Medium-scale Preparation of Drosophila Embryo Extracts for Proteomic Experiments
Summary
The goal of this protocol is to provide a straightforward and inexpensive approach to collecting Drosophila embryos at medium scale (0.5-1 g) and preparing protein extracts that can be used in downstream proteomic applications, such as affinity purification-mass spectrometry (AP-MS).
Abstract
Analysis of protein-protein interactions (PPIs) has become an indispensable approach to study biological processes and mechanisms, such as cell signaling, organism development, and disease. It is often desirable to obtain PPI information using in vivo material, to gain the most natural and unbiased view of the interaction networks. The fruit fly Drosophila melanogaster is an excellent platform to study PPIs in vivo, and lends itself to straightforward approaches to isolating material for biochemical experiments. In particular, fruit fly embryos represent a convenient type of tissue to study PPIs, due to the ease of collecting animals at this developmental stage and the fact that the majority of proteins are expressed in embryogenesis, thus providing a relevant environment to reveal most PPIs. Here we present a protocol for collection of Drosophila embryos at medium scale (0.5-1 g), which is an ideal amount for a wide range of proteomic applications, including analysis of PPIs by affinity purification-mass spectrometry (AP-MS). We describe our designs for 1 L and 5 L cages for embryo collections that can be easily and inexpensively set up in any laboratory. We also provide a general protocol for embryo collection and protein extraction to generate lysates that can be directly used in downstream applications such as AP-MS. Our goal is to provide an accessible means for all researchers to carry out the analyses of PPIs in vivo.
Introduction
Genetic screens and, more recently, genomic approaches have revolutionized the study of biological functions. However, important cellular information is encoded in proteins and their ensemble of interacting partners. While traditional genetic modifier screens can identify rate-limiting pathway components and recover indirect interactions, the strength of the proteomic approaches lies in their ability to identify complete immediate interaction networks of proteins of interest. Proteomics is thus a valuable orthogonal method to study biological systems, and complements genomics, transcriptomics, and traditional genetic screens. Affinity purification-mass spectrometry (AP-MS) has proven to be a powerful approach to study protein-protein interactions (PPIs) in their native environment in cells and tissues1,2. This method allows for the identification of direct or indirect interactions at specific developmental stages or tissue contexts, and has been successfully used to identify multiple novel PPIs in a variety of developmental pathways (reviewed in reference1). Despite the undisputed success of PPI studies, most of them have been carried out in cultured cells, in which the "bait" proteins of interest were overexpressed. There are two issues with studying PPIs in cell culture: first, a specific cell line may not provide a full complement of interactions due to lack of expression of certain proteins. Second, high overexpression usually employed in such analyses might lead to artefacts such as protein misfolding or identification of false positive interactions.
Both of these limitations can be overcome by analyzing PPIs in vivo. A limiting step in such experiments is the availability of the starting material for purifying protein complexes. Drosophila melanogaster has long been used as a model for functional analysis, and recently it has also been shown to be an excellent system for studying PPIs in vivo. Drosophila embryogenesis represents a particularly attractive tissue type to study PPIs, because embryos can be easily collected in large quantities, and also because most genes (>88%) are expressed over the course of embryogenesis, thus providing a rich in vivo environment for detecting relevant PPIs3.
Traditionally, biochemical studies in flies utilized very large-scale embryo collections (100-150 g), such as those necessary for purifying functional transcriptional lysates4,5. Previous AP-MS studies in Drosophila also needed large amounts of embryos (5-10 g), because they relied on a two-step purification approach such as tandem affinity purification (TAP), with the associated loss of material at each step6. Large amounts of starting material necessitated setting up embryo collections in large population cages, which can be both expensive (when purchased commercially) and time-consuming to maintain and clean7,8,9.
Recent advances in the development of single-step affinity purification approaches, as well as the increasing sensitivity of mass spectrometers, have reduced the necessary amount of starting material by an order of magnitude. Using tags such as the streptavidin-binding peptide (SBP) or green fluorescent protein (GFP) and starting from less than 1 g of embryos, it is possible to isolate the amounts of the bait protein and interacting components that would be sufficient for identification by mass spectrometry10,11.
