Experimental Design for Laser Microdissection RNA-Seq: Lessons from an Analysis of Maize Leaf Development
Summary
Many developmentally important genes have cell- or tissue-specific expression patterns. This paper describes LM RNA-seq experiments to identify genes that are differentially expressed at the maize leaf blade-sheath boundary and in lg1-R mutants compared to wild-type. The experimental considerations discussed here apply to transcriptomic analyses of other developmental phenomena.
Abstract
Genes with important roles in development frequently have spatially and/or temporally restricted expression patterns. Often these gene transcripts are not detected or are not identified as differentially expressed (DE) in transcriptomic analyses of whole plant organs. Laser Microdissection RNA-Seq (LM RNA-Seq) is a powerful tool to identify genes that are DE in specific developmental domains. However, the choice of cellular domains to microdissect and compare, and the accuracy of the microdissections are crucial to the success of the experiments. Here, two examples illustrate design considerations for transcriptomics experiments; a LM RNA-seq analysis to identify genes that are DE along the maize leaf proximal-distal axis, and a second experiment to identify genes that are DE in liguleless1-R (lg1-R) mutants compared to wild-type. Key elements that contributed to the success of these experiments were detailed histological and in situ hybridization analyses of the region to be analyzed, selection of leaf primordia at equivalent developmental stages, the use of morphological landmarks to select regions for microdissection, and microdissection of precisely measured domains. This paper provides a detailed protocol for the analysis of developmental domains by LM RNA-Seq. The data presented here illustrate how the region selected for microdissection will affect the results obtained.
Introduction
The maize leaf is an ideal model to study the formation of developmental fields during morphogenesis, as it has a distinct boundary between the blade and sheath that is amenable to genetic dissection (Figure 1A). During the early stages of leaf development, a linear band of smaller cells, the preligule band (PLB), subdivides the leaf primordium into pre-blade and pre-sheath domains. A fringe-like ligule and triangular auricles develop from the PLB (Figure 1A, C, D). Genetic screens have identified mutations that disrupt the blade-sheath boundary. For example, recessive liguleless1 (lg1) mutations delete the ligule and auricles1,2,3,4 (Figure 1B). In situ hybridization revealed that lg1 transcript accumulates at the PLB and emerging ligule, making it an excellent marker for ligule development5,6 (Figure 1E).
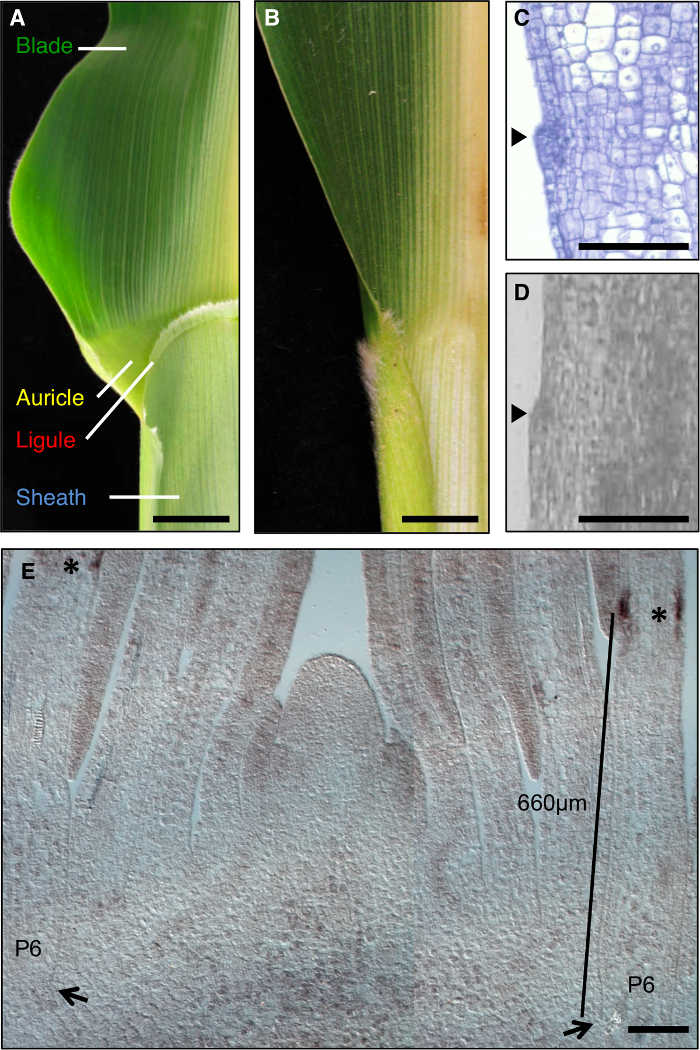
Figure 1: Wild-type and liguleless1-R maize leaves. (A) Blade-sheath boundary region of mature wild-type leaf showing ligule and auricle structures. (B) Blade-sheath boundary region of mature liguleless1-R leaf showing absence of ligule and auricle structures. Leaves in A and B have been cut in half along the midrib. (C) Longitudinal section through wild-type leaf primordium. Sample has been processed and stained for histological analysis. The initiating ligule is apparent as a bump protruding from the plane of the leaf (arrowhead). (D) Longitudinal section through wild-type leaf primordium. Sample has been processed for LM as described in the text. Arrowhead indicates initiating ligule. (E) lg1 in situ hybridization of shoot apex lateral longitudinal section. Asterisks indicate lg1 transcript accumulation at the PLB of the P6 leaf primordium. Arrows indicate base of P6 primordium. Bar indicates measurement from the base of the primordium to the PLB. Scale bars in A and B = 20 mm. Scale bars in C-E = 100 µm. This figure has been modified from reference6(Copyright American Society of Plant Biologists). Please click here to view a larger version of this figure.
