is not achieved, return the wrapped filter to the desiccator and repeat the process the following day.
- Open the shelter lid (Figure 2). Remove filter holder frame by loosening the four wing nuts, allowing the brass bolts and washers to swing down out of the way. Unfold the filter from the aluminum foil, and use cleaned forceps to carefully center it, rougher side up, on the supporting screen. Properly align the filter on the screen.
- Place the frame on the screen and secure it with the brass bolts and washers while applying sufficient pressure to avoid air leakage at the edges. Wipe any dust accumulation from around the filter holder with a clean cloth. Close shelter lid carefully and secure it.
- Make sure all cords are plugged into their appropriate receptacle sockets and the tubing between the blower motor pressure tap and the continuous flow recorder is connected.
- Prepare the flow recorder. Depress pen arm lifter to raise pen point and insert a new chart. Align the tab of the chart to the drive hub of the recorder and press with thumb to lower chart center onto hub. Set time by rotating the drive hub clock-wise until the correct time on chart is aligned with time index pointer.
- Manually switch on the sampler and start the collection. Monitor the recorder to be sure it is inking correctly and the elapsed time indicator to be sure it is working properly.
- At the end of the sampling period, switch off the sampler, record the elapsed time and retrieve the chart. Remove the frame to expose the filter. Remove the exposed filter from the supporting screen by holding it gently at the ends with the forceps. Fold the filter lengthwise so that sample touches sample twice and wrap it in the original aluminum foil.
- Store the filter in a desiccator with CaCO3 for at least 24 hr prior to weighing. Repeat step 1.2.2 to measure the weight of the filters after sampling. Place the filter in a zip plastic bag and store it in -80 °C until analysis.
- Analysis of collected total suspended particles sample
- Retrieve the stored filter, unfold the aluminum foil and transfer the filter on a clean Teflon surface. Using forceps and scalpel, cut outer (white) part of the filter and dispose.
- Next, cut the filter in 0.5" x 0.5" pieces and place pieces in a 100 ml flask. Add 50 ml of ultrapure water. Place the flask in an ultrasound bath and sonicate for 60 min at 33±3 kHz.
- Filter extract through a 0.45 µm polypropylene syringe filter to a 100 ml round bottom flask. Evaporate the water using a rotavap instrument under pressure at 50 °C until about 5 ml of extract remains in the flask.
- Evaporate the remaining water in a 15 ml pear-shaped flask of known weight. Weigh the flask with the dry extract and record the mass of the dry extract.
- Dissolve the dry sample with 400 µl of D2O by sonicating for 15 min and transfer into a 5 mm standard NMR tube. Centrifuge for 5 min at 12,000 x g at room temperature to remove any undissolved matter.
- Repeat with 300 µl of D2O. Add 10 µl of NaN3 1.4% (final conc.: 0.02% w/v) in the NMR tube. Add 10 µl (32 µg) of total suspended particles (TSP)-d4 (3.2 mg/ml) to D2O (final concentration 0.258 mM) in the NMR tube.
- Acquire 1H NMR spectra using a NMR spectrometer operating at 600.17 MHz.
- Insert NMR tube into the triple resonance cryoprobe. Calibrate the instrument in the following order: lock, tune, shim, 90° pulse length calibration and receiver gain adjustment.
- Measure spectra at 25 °C with a 1 D excitation sculpting sequence using 180 water-selective pulses in Digital Quad detection mode, 8,192 scans, 4 dummy scans, sweep width of 8012.57 Hz, frequency offset for water signal suppression set at 4.70 ppm, 1.70 sec acquisition time, 32,768 time domain data points (TD), 1 msec duration gradient (p16), 100 µsec recovery delay after gradient (d16).
- Process 1H NMR spectra by applying a line broadening of 1 Hz and zero filling by a factor of 2 (65,536 data points). Correct the phase and baseline manually and integrate. Set TSP-d4 peak at 0.0 ppm.