The goal of the protocol presented here is to help the researchers overcome a perceived barrier to biochemical analysis of PPIs in vivo. To that end, we provide a simple and inexpensive procedure to collect Drosophila embryos at medium scale (0.5-1 g), followed by one-step preparation of whole-cell protein extracts that are suitable for subsequent analysis by AP-MS or other approaches. Our method relies on the use of custom-made 1-L or 5-L population cages that can be easily produced by any laboratory. Furthermore, the extraction conditions presented here have been validated in several studies, both in cultured cells and in vivo10,12,13,14,15,16,17.
Protocol
1. Preparation of 5 L Fly Cages (to Fit 15 cm Plates)
- Obtain the materials needed: 5-quart (4.7-L) container with lid (Figure 1A), nylon mesh, and a razor blade.
- Mark a 12 cm diameter circle and cut a hole in the bottom of the container using the razor blade (Figure 1B). Cut a 15 cm diameter hole in the lid (Figure 1C).
- Cut nylon mesh into 25 x 25 cm squares.
- Place the mesh over the top rim of the container and press down with the lid (Figure 1C). Make sure the mesh is tight and the lid is not tilted. The cage is ready to be populated with flies (see step 5.3).
2. Preparation of 1 L Fly Cages (to Fit 10 cm Plates)
- Obtain the materials needed: an electric drill, a 3 inch (76 mm) cutting tool (Figure 1E and Figure 1F), a straight-sided 1 L plastic jar with lid, nylon mesh, a razor blade, sandpaper, goggles, and work gloves.
- Cut 3 inch (76 mm) holes in the bottom and the lid of the plastic jar (Figure 1G), with the lid tightly screwed onto the container while drilling. Position the cutting tool in the center of the circle when starting to make a hole.
Caution: Wear eye protection and work gloves during this step. - Smoothen the edges of the holes with a razor blade and sandpaper.
- Cut nylon mesh into 18 x 18 cm squares.
- Place the mesh over the top rim of the jar and tighten by screwing the lid on (Figure 1G). Make sure the mesh is tight and the lid is not tilted. The cage is ready to be populated with flies (see step 5.3).
3. Preparation of Apple Juice (AJ) Plates
- Add 19 g agar and 1 L of 18.2 MΩ·cm water into a 2 L flask. Add a stir bar, and mix briefly.
- Autoclave for 30 min. Meanwhile, thaw 100% apple juice concentrate from frozen stock, and prepare 10 mL of 10% (weight/volume) methyl 4-hydroxybenzoate solution in ethanol.
- After autoclaving, start mixing the agar on a stir plate, and add 80 mL of 100% apple juice concentrate and 10 mL of 10% (weight/volume) methyl 4-hydroxybenzoate solution to the flask. Let cool at room temperature with constant mixing.
- Pour 15 cm Petri dishes (for 5 L cages) or 10 cm Petri dishes (for 1 L cages) so that the level of agar in each dish is approximately 5 mm thick. Allow to solidify at room temperature, and wrap in plastic bags. AJ plates can be stored at 4 °C for two weeks.
Note: Petri dishes can be washed and reused. - Prepare wet yeast paste: pour some active dry yeast into a 100 mL container with a lid that would allow easy mixing, and add water in small amounts while stirring, to obtain a thick but thoroughly mixed paste. Store at 4 °C for up to 2 weeks.
4. Preparation of Lysis Reagents
- Prepare 5x lysis buffer: 250 mM Tris pH 7.5, 25% glycerol, 1% octylphenoxy poly(ethyleneoxy)ethanol (IGEPAL CA-630), 7.5 mM MgCl2, 625 mM NaCl, 125 mM NaF, 5 mM Na3VO4.
- To make 200 mL: completely dissolve 1 g NaF powder and 184 mg Na3VO4 powder in 71 mL water with constant stirring. Add 50 mL 1 M Tris pH 7.5, 2 mL 100% IGEPAL, 1.5 mL 1 M MgCl2, and 25 mL 5 M NaCl. Then add 50 mL glycerol (add last) and continue stirring for 1 h.