In this study, LM RNA-Seq was employed to identify a suite of genes that are differentially expressed (DE) at the blade-sheath boundary relative to other parts of the leaf primordium and to identify genes that are DE in lg1-R mutants relative to wild-type siblings. LM RNA-Seq is a method of quantifying transcript accumulation in specific cells or cellular domains7. LM systems combine a laser and a microscope with a digital camera. Sectioned tissue is mounted on slides and viewed through the microscope. The LM software typically includes drawing tools that allow the user to outline any selected region for microdissection. The laser cuts along the line, and the selected tissue is catapulted off the slide and into a tube suspended above the slide. LM allows the user to microdissect precise domains, including specific cell layers and even single cells8,9. RNA can then be extracted from the microdissected tissue. Subsequently, the RNA-Seq component utilizes next-generation sequencing to sequence cDNA libraries generated from the extracted RNA10,11.
Key advantages of LM RNA-seq are the ability to quantify transcript accumulation in precisely defined domains and the capacity to profile the entire transcriptome simultaneously7. The technique is particularly suited to probing early developmental events where the region of interest is often microscopic. Previous studies have utilized LM combined with microarray technology to study developmental processes in plants9,12,13. RNA-Seq has the advantage of quantifying transcripts across a broad dynamic range, including low-expressed genes, and prior sequence information is not required10,11. Moreover, LM RNA-Seq has the potential to highlight developmentally important genes that may be missed in mutagenesis screens due to genetic redundancy or to lethality of the loss-of-function mutant.
Developmentally important genes, such as narrow sheath1 (ns1) and cup-shaped cotyledon2 (cuc2), often have specific expression patterns of just one or a few cells17,18,19,20. Many are expressed only during early developmental stages and not in the mature organ. When whole organs or large domains are analyzed, these cell-specific transcripts are diluted and may not be detected in more conventional analyses. By permitting analyses of precisely defined domains, LM RNA-Seq enables these tissue-specific genes to be identified and quantified.
Crucial factors in the success of the experiments described here were a thorough histological analysis that guided selection of the appropriate developmental stage and domain for analysis, and precise measurement of cell-tissue domains for LM. To ensure that equivalent domains were sampled for all replicates, tissue was collected from leaf primordia at the same developmental stage and the microdissected domains were measured relative to morphological landmarks such as the emerging ligule (Figure 2). It is known that some genes are expressed in a gradient from the tip to the base of the leaf. By measuring precise domains, variation due to sampling from different locations along the leaf proximal-distal axis was kept to a minimum (Figure 3A). By microdissecting domains of the same size, variation due to differential dilution of cell-specific transcripts was also reduced (Figure 3B). Lateral longitudinal sections of the shoot apex were used for all microdissections. These are sections that are perpendicular to the midrib-margin axis (Figure 4). Using only sections that include the SAM ensures that equivalent lateral regions of leaf primordia are analyzed.
In samples processed and sectioned for LM, the first morphological sign of ligule outgrowth is a bump on the adaxial side due to periclinal cell divisions in the adaxial epidermis (Figure 1D, Figure 2). It was determined that the emerging ligule could be reliably identified at plastochron 7 stage leaf primordia. We were interested in genes expressed in the entire ligule region, including the emerging ligule and the cells immediately distal that will form the auricle. In order to ensure that equivalent tissue selections were made, the ligule bump was used as a morphological landmark and a 100 µm rectangle centered on the ligule bump was selected for LM (Figure 2A, 2B). Equivalent sized rectangles of pre-blade and pre-sheath were selected from the same leaf primordia.
Analyses of liguleless mutant plants presented a different challenge; lg1-R mutants do not form a ligule, therefore this morphological feature could not be used to select the region for LM. Instead, the domain of lg1 transcript accumulation in wild-type leaf primordia was determined, and a region that would encompass this domain was defined. These preliminary analyses were performed on seedlings from the same planting as were used for the final analysis, since previous work has shown that the location of the PLB varies depending on growth conditions. In situ hybridization indicated that lg1 transcripts accumulate in the PLB of P6 leaf primordia (Figure 1E). We selected a domain 400-900 µm from the base of the leaf primordia that encompassed the domain of lg1 expression (purple rectangles, Figure 2A) and captured these equivalent regions from wild-type and lg1-R plants. To minimize variation in genetic background and growth conditions when comparing transcript accumulation in lg1-R and wild-type plants, segregating families of mutants and wild-type siblings were used.
Protocol
NOTE: Fix tissue for histological analysis at the same time that tissue is fixed for LM. Examine stained sections for morphological features that will guide later LM. When comparing mutant to wild-type, perform in situ hybridization or immunolocalization to define the domain where the gene of interest is expressed (in this case lg1).
1. Tissue Fixation and Processing
- Grow flats of maize seedlings to two weeks old under standard conditions6.
- Dissect shoot apices for lateral sections (Figure 4).
- Excise seedling just below the soil line.
- Using a razor blade, remove thin slices from the base of the stem (cuts 1, Figure 4A) until an oval of culm encircled by one or two mature leaves is visible (Figure 4B).
- Make another cut approximately 10 mm above the base (cut 2, Figure 4A). This 10 mm segment will contain the SAM and young leaf primordia.
- Turn the 10 mm segment so the base is facing up. Make two cuts parallel to the lateral axis so that a slice of tissue 2-3 mm thick is obtained (cuts 3, and 4, Figure 4B). Discard the outer two portions and retain the central slice for fixation and embedding.
NOTE: Outer leaves may be trimmed and discarded.
- Fix tissue and process for embedding.
- Ensure that all materials to be used in subsequent steps are RNase free. Treat solutions with diethyl pyrocarbonate (DEPC) (1 ml DEPC per liter of solution. Incubate overnight with occasional shaking, and autoclave). Bake glassware in an oven at 200 °C or higher for at least 6 hr and treat plastic ware with RNase decontamination solution.
- Day 1: Immerse tissue slices in ~10 ml of Farmer's fix (3:1 Ethanol: Acetic acid) in glass vial on ice. After all samples have been dissected, apply vacuum to remove air bubbles and aid penetration of fixative. Hold under vacuum for 10 min then release the vacuum slowly. Replace fixative and incubate at 4 °C overnight with gentle shaking.
- Day 2: Incubate in the following series of solutions, ~10 ml each, 1 hr each, all with gentle shaking; 85% ethanol at 4 °C, 95% ethanol at 4 °C, 100% ethanol at 4 °C, 100% ethanol at 4 °C, 100% ethanol at 4 °C, 1:1 ethanol: xylenes at room temperature, 100% xylenes at room temperature, 100% xylenes at room temperature.