- Retrieve the stored filter, unfold the aluminum foil and transfer the filter on a clean Teflon surface. Using forceps and scalpel, cut outer (white) part of the filter and dispose.
2. Particle Exposure
- Culture set up
- Warm complete growth media containing 10% fetal bovine serum (FBS) and penicillin-streptomycin solution (100 U/ml), 0.25% trypsin solution and calcium magnesium free phosphate buffer saline (PBS) in a water bath (at 37 °C) for at least 30 min before culture initiation.
- Take out a T75 flask from 5% CO2 incubator containing cultured RAW264.7 cells and wash the cells with 2 ml of pre-warmed PBS two times.
- Dislodge adherent RAW264.7 cells from the bottom of tissue culture vessel using 1 ml of pre-warmed trypsin solution followed by scraping.
- Collect the cells from tissue culture vessel and make a single cell suspension by repeated pipetting. Count the number of cells with trypan blue and plate at a density of 8,000/cm2 (500,000 cells total) in a 100 mm tissue culture dish with pre-warmed complete media. Adjust media volume to 10 ml for a final concentration of 50,000 cells per ml.
NOTE: Double the number of plates if DNA methylation analysis is also performed. - Place the newly plated RAW264.7 cells back in 5% CO2 incubator at 37 °C and allow to attach for 24 hr.
- Also in a biosafety cabinet, resuspend filtered particles. For example, dilute particles at a concentration of 0.5 mg/ml in sterile PBS for a 5 µg/ml treatment dose and at 5 mg/ml for a 50 µg/ml treatment dose. Dilutions can be prepared up to month in advance and stored at -80 °C.
- Treatment with particulate matter
- Take out the cells from 5% CO2 incubator and check under 10x magnification using an inverted microscope for cell growth, cell morphology, culture condition, and contamination. One should expect to have at least 30% confluent cells at this stage.
- Aseptically, aspirate the media using sterilized pipette and wash the culture vessel with 2 ml 37 °C PBS two times to completely remove the media and floating or dead cells.
- Add pre-determined dose of particulate matter with fresh media in the culture vessels and incubate for desired time at 37 °C in 5% CO2 incubator. In this study, treat RAW264.7 cells with PM for 72 hr to parallel the exposure used for concurrent genotoxic and epigenotoxic analysis.
NOTE: Cytotoxicity may negatively affect the quality of the cytogenetic analysis. Therefore, additional assays such as cytotoxicity, DNA methylation, Western blotting, or gene expression analysis are typically performed on treated cells. Expose cells as described and proceed with the specific assay desired. Inexperienced users may also want to include a positive control such as methyl methanesulphonate to test their ability to detect CAs.
3. Cytogenetic Assay
- Treatment to arrest cells at metaphase
- Warm the colcemid solution (10 µg/ml) in a 37 °C water bath for 30 min. Wipe the lid with 70% alcohol under a biosafety cabinet to minimize the risk of contamination
- Completely remove the media aseptically and wash the culture vessels with 2 ml pre-warmed PBS two times.
- Add pre-warmed fresh media (2 ml media/106 cells) containing colcemid solution (5 µl/ml) in the culture vessel and incubate for 2 hr.
- Cell harvest
- Warm trypsin solution and PBS in a water bath maintained at 37 °C for 30 min. From this step onwards, there is no need to maintain aseptic condition.
- Completely remove media, wash the culture vessels with 2 ml 37 °C PBS for two times, add required volume of pre-warmed trypsin solution (usually 2 ml for 60 mm culture dish or T25 flask and 4 ml for 100 mm dish or T75 flask), and place the culture vessel back in 5% CO2 incubator at 37 °C for 1 min.
- Take out the culture vessel, detach the cells using a scraper, and add 10 ml of PBS to stop trypsin action.
- Pipette up and down at least ten times to make single cell suspension and transfer cells in a 15 ml conical bottom centrifuge tube.
- Centrifuge cell suspension at 400 x g for 5 min in a swing out centrifuge at room temperature.