- Filter-sterilize using 250-mL filter units. This step can be slow, but filtering the solution is important to remove insoluble particulates. Aliquot by 10 mL in 50 mL tubes and store at -80 °C.
- Prepare 1 M dithiothreitol (DTT) solution in water. Store at -20 °C in 1 mL aliquots.
- Obtain protease inhibitor cocktail tablets, with ethylenediaminetetraacetic acid (EDTA). Store at 4 °C.
- Obtain 10 mL syringes and syringe filters: 26 mm diameter, surfactant-free cellulose acetate (SFCA) membrane, 0.45 µm pore size.
5. Collection of Fly Embryos
- Amplify the transgenic fly lines carrying the tagged protein of interest in bottles (approximately 100 flies per bottle), and transfer the bottles every 2 days until 25-30 bottles are accumulated for each fly line. Amplify the control stock (e.g. the y w line) in a similar way.
- Warm AJ plates to room temperature (about 1-2 h, can be done overnight). Using a gloved finger, spread some wet yeast paste in the center of AJ plates, to make an approx. 6-cm circle for a 15 cm plate and a 4 cm circle for a 10 cm plate.
- Anesthetize the flies with a flow of carbon dioxide using a standard Drosophila stage setup, and transfer into the cages (approx. 15 well-grown bottles of flies per 5 L cage, 5 bottles per 1 L cage).
- Cover the cage bottom with prepared AJ plates from step 5.2, and tape the AJ plate to the cage using two strips of tape (Figures 1D and 1H). It is useful to fold over the tips of the tape that go over the plate for easy peeling when changing plates.
- After the flies recover from anesthesia, keep the cages with the plates at the bottom and mesh on top.
Note: It is important to wait until the flies fully recover, as they will otherwise stick to the AJ plate and yeast paste. - At the end of the experiment, to prepare the cage for cleaning, place the cage in the freezer for several hours. Soaking the mesh in water for a few hours will facilitate cleaning.
- Incubate the cages with flies at room temperature for 2-3 days and change AJ plates twice a day. To change the plates, turn the cage upside down, peel the tape from the plate but hold the plate to the cage, tap the whole cage gently on the bench, and quickly swap the old plate for a new one, trying not to allow flies to escape.
Note: Flies need to adjust to the cage and will lay best starting on day 3. They will continue laying at peak level for about 4-5 days. Two 5 L cages at top laying capacity can produce up to 1 g of embryos in an overnight collection. Use 1 L cages for smaller-scale experiments. - Allow flies to lay embryos on fresh AJ plates overnight (approx. 16 h), or for any other desired length of time.
- The next morning, mark and weigh mesh containers for embryo collection, one per each fly line used.
- Prepare 1x lysis buffer, using reagents from section 4 (Note: ready-to-use 1x lysis buffer has to be prepared right before extraction): add 40 mL water to 10 mL of 5x concentrated lysis buffer (stored at -80 °C) in a 50 mL tube.
- Add 50 µL of 1 M DTT to a final concentration of 1 mM, mix well and separate into two 50 mL tubes, 25 mL in each. To one of the tubes, add one protease inhibitor tablet and rotate at 4 °C for 30 min. While the tablet is dissolving, collect and dechorionate embryos (steps 5.8 to 6.6). Check to make sure the tablet has fully dissolved, and keep the buffer on ice.
Note: The second tube of 1x buffer with DTT can be stored at -80 °C and will only require addition of the tablet before the next experiment. 25 mL of lysis buffer is sufficient for up to 2 samples, taking into account subsequent washes during protein purification steps.
- Add 50 µL of 1 M DTT to a final concentration of 1 mM, mix well and separate into two 50 mL tubes, 25 mL in each. To one of the tubes, add one protease inhibitor tablet and rotate at 4 °C for 30 min. While the tablet is dissolving, collect and dechorionate embryos (steps 5.8 to 6.6). Check to make sure the tablet has fully dissolved, and keep the buffer on ice.
- Mark the AJ plates on the cages with the genotypes of the fly lines used, and swap the plates for fresh ones.