NOTE: Xylenes are toxic by contact and inhalation. Work in a fume hood and use appropriate gloves. - Add paraffin tissue embedding medium pellets to approximately half volume of xylenes and incubate overnight at room temperature with gentle shaking.
- Day 3: Transfer the vial to 60 °C oven until the pellets melt. Pour off solution and replace with fresh melted tissue embedding medium. Change the medium two more times during the day.
- Day 4: Change tissue embedding medium once in the morning. Return to 60 °C oven until afternoon.
- Cast blocks
- Place embedding molds on hot plate of tissue embedding station. Use forceps to transfer the tissue samples to the embedding molds with the cut surface facing down. Top up the mold with melted paraffin and place the embedding ring on top of the mold. Transfer to a cold plate until paraffin has solidified. Store the paraffin blocks at 4 °C in an airtight container with silica gel.
2. Sectioning and Slide Preparation
- Cut 10 µm sections on a microtome25.
- Examine ribbons and choose median sections. Median sections are those that include the SAM, which appears as a dome of cells surrounded by leaf primordia.
- Mount sections on slides.
- Place slides that are suitable for LM (either RNase free or baked) on 42 °C slide warmer and apply several drops of 50% ethanol solution to cover the slide.
- Float sections on ethanol solution until the sections have expanded.
NOTE: Floating sections on ethanol solution rather than water keeps RNA in a precipitated state reducing RNA degradation. - Tilt slide and remove excess ethanol solution by aspiration with a disposable transfer pipette. Use lint-free wipes to wick away any additional ethanol solution.
- Dry slides at 42 °C for several hours or overnight. Store slides at 4 °C in an airtight container with silica gel.
- Deparaffinize slides on the day of use.
- Prepare three glass Coplin jars containing; 100% xylenes (xylenes I), 100% xylenes (xylenes II), and 100% ethanol (~50 ml of each solution).
- Using clean forceps to transfer slides, immerse slides in xylenes I for 2 min, xylenes II for 2 min, and 100% ethanol for 1 min.
- Drain slides on lint-free wipes and air dry at room temperature.
3. Microdissection of Blade, Ligule and Sheath Samples from Plastochron 7 Leaf Primordia
- Secure the slides on stage of LM microscope. Use five or six slides for each replicate, utilizing five sections per slide.
NOTE: Tissue pooling for a single replicate is illustrated in Figure 5. - Examine slides and identify the five most median sections on each slide, using the SAM apex as the central reference point.
NOTE: This can be done at low magnification, usually a 5X objective is sufficient. - Using 10X or 20X objective, identify the position of the ligule on the plastochron 7 leaf primordium of each section. The ligule will be visible as a bump protruding from the adaxial surface of the leaf primordium. Mark this position using the drawing tool of the LM software; select the pencil icon, move the cursor to the appropriate position and click and drag the mouse to draw.
NOTE: A 10X or 20X objective is appropriate for this and subsequent steps. When using lateral sections, the two sides of each leaf primordium will be present in each section (Figure 2A). - Using the ruler tool and the rectangular drawing tool, measure 100 µm high rectangles centered on the ligule of each section (red rectangles, Figure 2A, 2B). These will be the "Ligule" sample.
- To use the ruler tool; select the ruler icon, move the cursor to one end of the object to be measured, click and drag to measure the object. The length of the ruler will be shown on the screen.
- To draw a rectangle; select the rectangle icon, move the cursor to a point that will be one corner of the rectangle, click and drag to draw a rectangle of the appropriate size. Alternatively, select the straight line drawing tool and draw four straight lines.
- Measure 100 µm rectangles positioned 50 µm above and below the "Ligule" rectangle.
NOTE: These will be the "Blade" and "Sheath" samples, respectively (green and blue rectangles, Figure 2A, 2B). Based on our histological data, a 100 µm rectangle encompasses the entire ligule region. Equivalent sized portions of blade and sheath were chosen to ensure that similar amounts of tissue were collected for each. Spacers of 50 µm were used to ensure that no ligule region tissue is inadvertently included in the blade or sheath microdissections.
- Microdissect measured rectangles (Figure 2D–2F)7,8,9, collecting Ligule, Blade and Sheath samples in separate tubes. Use the laser cut function to cut through the tissue section along the outline of the selected domain. Use the catapult function to propel the rectangle of tissue off the slide and into the lid of the tube (Figure 2D–2F).
4. Microdissection of Blade, Ligule and Sheath Adaxial Epidermal Samples from Plastochron 7 Leaf Primordia
- Select sections and use the ruler tool to measure 100 µm high segments centered on the plastochron 7 ligule, as described in section 3 (above).
- Select only the adaxial epidermal cells of each 100 µm high "Blade" and "Sheath" segment (green and blue selections, Figure 2C) by outlining with the drawing tool. The epidermis is the outer cell layer; the adaxial side is the one closest to the SAM.
- For the "Ligule" sample, select only the cells of the emerging ligule bump as described in Section 3.3 (red selection, Figure 2C).
- Microdissect selected regions, collecting Blade, Ligule and Sheath epidermis samples in separate tubes, as described in section 3.5.
5. Microdissection of Plastochron 6 Leaf Primordia from lg1-R and Wild-type Siblings
- Grow segregating families of mutant (lg1-R) and wild-type plants.
- Fix and process shoot apices for LM, as described in sections 1.2-1.4. Fix wild-type and mutant shoot apices in separate vials in order to keep them separate. Samples from the same planting should be fixed and processed for in situ hybridization.
- Determine where lg1 is transcribed in wild-type siblings, by performing lg1 in situ hybridization6,26,27. Measure position of lg1 transcript accumulation from base of leaf primordium in multiple samples (Figure 1E).
- Based on in situ hybridization data, choose portion of leaf primordium that encompasses the region where lg1 is transcribed. In this case, 400-900 µm from the base of plastochron 6 leaf primordia (purple rectangles, Figure 2A).
- Microdissect selected portion of leaf primordia, as described in section 3.5, collecting lg1-R and wild-type samples in separate tubes.