- Remove the supernatant, break the cell pellet by gentle tapping, add 10 ml of PBS, and again centrifuge for five min.
- Hypotonic treatment and fixation
- Pre-warm 0.075 M potassium chloride solution in a 37 °C water bath at least for 30 min.
- Remove supernatant without disturbing the cell pellet and leave approximately 0.5 ml of PBS.
- Gently break the cell pellet and make single cell suspension using a P200 pipette.
- Add 4 ml of pre-warmed potassium chloride solution drop by drop with gentle shaking.
- Incubate the cells in hypotonic potassium chloride solution in a 37 °C water bath for 20 min.
- Make fresh acetomethanol solution by adding 3-parts methanol with 1-part glacial acetic acid. Keep this freshly prepared fixative at room temperature.
- After hypotonic treatment, add an equal volume (4 ml) of fixative in the tube and gently mix the solution by inverting the tube.
- Centrifuge at 400 x g for 5 min at room temperature, and remove the supernatant. At this stage cell pellet size increases, become whitish and more visible.
- Remove supernatant, add 4 ml fresh fixative and keep at room temperature for 30 min.
- Centrifuge at 400 x g for 5 min, remove supernatant, and give two more washes with 4 ml fixative.
- Slide cleaning and metaphase spread preparation
- Immerse glass slides completely in 10% liquid detergent for 30 min.
- Rinse the slides thoroughly in cold running tap water for 30 min.
- Rinse slides three times with distilled water to completely remove detergent, immerse in distilled water, and store in the refrigerator for future use.
NOTE: Cleaning slides before use facilitates uniformity of spreading and increases metaphase quality. - Drop 10 µl cell suspension on chilled wet slide and allow it to air dry overnight at room temperature.
- Staining and mounting
- Prepare 50 ml Giemsa staining solution by mixing one part of Giemsa stain solution with one part of PBS and pour in a Coplin jar.
- Immerse the slides in staining solution for 20 min.
- Rinse the slides in distilled water to remove excess stain and immediately dry the slides using a hair-dryer.
- Leave the slides at room temperature overnight.
- Apply a single drop of mounting medium on the slide, gently place a coverslip on the mounting medium, allow the medium to spread over the slide slowly, and remove the excess medium with a paper towel. Allow the mounted slide to dry for at least 1 hr prior to moving or viewing under a microscope to avoid movement of the coverslip.
Caution: It is important to place the coverslip carefully without forming any bubbles during this step. The use of excessive heat to remove bubbles is not recommended.
- Microscopic observation and aberration types
- Examine the structural CAs in metaphase spreads at 100X magnification using a good-quality bright-field microscope.
- Score at least 50 to 100 metaphase spreads for each treatment group for treatments that induce a large number of CAs. For smaller effect sizes, the analysis of up to 300 metaphase spreads is advised.
Representative Results
Great care should be taken in choosing the location of the TSP sampler, as well as the time of the year when collection is performed. The chemical composition as well as the size of particles may substantially influence the results. The material collected should be visible against the white filter. A normal mouse metaphase spread will have 40 acentric chromosomes. The goal of the technique is to demonstrate a change (or absence thereof) in the proportion of abnormal chromosomes in treated cells versus controls. That change can then be quantified (number of abnormal chromosomes) and qualified (type of abnormalities).
Normal mouse metaphase spread will have 40 acentric chromosomes (Figure 3A). Treatment with particulate matters induces various CAs as shown in Figure 3, such as chromatid break (3B), acentric fragment (3C), ring chromosome (3D), dicentric chromosome (3E), double min (3F), Robertsonian translocation (3G), and endoreduplication (3H). For full results, please refer to our published study12.

Figure 1. The high volume total suspended particles (TSP) sampler. The picture shows the TSP sampler installed in a roof in an urban area to collect ambient particles. Please click here to view a larger version of this figure.

Figure 2. Loaded quartz fiber filter following collection using the high volume TSP sampler. Close-up on the filter installed on the TSP sampler, after use. Note that the color of the filter changed from white to gray. Please click here to view a larger version of this figure.