Note: Excess yeast paste can be scraped off the plate at this step. Embryos can be aged on the plates at 25 °C or room temperature to obtain the desired window of developmental time. - Add enough water to cover each AJ plate. Gently dislodge the embryos from the plate with a soft paint brush (flat head approx. 10 mm wide works well for large plates). Pour the embryos into the previously weighed and marked mesh container, and drain excess water.
- If needed, perform a second rinsing and brushing of the plate to remove all the embryos. Combine the embryos from additional plate(s) for the same fly line into the same mesh container.
- Briefly blot the mesh on paper towels to remove excess water.
6. Embryo Dechorionation
- Half-fill a 6 cm Petri dish with 50% bleach (vol/vol solution in water).
Note: See important comment about bleach brand in the Table of Materials and Reagents.
Caution: Wear gloves and avoid getting bleach on clothes. - Prepare 2 larger Petri dishes with water.
- Immerse the mesh container with embryos in 50% bleach for 90 s. Tap the mesh gently during incubation a couple of times to suspend the embryos in the bleach solution.
- At the end of the 90 s incubation, wash the mesh container in each of the 2 dishes with water for about 10 s.
- Rinse the mesh container thoroughly with 18.2 MΩ·cm water using the squirt bottle. Also, wash the sides and the outside of the mesh container.
Note: No bleach smell should be detected after a thorough wash. - Blot the mesh container on paper towels. Weigh the mesh container with the embryos and subtract the weight of the empty container obtained in step 5.6. Record the resulting weight of the embryos. A good amount of embryos to aim for is 0.5 – 1 g.
7. Preparation of Embryo Extract
- Add 1 mL of lysis buffer with protease inhibitors (from step 5.7) to a dounce glass homogenizer and keep it on ice. Transfer the embryos from the mesh container into the homogenizer.
Note: Embryos can be picked up with a flat end of a metal spatula or a brush and transferred directly into the buffer. Perform all steps in this section on ice. - Wash off the remaining embryos on the mesh with an additional 1 mL lysis buffer and add to the material in the homogenizer. Let the embryos settle down to the bottom of the homogenizer, and carefully aspirate the supernatant.
- Add lysis buffer with protease inhibitors (from step 5.7) to the homogenizer, aiming for 5-6 mL of lysis buffer per gram of embryos.
- Homogenize the embryos by 4-6 strokes with a loose fitting pestle.
Note: During the first strokes, there will be more resistance. Try to move the pestle in a continuous motion to avoid splashing the solution. - Homogenize the embryos by 8-10 strokes with a tight fitting pestle.
- Incubate the homogenate on ice for 20 min.
- Transfer the homogenate into microcentrifuge tubes and centrifuge at 4 °C for 15 min at top speed (approx. 13,000 x g).
- Avoiding the pellets and the very top lipid layer, transfer the supernatants into fresh microcentrifuge tubes and centrifuge at 4 °C for 15 min at top speed (approx. 13,000 x g).
- Carefully aspirate supernatants with 1 mL tip and load into chilled 10 mL syringes with 0.45 µm filters attached (combine aliquots of the same sample into one syringe). Push all of the solution into a labeled 15 mL tube on ice. The extract can be immediately used for subsequent protein purification experiments or snap-frozen in liquid nitrogen and kept at -80 °C for future use.
Note: It is better to freeze the extracts rather than the embryos, because during subsequent thawing of embryos proteases may become activated and lead to protein degradation.
Representative Results
To illustrate the use of this protocol in a protein complex purification experiment, we generated a homozygous viable fly line, arm-EGFP-ERK, expressing EGFP-tagged Drosophila extracellular signal-regulated kinase (ERK, encoded by the rolled gene) under the control of a ubiquitously expressed armadillo (arm) promoter18,19. The arm promoter is active during most stages of Drosophila embryogenesis19. Lysates were prepared using the described protocol, and the expression of protein from the arm-EGFP-ERK transgene was confirmed by western blotting for total ERK protein to simultaneously detect the levels of endogenous and transgenic EGFP-ERK, migrating at 42 kDa and 69 kDa, respectively (Figure 2A). A single band corresponding to untagged endogenous ERK (42 kDa) was detected in the y w control line. This result confirms a successful extraction of ERK and EGFP-ERK from embryos.