6. Apply RNA Extraction Buffer
- Apply 50 µl RNA extraction buffer to microdissected tissue and proceed with RNA extraction. Continue with RNA extraction, RNA amplification, library construction, sequencing and bioinformatics analysis as described in reference6.
Representative Results
Using the LM scheme outlined in Figure 2, approximately 1,000,000-1,500,000 µm2 of tissue was collected for each replicate in the all-cell-layers LM (Figure 5), and 200,000 µm2 per replicate for the adaxial epidermis LM. Approximately 2,500,000 µm2 of tissue was collected for each replicate in the LM of lg1-R and wild-type leaf primordia. Two rounds of linear RNA amplification yielded microgram quantities of RNA for each replicate. The amount of RNA obtained from any given microdissection will depend on cell size and transcriptional activity as well as the amount of tissue captured. The integrity of the RNA will affect the efficiency of the RNA amplification.
Transcript accumulation was analyzed in all cell layers of the blade, ligule and sheath regions and a total of 2,359 DE genes were found, with a false discovery rate (FDR) of <0.05 (Figure 6A). Specifically, 1,714 genes were DE between ligule and blade, 1,044 genes were DE between ligule and sheath, and 657 genes were DE between blade and sheath (some genes are DE in more than one comparison). Genes were classified as upregulated in the ligule region if they were significantly upregulated in both ligule compared to blade and ligule compared to sheath. In the all cell layers LM, 373 genes were significantly upregulated in the ligule region.
Transcript accumulation was analyzed in the adaxial epidermis blade, ligule and sheath regions and a total of 3,128 DE genes were found; 1,971 genes were DE between ligule and blade, 2,032 genes were DE between ligule and sheath, and 871 genes were DE between blade and sheath (Figure 6B). 287 genes were significantly upregulated in the ligule compared to blade and sheath epidermis.
Data for selected genes that are upregulated in the ligule region are presented in Table 1 and Figure 7. An initial in situ hybridization analysis indicated that lg1 transcripts accumulate specifically in the PLB and the emerging ligule of wild-type leaf primordia, and that lg1 is more highly transcribed in the epidermis than in internal cell layers (Figure 6C). The LM RNA-seq data that were obtained are consistent with this result; lg1 transcript accumulation is significantly higher in the ligule than in either blade or sheath in both the epidermal LM and LM of all cell layers (Table 1, Figure 7A). lg1 has a higher CPM in the epidermal analysis than in the analysis of all cell layers, similar to the scenario illustrated in Figure 8C.
ns1 was significantly upregulated in the ligule region in both the all cell layers LM and the adaxial epidermis LM (Table 1, Figure 7B). This finding was confirmed by in situ hybridization, which shows that ns1 transcript accumulates specifically in the tip of the emerging ligule (Figure 6D). ns1 has a much higher read count in the epidermis-only LM analysis than in LM of all cell layers (19.6 CPM and 2.7 CPM, respectively) due to dilution of the transcript in the all layers LM. This dilution effect is illustrated in Figure 8A. Similarly, GRMZM2G101682, a gene of unknown function, had a higher mean read count in the ligule epidermal LM than in the all layers LM (Table 1, Figure 7C). In situ hybridization revealed that this gene transcript also accumulates most strongly in epidermal cells (Figure 6E).
Zm PIN1a was not significantly DE in the analysis of all cell layers, but was significantly upregulated in the ligule in the epidermis-only analysis (Table 1, Figure 7D). This is most likely because Zm PIN1a is highly expressed in vascular tissues and this L2-derived vascular expression confounds differences in Zm PIN1a accumulation in the epidermis when all cell layers are collected (Figure 6F). A similar scenario is depicted in Figure 8B.
To identify genes that are DE in lg1-R mutants, transcript accumulation was compared in a defined region of wild-type and lg1-R P6 leaf primordia. Ninety-six genes were DE in lg1-R (FDR <0.05); 59 were downregulated in lg1-R mutants, and 37 were upregulated. Data for selected genes that are DE in lg1-R mutants are presented in Table 2. bel14 is one of 34 genes that are both upregulated in the ligule region in wild-type leaf primordia and downregulated in lg1-R mutants. In situ hybridization confirmed that bel14 transcripts accumulate in the preligule region in wild-type plants but not in the corresponding region of lg1-R leaf primordia6.
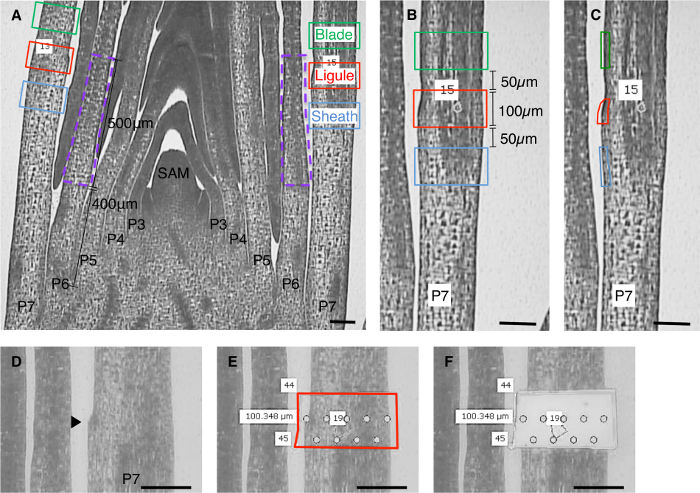
Figure 2: Scheme for laser microdissection of leaf primordial domains. (A) Lateral longitudinal section of maize shoot apex processed for LM. Boxes indicate regions selected for microdissection. Ligule region tissue (red box) was microdissected from 100 µm high rectangles, centered on the PLB. Pre-blade (green) and pre-sheath (blue) tissue was taken from 100 µm rectangles 50 µm above and below the preligule selection respectively. For the comparison of wild-type and lg1-R transcriptomes, tissue between 400-900 µm from the base of P6 leaf primordia was microdissected from lateral sections (purple dashed line). (B) Close-up of P7 primordium and regions selected for microdissection of all cell layers. (C) Close-up of P7 primordium and regions selected for microdissection of adaxial epidermis. (D) Preligule region of P7 primordium before microdissection. Arrowhead indicates position of initiating ligule. (E) Red line indicates region selected for microdissection. The laser will cut along this line. Circles indicate points where laser pulses will catapult tissue. (F) Primordium after microdissection. Circles indicate points where laser pulses have catapulted tissue. SAM = shoot apical meristem. P indicates plastochron number. Scale bars = 100 µm. This figure has been modified from reference6(Copyright American Society of Plant Biologists). Please click here to view a larger version of this figure.