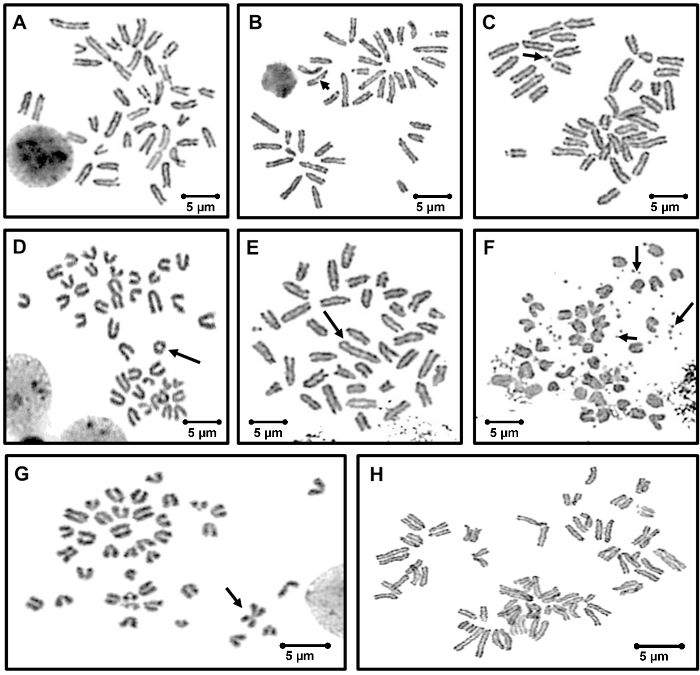
Figure 3. Photomicrograph showing representative examples of different types of structural aberrations induced in RAW264.7 cells after treating with different concentrations of particulate matter. The aberrations are indicated by arrows. (A) normal metaphase spread with 40 acrocentric chromosomes, (B) chromatid-type break (CTB), (C) acentric fragment (acentrics), (D) ring chromosome, (E) dicentric chromosome (Dic), (F) double min (Dimn), (G) Robertsonian translocation, and (H) endoreduplication (duplicated chromosomes are held together). Original magnification 100X, scale bar = 5 µm. Please click here to view a larger version of this figure.
Discussion
Cytogenetic study or microscopic analysis of the numbers or structures of chromosomes, primarily in metaphase spreads, provides information crucial for prognosis, risk assessment, and treatment for various diseases. It is now well-established that cytogenetic abnormalities are linked with the progression and development of several diseases, including cancer. To date, CAs have been found in all major tumor types. CAs may arise spontaneously or by either external or internal stimuli, such as ionizing radiation (IR) — one of the major external risk factors that can induce various types of cytogenetic alterations16. Analysis of CAs aids in assessment of the absorbed radiation dose (called cytogenetic radiation biodosimetry). Moreover, the analysis of frequency and type of specific aberration also provides information important for determining the quality of radiation being absorbed17, 18. Cytogenetic study provides a direct image of the impact of damaging agents on DNA. This is in contrast to indirect techniques of such as the comet assay. However, cytogenetic study has been under-utilized in fields outside of radiation, partly because of the numerous hands-on steps involved.
Although exposure to organic carbon present in the PM is known to adversely affect our health, few studies reporting biological effects include their composition9. This is due to the costly and cumbersome chromatographic methods that require labor-intensive extraction, processing, concentrations, and derivatization protocols, while they only provide insights for a small subset of a very specific type of compounds each time. Moreover, the protocols result in the substantial manipulation or destruction of the original sample, thus, additional testing cannot be performed. Advantages of nuclear magnetic resonance (NMR) analysis include that NMR is a non-destructive method; it requires a very limited number of steps (extraction, concentration) prior to analysis, and high-frequency NMR instruments (from 400 MHz to 900 MHz) are available in many, if not all, universities within the core facility. A comprehensive review of atmospheric aerosol NMR studies has been recently published19. Finally, because of its ability to provide structural information and bonds between groups, NMR is superior to other spectrometric methods such as FT-IR, UV-Vis, and Raman spectrometry that only provide data on the type of functional groups. If needed, morphological characterization of PM can be performed in addition to NMR spectrometry with the use of scanning electron microscopy.