To test whether our extraction protocol is suitable for subsequent purification of a tagged protein, extracts from the y w and arm-EGFP-ERK flies were subjected to affinity purification using GFP-agarose beads following established protocols10,11. Silver staining of samples run on a gradient SDS-PAGE showed a successful purification of the bait protein, EGFP-ERK (Figure 2B). Besides EGFP-ERK itself, the experimental lane contained additional bands that were not observed in the control y w sample, suggesting that the purification procedure recovered potential ERK-interacting proteins.
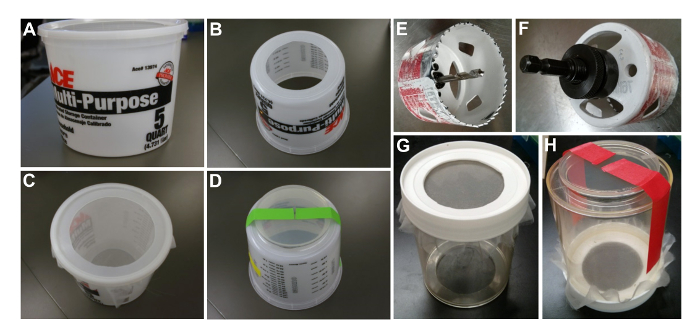
Figure 1: Preparation of Population Cages. (A–D) A 5-L cage made from a plastic container. (A) A starting container. (B) Bottom view of the 5 L cage. (C) Top view of 5 L cage with mesh attached. (D) Assembled 5 L cage with a 15 cm apple juice plate, bottom view. (E and F) A 3 inch (76 mm) drill bit used for drilling holes in the 1 L container. (G, H) A 1 L cage. (G) Top view of a 1 L cage with mesh attached. (H) Assembled 1 L cage with a 10 cm apple juice plate attached, bottom view. Please click here to view a larger version of this figure.
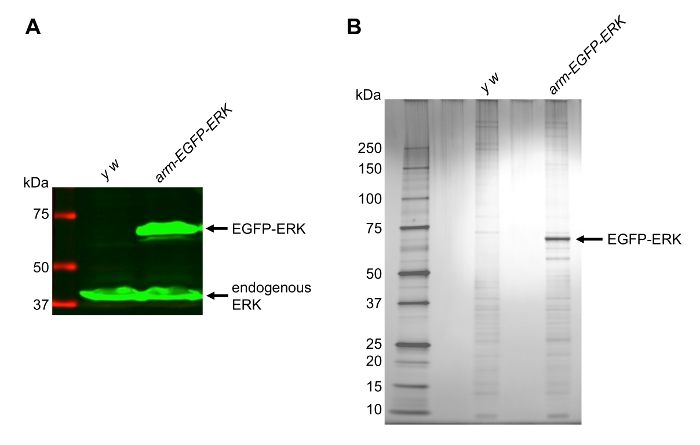
Figure 2: Extraction and Purification of EGFP-ERK from Embryos. (A) Western blot showing expression of EGFP-ERK and endogenous ERK in an extract prepared from the arm-EGFP-ERK transgenic line. The y w line was used as a control. Green, total ERK antibody; red, molecular weight marker. (B) Silver stained gel showing purified EGFP-ERK and associated proteins. Please click here to view a larger version of this figure.
Discussion
The protocol presented here is a simple and general procedure for setting up Drosophila population cages at medium scale and making whole-cell protein extracts from embryos. The resulting extracts can be used in a variety of downstream applications, such as purification of protein complexes on affinity resins. It is critical to perform the extraction steps on ice and use strong protease inhibition, to minimize protein degradation. As described, the protocol is suitable for isolation of most cytoplasmic and some membrane and soluble nuclear proteins6,10,12,15. It can be easily adapted for a more focused purification of proteins from other compartments, such as isolation of less soluble nuclear proteins using high salt extraction5. As mentioned in step 5.8, collection times and embryo aging periods can be set up in precise intervals to obtain different developmental stages.