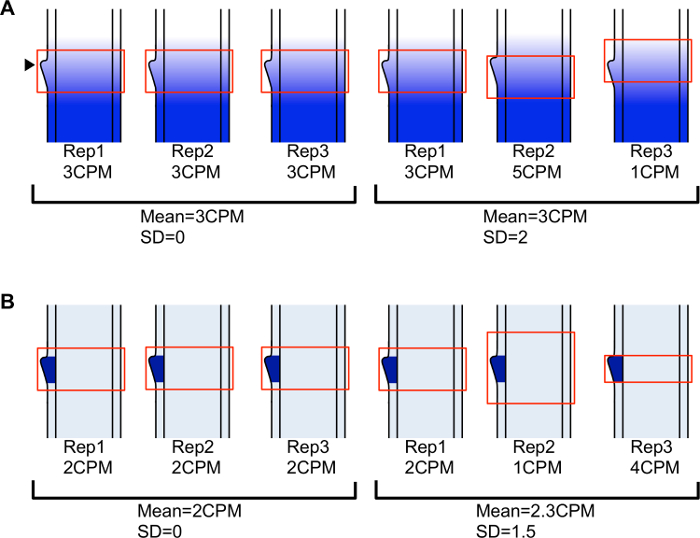
Figure 3: The region selected for LM affects read counts for genes that are DE along the leaf primordium. (A) Accurate positioning of the region selected for LM relative to morphological landmarks reduces variation for genes expressed in a gradient along the leaf axis. In replicates 1-3 on left, selections are centered on the emerging ligule resulting in low variation. In replicates 1-3 on right, positioning of region selected for LM is not consistent resulting in increased variation. (B) Microdissecting a consistently sized selection reduces variation (replicates 1-3 on left) compared to microdissecting different sized selections (replicates 1-3 on right) for genes with ligule-specific expression patterns. Transcripts are more diluted in the larger selection, resulting in a lower read count, and more concentrated in the smaller selection, resulting in a higher read count. Cartoons represent longitudinal sections through leaf primordia in the ligule region. Arrowhead indicates initiating ligule. CPM = counts per million. SD = standard deviation. Please click here to view a larger version of this figure.
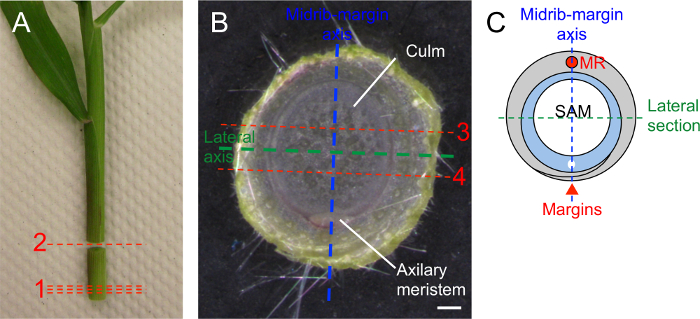
Figure 4: Dissection of maize seedling for lateral sections. (A) Two week-old maize seedling excised at root-shoot junction. Remove thin slices from the base of the shoot (1) until a small oval of culm is visible at base of shoot. Make transverse cut approximately 10 mm above base (2) and retain this cylinder. (B) Cylinder of tissue oriented so base is facing up. Note oval of culm encircled by leaf. Make two longitudinal cuts parallel to the lateral axis (3 and 4). Retain and fix central slice of tissue, this portion will include the SAM and young leaf primordia. (C) Cartoon illustrating plane of lateral section (green dashed line) through shoot apex. Red circle indicates midrib, red arrowhead indicates margins of grey leaf primordium. Blue dashed line: midrib-margin axis. Scale bar = 1 mm. Please click here to view a larger version of this figure.
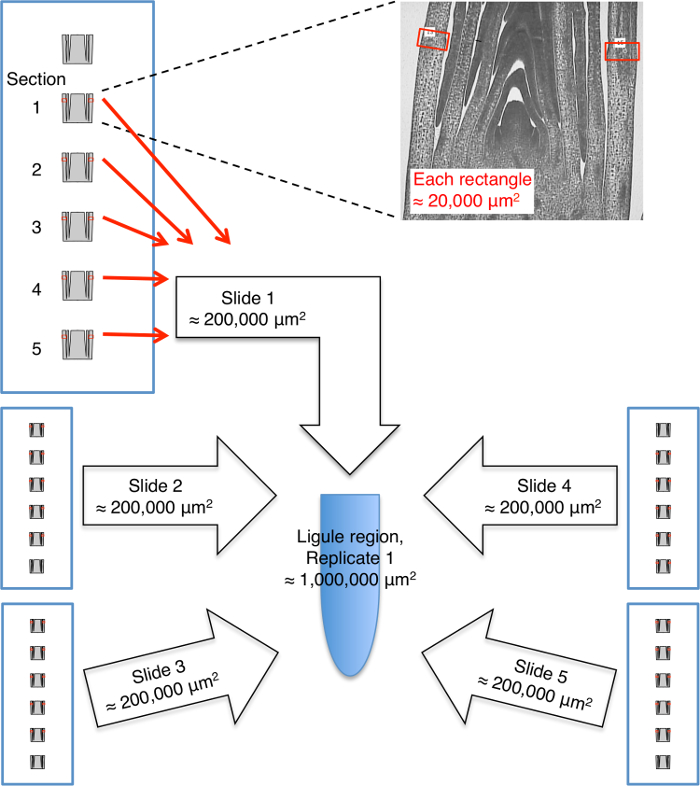
Figure 5: Cartoon illustrating pooling of laser microdissected tissue for one replicate of the ligule region. Tissue microdissected from five to six slides is pooled for each replicate. Five median sections from each slide are used and two selections (red rectangles) are made from each section. Each rectangle is approximately 20,000 µm2. Tissue for each replicate is collected in a separate tube (blue oval). Please click here to view a larger version of this figure.