Even from a single location, the composition of the PM collected may vary depending on the time of the year. A large enough amount of material should be processed and kept at -80 °C for repeat experiments or complementary assays. Steps 2.1.1 through 3.1.3 should be performed aseptically in a biosafety cabinet. If contamination or compromised cellular morphology is observed, do not proceed with the rest of the protocol. The trypsin treatment time needs to be adjusted based on cell type. Here, macrophages are very adherent and we have found that a combination of 1 min trypsin and scraping works best, but trypsin alone for 5 min works for most cell types. Hypotonic treatment time also varies depending on cell type and needs to be standardized. The dose and treatment time of colcemid are the most important parameters for chromosome preparation; an over-dose can kill the cells or stall the cell cycle. We describe here conditions for RAW264.7 cells, and optimization is required for other cell types. Failure to find ideal conditions in this step may adversely affect the mitotic index as well as chromosome morphology, affecting the results. It is also recommended to assess the levels of cytotoxicity of the PM prior to cytogenetic analysis and select the doses accordingly.
Conceptually, in vitro toxicological assessments are limited by the type of used cells, exposure magnitude, and particle concentrations. Furthermore, in this study, we utilized the water soluble extract of PM and, therefore, the potential of water-insoluble species to cause cytogenetic aberrations may not be accounted. Other limitations are associated with the presence of "polymorphic variants" – variations in the centromeric regions and short arms of chromosome; some submicroscopic or cryptic rearrangements that may be misinterpreted and the presence of complex karyotypes.
Here, we demonstrate that the introduction of cytogenetic analysis may aid in the identification of PM-induced damage to DNA and may potentially serve as a predictive endpoint in assessment of the health effects caused by exposure to ambient PM. Our study is, to our knowledge, the first one to indicate CAs in response to PM. Replication of these results is desirable to confirm our findings. This protocol could also be adapted to assess almost any type of suspected DNA damaging agent.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The work was supported, in part, by the National Institute of Health Center of Biological Research Excellence [grant number 1P20GM109005], the Arkansas Space Grant Consortium through National Aeronautics and Space Administration [grant number NNX15AK32A], and the National Institute for Occupational Safety and Health (NIOSH) [grant number 2T420H008436]. The authors would like to thank Christopher Fettes for proofreading and editing this manuscript.
Materials
| Total suspended particulate sampler | Tisch Environmental | TE-5170 | |
| Bruker Avance III NMR spectrometer | Bruker | NA | |
| TopSpin 3.5/pl2 software | Bruker | NA | |
| ACD/NMR Processor Academic Edition | ACD/Labs | NA | |
| RAW264.7 murine macrophages | ATCC | ATCC TIB-71 | |
| High glucose DMEM GlutaMAX media | ThermoFisher | 10569010 | Warm in a 37°C waterbath before use |
| Fetal Bovine Serum | ThermoFisher | 16000044 | Warm in a 37°C waterbath before use |
| Penicillin-Streptomycin (10,000 U/mL) | ThermoFisher | 15140163 | Warm in a 37°C waterbath before use |
| Trypsin-EDTA (0.25%) | ThermoFisher | 25200056 | Warm in a 37°C waterbath before use |
| PBS, pH 7.4 | ThermoFisher | 10010049 | Warm in a 37°C waterbath before use |
| KaryoMAX Colcemid Solution in PBS | ThermoFisher | 15212012 | Warm in a 37°C waterbath before use |
| KaryoMAX Potassium Chloride Solution | ThermoFisher | 10575090 | Warm in a 37°C waterbath before use |
| Methanol (HPLC) | Fisher Scientific | A452N1-19 | |
| Acetic Acid, Glacial | Fisher Scientific | BP1185-500 | |
| Decon Contrad 70 Liquid Detergent | Fisher Scientific | 04-355-1 | |
| Wright and Wright-Giemsa Stain Solutions | Fisher Scientific | 23-200733 | |
| Permount Mounting Medium | Fisher Scientific | SP15-100 | |
| Axio Imager 2 | Zeiss | NA |
References
- Lim, S. S., et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 380, 2224-2260 (2012).