It is often desirable to ascertain that the tagged protein of interest is expressed at levels closely matching the endogenous levels of expression, as we have shown using anti-total ERK antibody (Figure 2A). If the antibody against endogenous protein is not available, the functionality of the constructs can be verified by other assays, e.g. in a genetic rescue experiment. In most cases however, a mild overexpression of the tagged protein should still result in isolation of meaningful protein complexes, and may in fact facilitate extraction and purification of "difficult" proteins.
The main advantage of this protocol is its simplicity and a possibility to set up the whole procedure at moderate cost. An additional advantage of using smaller cages is that multiple transgenic lines can be set up and analyzed in parallel, which may not be feasible when using larger cages. The fly cages we describe here can be easily cleaned and reused for years. Instead of making 1-L cages from Nalgene jars as described, it is possible to use other available containers, such as a smaller version of the 5-L container.
We have successfully used this protocol (or its variations with small modifications) for purifying complexes containing various signaling proteins and associated components, followed by analysis by mass spectrometry6,10,15. These approaches have led to the identification of novel PPIs that were further validated in functional studies10,15. Variations of the protocol presented here have been previously used to analyze the phosphoproteome in Drosophila embryogenesis20, and to study the landscape of ubiquitination in neural development21,22. This procedure therefore represents a convenient gateway to analyzing PPIs in vivo, and is usable for other proteomics applications.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank members of the Veraksa laboratory for helpful comments on the manuscript and suggestions for protocol improvements. A.V. was supported by the NIH grants GM105813 and NS096402. L.Y. was supported by the University of Massachusetts Boston Sanofi Genzyme Doctoral Fellowship.
Materials
| Leaktite 5-Qt. Natural Multi Mix Container (Pack of 3) | Home Depot | 209330 | For making 5-L cages. |
| Leaktite 5-Qt. Natural Multi Mix Lid (Pack of 3) | Home Depot | 209320 | Diameter 8.37 in. Lids for 5-L cages. |
| Fisherbrand Petri dishes with clear lid, 100 x 15 mm | Fisher Scientific | FB0875712 | Material: Polystyrene. To be used with 1-L cages. |
| Fisherbrand Petri dishes with clear lid, 150 x 15 mm | Fisher Scientific | FB0875714 | Material: Polystyrene. To be used with 5-L cages. |
| Fisherbrand Petri dishes with clear lid, 60 x 15 mm | Fisher Scientific | FB0875713A | Material: Polystyrene. Used during embryo dechorionation. |
| Sefar NITEX nylon mesh | Sefar | 03-180/44 | 180 micron, 44% open area, used for fly cages. |
| Milwaukee 3 in. Hole Dozer Hole Saw with Arbor | Home Depot | 49-56-9670 | 3 inch cutting tool for making 1-L cages. |
| Nalgene Wide-Mouth Straight-Sided PMP Jars with White Polypropylene Screw Closure | Fisher Scientific | 11-823-33 | For making 1-L cages. Thermo Scientific cat # 21171000. Alternative 1-L containers can be used. |
| Red Star Active Dry Yeast, 2 pound pouch | Red Star | can be purchased from Amazon.com or other suppliers. | |
| Frozen 100% Apple Juice Concentrate | can be purchased from a grocery store. Has to say "100% apple juice" on the can. | ||
| Methyl 4-hydroxybenzoate | Acros Organics | AC126965000 | also known as methylparaben or Tegosept. Used as a preservative in AJ plates. |
| IGEPAL CA-630 for molecular biology, 100 ml | Sigma | I8896 | used for preparing lysis buffer. |
| Nalgene Rapid-Flow Sterile Disposable Filter Units with CN Membrane | Fisher Scientific | 09-740-2A | 0.2 μm pore size. Used for filtering 5x lysis buffer. Thermo Scientific cat # 1260020. |
| CO-RO ROCHE cOmplete Protease Inhibitor Cocktail | Sigma | 11697498001 | Vial of 20 tablets. Used for protease inhibition in lysis buffer. |
| Corning SFCA syringe filters | Fisher Scientific | 09-754-21 | SFCA membrane, diameter 26 mm, pore size 0.45 μm. Used for filtering final extract samples. |
| BD Luer-Lok Disposable Syringes without Needles | Fisher Scientific | 14-823-2A | for filtering final extract samples. BD cat # 309604. |
| Bleach | Clorox | used for embryo dechorionation at 50% (vol/vol) in water, can be purchased at any supermarket. It is important to use the Clorox brand, as other brands may result in incomplete dechorionation or may be toxic for embryos. | |
| Corning Costar Netwell Plates | Fisher Scientific | 07-200-213 | mesh containers for embryo collections. 74 μm mesh size, 6-well. |
| Wheaton Dounce Tissue Grinders, capacity 15 ml | Fisher Scientific | 06-435B | homogenizer for embryos, with loose and tight pestles. Wheaton cat # 357544. |
References
- Veraksa, A. Regulation of developmental processes: insights from mass spectrometry-based proteomics. Wiley Interdiscip Rev Dev Biol. 2 (5), 723-734 (2013).