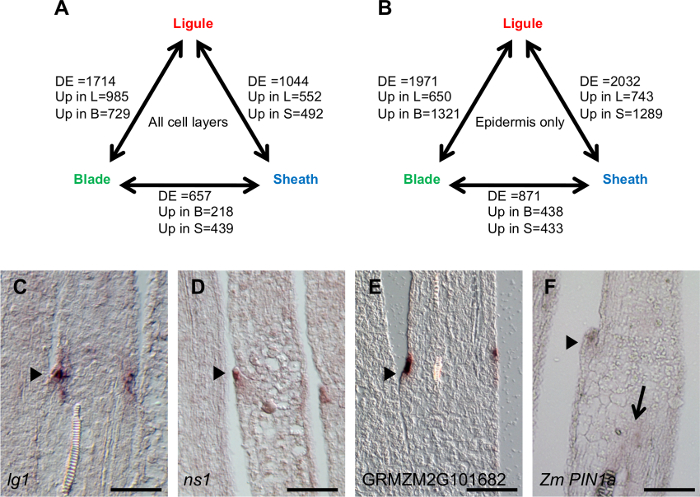
Figure 6: Number of DE genes in pairwise comparisons between leaf regions and in situ hybridization of selected DE genes. (A) Number of DE genes in pairwise comparison between blade, ligule and sheath regions, LM of all cell layers. (B) Number of DE genes in pairwise comparison between blade, ligule and sheath regions, LM of adaxial epidermis only. (C) lg1 in situ hybridization. (D) ns1 in situ hybridization. (E) GRMZM2G101682 in situ hybridization. (F) ZmPIN1a in situ hybridization. DE: differentially expressed. Up in L: up in ligule, Up in B: up in blade. Up in S: up in sheath. Arrowheads in C-F indicate emerging ligule. Arrow in F indicates transcript accumulation in vascular tissues. Scale bars = 100 µm. This figure has been modified from reference6 (Copyright American Society of Plant Biologists). Please click here to view a larger version of this figure.
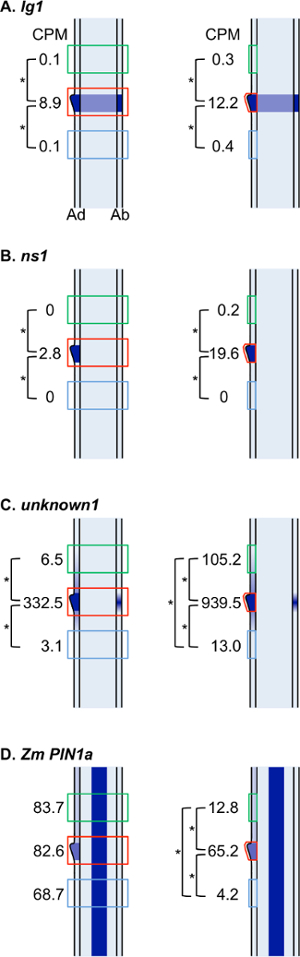
Figure 7: Graphical representation of data for selected DE genes. Figure illustrates data presented in Table 1 and in situ hybridization results. Cartoons represent longitudinal sections through leaf primordia. Dark blue indicates transcript accumulation for a particular gene. Green, red and blue boxes indicate region selected for microdissection for blade, ligule and sheath samples. Cartoons on the left illustrate LM of all cell layers. Cartoons on the right illustrate LM of adaxial epidermis only. Ad: adaxial epidermis, Ab: abaxial epidermis, CPM: counts per million, asterisk indicates significant difference between two domains. Please click here to view a larger version of this figure.
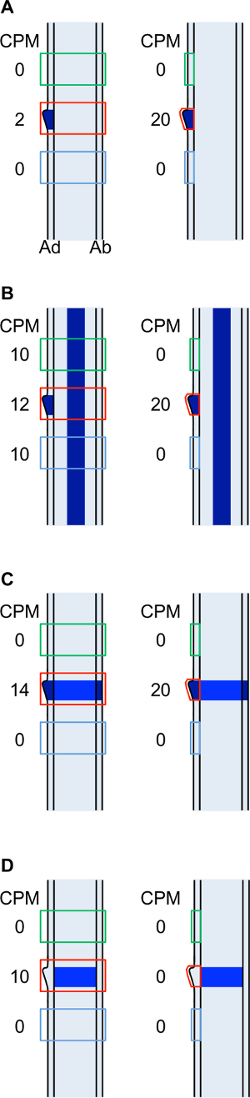
Figure 8: Cartoon illustrating how read counts for hypothetical DE genes differ depending on the precise regions selected for microdissection. (A) Gene with ligule-specific expression. (B) Gene expressed in the ligule and in vascular tissues. (C) Gene expressed specifically in the ligule region in all cell layers but with higher expression in the epidermis. (D) Gene expressed in the ligule region, L2-derived cell layers only. Cartoons represent longitudinal sections through leaf primordia. Dark blue indicates transcript accumulation for a particular gene. Green, red and blue boxes indicate region selected for microdissection for blade, ligule and sheath samples. Cartoons on the left illustrate LM of all cell layers. Cartoons on the right illustrate LM of adaxial epidermis only. Ad: adaxial epidermis, Ab: abaxial epidermis, CPM: counts per million. Please click here to view a larger version of this figure.
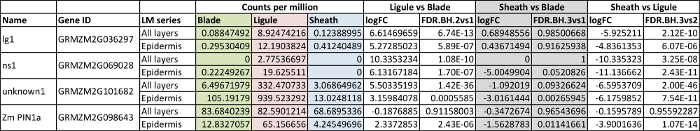
Table 1: Selected genes upregulated in the primordial ligule. Counts per million: counts per million total transcripts, mean of three replicates, logFC = log base 2 Fold Change, FDR.BH: false discovery rate according to Benjamini and Hochberg method. All layers: LM of all cell layers. Epidermis: LM of adaxial epidermis only. This figure has been modified from reference6 (Copyright American Society of Plant Biologists). Please click here to view a larger version of this table.