- . . IARC: Outdoor air pollution a leading environmental cause of cancer deaths. , (2013).
- Putaud, J., et al. A European aerosol phenomenology-2: chemical characteristics of particulate matter at kerbside, urban, rural and background sites in Europe. Atmos. Environ. 38 (16), 2579-2595 (2004).
- Hand, J., Schichtel, B., Pitchford, M., Malm, W., Frank, N. Seasonal composition of remote and urban fine particulate matter in the United States. J Geophys Res-Atmos. 117 (5), (2012).
- Schauer, J. J., et al. Source apportionment of airborne particulate matter using organic compounds as tracers. Atmos. Environ. 30 (22), 3837-3855 (1996).
- Simoneit, B. R. A review of biomarker compounds as source indicators and tracers for air pollution. Environ. Sci. Pollut. Res. 6 (3), 159-169 (1999).
- Kavouras, I. G., Stephanou, E. G. Particle size distribution of organic primary and secondary aerosol constituents in urban, background marine, and forest atmosphere. J Geophys Res-Atmos. 107 (8), (2002).
- Graber, E., Rudich, Y. Atmospheric HULIS: How humic-like are they? A comprehensive and critical review. Atmos. Chem. Phys. 6 (3), 729-753 (2006).
- Pöschl, U., et al. Atmospheric aerosols: composition, transformation, climate and health effects. Angew. Chem. Int. 44 (46), 7520-7540 (2005).
- Hou, L., et al. Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ. Mol. Mutagen. 55 (3), 256-265 (2014).
- Miousse, I. R., et al. Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ. Mol. Mutagen. 55 (5), 428-435 (2014).
- Miousse, I. R., et al. In Vitro Toxicity and Epigenotoxicity of Different Types of Ambient Particulate Matter. Toxicol. Sci. 148 (2), 473-487 (2015).
- Ehrlich, M. Genomic instability in cancer development. Genome Instability in Cancer Development. , 363-392 (2005).
- Michael, S., Montag, M., Dott, W. Pro-inflammatory effects and oxidative stress in lung macrophages and epithelial cells induced by ambient particulate matter. Environ Pollut. 183, 19-29 (2013).
- Li, N., et al. Induction of heme oxygenase-1 expression in macrophages by diesel exhaust particle chemicals and quinones via the antioxidant-responsive element. J Immunol. 165 (6), 3393-3401 (2000).
- Pathak, R., Ramakumar, A., Subramanian, U., Prasanna, P. G. Differential radio-sensitivities of human chromosomes 1 and 2 in one donor in interphase- and metaphase-spreads after 60Co gamma-irradiation. BMC Med. Phys. 9, 6649 (2009).
- Pathak, R., Dey, S. K., Sarma, A., Khuda-Bukhsh, A. R. Genotoxic effects in M5 cells and Chinese hamster V79 cells after exposure to 7 Li-beam (LET= 60keV/µm) and correlation of their survival dynamics to nuclear damages and cell death. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 628 (1), 56-66 (2007).
- Pathak, R., Dey, S. K., Sarma, A., Khuda-Bukhsh, A. R. Cell killing, nuclear damage and apoptosis in Chinese hamster V79 cells after irradiation with heavy-ion beams of 16 O, 12 C and 7 Li. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 632 (1), 58-68 (2007).
- Chalbot, M. C., Kavouras, I. G. Nuclear magnetic resonance spectroscopy for determining the functional content of organic aerosols: a review. Environ. Pollut. 191, 232-249 (2014).