- Gavin, A. C., Maeda, K., Kuhner, S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 22 (1), 42-49 (2011).
- Arbeitman, M. N., et al. Gene expression during the life cycle of Drosophila melanogaster. Science. 297 (5590), 2270-2275 (2002).
- Soeller, W. C., Poole, S. J., Kornberg, T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 2 (1), 68-81 (1988).
- Kamakaka, R. T., Kadonaga, J. T. The soluble nuclear fraction, a highly efficient transcription extract from Drosophila embryos. Methods Cell Biol. 44, 225-235 (1994).
- Kyriakakis, P., Tipping, M., Abed, L., Veraksa, A. Tandem affinity purification in Drosophila: The advantages of the GS-TAP system. Fly (Austin). 2 (4), 229-235 (2008).
- Shaffer, C. D., Wuller, J. M., Elgin, S. C. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 44, 99-108 (1994).
- Sisson, J. C. Culturing large populations of Drosophila for protein biochemistry. CSH Protoc. 2007, (2007).
- Caravaca, J. M., Lei, E. P. Maintenance of a Drosophila melanogaster Population Cage. J Vis Exp. (109), (2016).
- Yang, L., et al. Minibrain and Wings apart control organ growth and tissue patterning through down-regulation of Capicua. Proc Natl Acad Sci U S A. 113 (38), 10583-10588 (2016).
- Neumuller, R. A., et al. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. 유전학. 190 (3), 931-940 (2012).
- Veraksa, A., Bauer, A., Artavanis-Tsakonas, S. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev Dyn. 232 (3), 827-834 (2005).
- Mukherjee, A., et al. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 7 (12), 1191-1201 (2005).
- Gilbert, M. M., Tipping, M., Veraksa, A., Moberg, K. H. A Screen for Conditional Growth Suppressor Genes Identifies the Drosophila Homolog of HD-PTP as a Regulator of the Oncoprotein Yorkie. Dev Cell. 20 (5), 700-712 (2011).
- Anjum, S. G., et al. Regulation of Toll Signaling and Inflammation by beta-Arrestin and the SUMO Protease Ulp1. 유전학. 195 (4), 1307-1317 (2013).
- Degoutin, J. L., et al. Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat Cell Biol. 15 (10), 1176-1185 (2013).
- Zhang, C., et al. The ecdysone receptor coactivator Taiman links Yorkie to transcriptional control of germline stem cell factors in somatic tissue. Dev Cell. 34 (2), 168-180 (2015).
- Lemmon, M. A., Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell. 141 (7), 1117-1134 (2010).
- Vincent, J. P., Girdham, C. H., O’Farrell, P. H. A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev Biol. 164 (1), 328-331 (1994).
- Zhai, B., Villen, J., Beausoleil, S. A., Mintseris, J., Gygi, S. P. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 7 (4), 1675-1682 (2008).
- Franco, M., Seyfried, N. T., Brand, A. H., Peng, J., Mayor, U. A novel strategy to isolate ubiquitin conjugates reveals wide role for ubiquitination during neural development. Mol Cell Proteomics. 10 (5), (2011).
- Ramirez, J., et al. Proteomic Analysis of the Ubiquitin Landscape in the Drosophila Embryonic Nervous System and the Adult Photoreceptor Cells. PLoS One. 10 (10), 0139083 (2015).