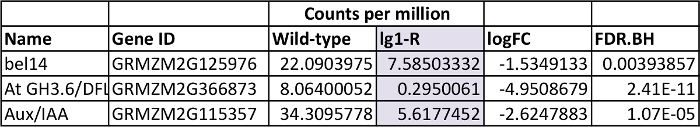
Table 2: Selected genes DE in lg1-R mutants. Counts per million: counts per million total transcripts, mean of three replicates, logFC = log base 2 Fold Change, FDR.BH: false discovery rate according to Benjamini and Hochberg method. This figure has been modified from reference6 (Copyright American Society of Plant Biologists). Please click here to view a larger version of this table.
Discussion
Experimental design is a critical factor in RNA-seq experiments. Key considerations are the precise domain(s) and developmental stage(s) to be analyzed, and what comparisons will be made. It is crucial to think in terms of comparisons, since the output is typically a list of genes that are DE between two or more conditions. As with all experiments, it is important to alter only one variable at a time. For example, when comparing different leaf domains, leaves of the same age and developmental stage, grown under the same conditions should be compared.
The aim of these experiments was to identify genes that are specifically up- or down-regulated in the ligule region of wild-type leaf primordia, and to identify genes that are DE in lg1-R mutants compared to wild-type. We first studied the morphology of developing leaf primordia and the pattern of lg1 transcript accumulation. These morphological and molecular landmarks were used to select precise domains for LM14,15,16. In the first experiment, transcript accumulation in the ligule region was compared to other regions of the same leaf primordia, thus eliminating differences due to growth conditions, genetic background, developmental stage, and plant-to-plant variation. To compare mutant to wild-type, it is necessary to determine where and when the gene of interest is expressed. We recommend analyzing the expression pattern by either in situ hybridization or immunolocalization and selecting a region for LM that encompasses the domain of gene expression. It is important that equivalent domains are compared and that the genetic background is the same for both mutant and wild-type.
The accuracy of microdissections may be verified by checking read counts for genes that are expressed specifically in the domain of interest, if such genes are known. This can also be done by performing RT-PCR on RNA extracted from LM tissue prior to making libraries for sequencing. In the experiments described here, lg1 served as a control for LM of the ligule region, since in situ hybridization analyses had shown that lg1 expression is specific to the PLB and emerging ligule5,6.
Unlike methods such as RT-qPCR, where the candidate gene(s) must be known, RNA-seq quantifies accumulation of all transcripts in a given tissue sample. Thus, LM RNA-seq is a useful method to identify candidate genes. An example from this study is GRMZM2G101682, a gene of unknown function that has a striking ligule-specific expression pattern (Figure 6E). This study also identified genes that are DE in lg1-R, such as bel14, which had not previously been implicated as acting downstream of lg1. In this instance, the comparison of multiple data sets (upregulated in the wild-type ligule region and DE in lg1-R) was particularly informative. It is important to note that the expression pattern of a gene does not demonstrate its function: subsequent genetic and/or biochemical analyses are required to establish gene function.
Transcriptomic analyses that are focused on small, discrete developmental domains, which are differentially distinct and easily delineated, are the most successful. LM RNA-seq is an appropriate method for such analyses and has the potential to be applied to many developmental phenomena. It is particularly suited to identify differential gene expression in mutants of genes that have spatially restricted expression patterns, or mutants that affect development of specific structures. It may also be applied to processes that occur in specific and identifiable cells or tissues, such as the epidermis or vasculature, and has been used in a transcriptomic analysis of maize shoot apical meristem (SAM) ontogeny16. It is less appropriate to studying the function of genes that are expressed ubiquitously or to processes that occur in all cells. The methods presented here have been successfully applied to a variety of plant species (maize, Sorghum, Ocimum, Mentha and Antirrhinum) and structures (leaf primordia, meristems, rhizomes and trichomes).
LM is not required when an analysis involves relatively large domains. Indeed, RNA-Seq on hand-dissected or whole organs has been used successfully to identify DE genes in plants14,15. This approach has the advantage of being less labor intensive and no experience with histological techniques is required. However, there are limitations to the precision of hand dissections, meaning that hand dissection is less suitable for studying early developmental processes.
For successful LM RNA-Seq analyses of developmental phenomena, the developmental context must be well characterized. We recommend undertaking a thorough histological examination of the process or domain of interest before planning an experiment. The histology of samples processed for LM is generally poor, since the tissues are unstained and are not mounted with cover slips that provide depth-of-field. Therefore, it is useful to fix separate samples for staining and observation at the same time as LM samples are fixed (Figure 1C, and D).
RNA obtained from LM material is generally fragmented and the total yield is low8. An amplification step is required to generate sufficient RNA for library preparation28. RNA fragment length should be assessed after amplification and prior to library generation. An average fragment length of several hundred base pairs is generally considered good, 500 base pairs is excellent. Poor RNA yield may result if tissue is not well fixed, if degradation due to RNase contamination occurs, or if paraffin blocks are incorrectly stored. It is helpful to test the quality of fixed tissue by undertaking a trial LM of a relatively large area before devoting a lot of time to microdissecting very small domains. Because the RNA obtained from LM cells is fragmented and amplification using an oligo dT primer introduces a strong 3' bias, this method is not suitable for detecting alternative transcripts such as splice variants. Nor is LM suitable for detecting small RNAs.
An alternative method to quantify transcript accumulation in specific tissues or cellular domains is RNA-seq of cells separated by Fluorescence Activated Cell Sorting (FACS)21. This method generally requires identification of a suitable marker gene for the region of interest, the generation of transgenic plants expressing a fluorescent-tagged protein and protoplasting. FACS combined with microarray has been used to generate an expression map of the Arabidopsis root by separating cells expressing cell type-specific promoters in the root22,23. FACS of shoot tissues is more challenging than root tissues due to chlorophyll autofluorescence24. A limitation of LM is that the domain of interest must be identifiable in histological sections. FACS may be a more appropriate method in cases where a suitable marker gene is available. The choice of which cells or tissues to isolate and compare is an important consideration in transcriptomic analyses that use either FACS or LM.
The data presented here illustrate that different results will be obtained depending on the precise domains selected for LM (Figure 3, Figure 8). Transcripts of genes that are expressed in one or a few cells, such as ns1, will be diluted when large tissue domains are analyzed. It is likely that if whole leaf primordia were sampled, ns1 transcript would not be detected. ZmPIN1a is significantly upregulated in the ligule compared to blade and sheath epidermis but this difference is confounded by vascular expression when all cell layers are sampled. Conversely, genes that are expressed only in L2-derived cell layers will not be detected when only the epidermis is sampled (Figure 8D). This study demonstrates that, while LM RNA-Seq is a powerful tool for detecting differences in gene expression during morphogenesis, the domains selected for analysis are crucial to the success of the experiment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank S. Hake for ongoing collaboration and stimulating discussions about ligule development. This work is supported by National Science Foundation Grants MCB 1052051 and IOS-1848478.
Materials
| Diethyl pyrocarbonate | Sigma-Aldrich | 159220 | Used for RNase treatment of solutions |
| Razor blades | Electron Microscopy Sciences | 72000 | |
| RNase Zap | Sigma-Aldrich | R2020-250ML | RNase decontamination solution |
| Ethanol absolute 200 proof | Fisher Scientific | BP28184 | |
| Acetic acid, glacial | Sigma-Aldrich | A6283 | |
| Glass vials – 22mL | VWR | 470206-384 | |
| Xylenes, histological grade | Sigma-Aldrich | 534056-4L | |
| Paraplast plus | Sigma-Aldrich | P3683-1KG | |
| Disposable base molds, 15 x 15 x 5mm | VWR | 15154-072 | |
| Embedding rings | VWR | 15154-303 | |
| Silica gel packets | Electron Microscopy Sciences | 71206-01 | Desiccant for storage of paraffin blocks |
| Oven | Fisher Scientific | 15-103-0503 | Oven must maintain temperature of 60° C |
| Paraffin embedding station | Leica | EG1160 | |
| Microtome | Leica | RM2235 | |
| Slide warmer | Electron Microscopy Sciences | 71317-10 | |
| Coplin jars | Electron Microscopy Sciences | 70316-02 | |
| Laser microdissector | Zeiss | ||
| KIMWIPES™ Delicate Task Wipers | Kimberly-Clark Professional | 34120 | Lint-free wipes for wicking excess solutions from microscope slides |
| Membrane Slide 1.0 PEN | Zeiss | 415190-9041-000 | Slides for laser microdissection |
| Adhesive Cap 200 opaque | Zeiss | 415190-9181-000 | Tubes for laser microdissection |
| PicoPure RNA Isolation Kit | ThermoFisher Scientific | KIT0204 |
References
- Becraft, P. W., Bongard-Pierce, D. K., Sylvester, A. W., Poethig, R. S., Freeling, M. The liguleless-1 gene acts tissue specifically in maize leaf development. Dev Biol. 141 (1), 220-232 (1990).
- Sylvester, A. W., Cande, W. Z., Freeling, M. Division and differentiation during normal and liguleless-1 maize leaf development. Development. 110 (3), 985-1000 (1990).
- Moreno, M. A., Harper, L. C., Krueger, R. W., Dellaporta, S. L., Freeling, M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11 (5), 616-628 (1997).
- Emerson, R. A. The inheritance of the ligule and auricle of corn leaves. Neb. Agr. Exp. Sta. An. Rep. 25, 81-85 (1912).
- Moon, J., Candela, H., Hake, S. The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development. 140 (2), 405-412 (2013).
- Johnston, R., et al. Transcriptomic Analyses Indicate That Maize Ligule Development Recapitulates Gene Expression Patterns That Occur during Lateral Organ Initiation. Plant Cell. 26 (12), 4718-4732 (2014).
- Schmid, M. W., et al. A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS One. 7 (1), e29685 (2012).
- Kerk, N. M., Ceserani, T., Tausta, S. L., Sussex, I. M., Nelson, T. M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 132 (1), 27-35 (2003).
- Nakazono, M., Qiu, F., Borsuk, L. A., Schnable, P. S. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell. 15 (3), 583-596 (2003).
- Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M., Gilad, Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18 (9), 1509-1517 (2008).
- Wang, Z., Gerstein, M., Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10 (1), 57-63 (2009).
- Brooks, L., et al. Microdissection of shoot meristem functional domains. PLoS Genet. 5 (5), e1000476 (2009).
- Cai, S., Lashbrook, C. C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146 (3), 1305-1321 (2008).
- Li, P., et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 42 (12), 1060-1067 (2010).
- Eveland, A. L., et al. Regulatory modules controlling maize inflorescence architecture. Genome Res. 24 (3), 431-443 (2014).
- Takacs, E. M., et al. Ontogeny of the maize shoot apical meristem. Plant Cell. 24 (8), 3219-3234 (2012).
- Aida, M., Ishida, T., Tasaka, M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 126 (8), 1563-1570 (1999).
- Ishida, T., Aida, M., Takada, S., Tasaka, M. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41 (1), 60-67 (2000).
- Takada, S., Hibara, K., Ishida, T., Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 128 (7), 1127-1135 (2001).
- Nardmann, J., Ji, J., Werr, W., Scanlon, M. J. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development. 131 (12), 2827-2839 (2004).
- Bonner, W. A., Hulett, H. R., Sweet, R. G., Herzenberg, L. A. Fluorescence activated cell sorting. Rev Sci Instrum. 43 (3), 404-409 (1972).
- Birnbaum, K., et al. A gene expression map of the Arabidopsis root. Science. 302 (5652), 1956-1960 (2003).
- Brady, S. M., et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 318 (5851), 801-806 (2007).
- Carter, A. D., Bonyadi, R., Gifford, M. L. The use of fluorescence-activated cell sorting in studying plant development and environmental responses. Int J Dev Biol. 57 (6-8), 545-552 (2013).
- Ruzin, S. E. . Plant microtechnique and microscopy. , (1999).
- Jackson, D., Veit, B., Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 120, 405-413 (1994).
- Javelle, M., Marco, C. F., Timmermans, M. In situ hybridization for the precise localization of transcripts in plants. J Vis Exp. (57), e3328 (2011).
- Day, R. C., McNoe, L., Macknight, R. C. Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm. Int J Plant Genomics. , 61028 (2007).

