Detection of Small GTPase Prenylation and GTP Binding Using Membrane Fractionation and GTPase-linked Immunosorbent Assay
Summary
Here we describe a protocol to investigate the prenylation and guanosine-5′-triphosphate (GTP)-loading of Rho GTPase. This protocol consists of two detailed methods, namely membrane fractionation and a GTPase-linked immunosorbent assay. The protocol can be used for measuring the prenylation and GTP loading of different other small GTPases.
Abstract
The Rho GTPase family belongs to the Ras superfamily and includes approximately 20 members in humans. Rho GTPases are important in the regulation of diverse cellular functions, including cytoskeletal dynamics, cell motility, cell polarity, axonal guidance, vesicular trafficking, and cell cycle control. Changes in Rho GTPase signaling play an essential regulatory role in many pathological conditions, such as cancer, central nervous system diseases, and immune system-dependent diseases. The posttranslational modification of Rho GTPases (i.e., prenylation by mevalonate pathway intermediates) and GTP binding are key factors which affect the activation of this protein. In this paper, two essential and simple methods are provided to detect a broad range of Rho GTPase prenylation and GTP binding activities. Details of the technical procedures that have been used are explained step by step in this manuscript.
Introduction
Rho GTPases are a group of small proteins (21 – 25 kDa), which are well conserved throughout evolution, and form a unique subfamily in the Ras superfamily of small GTPases. In each subfamily within this superfamily, there is a shared G domain core that is involved in the GTPase activity and nucleotide exchange1. The difference between the Rho family and the other Ras subfamilies is the presence of a "Rho insert domain" within the 5th β strand and the 4th α helix in the small GTPase domain2.
Based on the recent classification, Rho GTPases are considered a family of signaling proteins that fit into the Ras GTPase superfamily3. Mammalian Rho GTPases have 22 members based on their specific function and general characterization4 in which RhoA, Rac1, and Cdc42 are among the most-studied members in this group. Rho GTPases are linked to intracellular signaling pathways via a tightly regulated mechanism which is dependent on molecular switches via protein posttranslational modifications5.
GTP loading and hydrolysis are essential mechanisms in the activation/deactivation cycle of small Rho GTPases and are regulated via GTPase-activating proteins (GAPs). GAPs are responsible for the GTP hydrolysis and work in concert with guanine nucleotide exchange factors (GEFs) which are responsible for the GTP-loading reaction. Rho GDP dissociation inhibitors (GDIs) provide further regulation of small Rho GTPases via binding to the GDP-bound Rho GTPases. This inhibits GDP dissociation and facilitates sequestering of small Rho GTPases away from the active intracellular membrane sites. There is also further regulation of Rho GTPase proteins involving the prenylation of GDIs which regulates both nucleotide hydrolysis and exchange and controls GDP/GTP cycling1,6,7,8.
Both GTP-loading and Rho GTPase prenylation are involved in the movement of Rho GTPase between cytosol and cell membranes by changing the lipophilic properties of these proteins1,9. The abovementioned regulators interact with phospholipids of the cell membrane and other modulating proteins of the GDP/GTP exchange activity10. Moreover, GDIs, dissociation inhibitors, block both the GTP hydrolysis and the GDP/GTP exchange. GDIs inhibit the dissociation of the inactive Rho proteins from GDP and, therefore, their interaction with downstream effectors. GDIs also regulate the cycling of GTPases between the cytosol and membrane in the cell. The activity of Rho GTPases depends to a great extent on their movement to the cell membrane; thus, GDIs are regarded as critical regulators that can sequester GTPases in the cytoplasm through hiding their hydrophobic region/domains11,12.
For Rho GTPase to have an optimum signaling and function in all stages of its activation cycle, the dynamic cycle of GTP-loading/GTP hydrolysis is crucial. Any kind of alterations in this process may result in subsequent changes in cell functions regulated by Rho GTPase, such as cell polarity, proliferation, morphogenesis, cytokinesis, migration, adhesion, and survival13,14.
The current protocol provides readers with a detailed method to monitor small RhoA GTPase activation via the investigation of their prenylation and GDP/GTP loading. This method can also be used to detect the prenylation and GTP binding of a wide range of small GTPases. The GTPase-linked immunosorbent assay can be used to measure the level of activation of other kinds of GTPases, such as Rac1, Rac2, Rac3,H-, K-, or N-Ras, Arf, and Rho15. The pharmacological agent simvastatin is used as an example, as it was recently reported to be involved in the regulation of small Rho GTPase prenylation and activity8,9,14,16.
Protocol
1. Determination of RhoA Localization Using Membrane/Cytosol Fractionation
- Cell culture and simvastatin treatment
- Seed 50,000 of U251 cells in a 100 mm dish and culture them in Dulbecco's modified Eagle's medium (DMEM) (high glucose, 10% fetal bovine serum [FBS]).
- When 30% confluent, treat the cells by removing the medium and adding simvastatin-containing medium to it (10 µM of simvastatin dissolved in dimethyl sulfoxide [DMSO]), and incubate for 36 h at 37 °C8. Use DMSO alone as a vehicle control.
NOTE: 10 million cells are needed for the cytosol and membrane fractionation of the cells.
- Collection of cells
- Remove the cells from the 37 °C incubator. Look at the cells under a microscope to confirm the confluency.
NOTE: The cells should be 70% – 80% confluent. - Aspirate the medium, wash the cells 1x with cold phosphate-buffered saline (PBS). Add 5 mL of ethylenediaminetetraacetic acid (EDTA) buffer (KCl: 400 mg/L, NaCl: 6800 mg/L, NaHCO3: 2200 mg/L, NaH2PO4.H2O: 140 mg/L, D-glucose: 1,000 mg, EDTA disodium: 373 mg/L) per plate and place the cells back into the 37 °C incubator for 5 min.
- After 5 min of incubation, collect the EDTA with the cells in a 15 mL tube containing the same amount of medium as EDTA.
NOTE: Having medium in the tube neutralizes the EDTA and prevents any further digestion of the cell membranes. - Place the tube in an ice box and proceed to the centrifuge.
- Set up the centrifuge to 1,500 x g at 4 °C and spin the cells for 5 min.
- Remove the supernatant without disturbing the pellet and add 1 mL of cold PBS. Mix the cells well.
- Transfer the cell mixture (solution) to a new 1.5 mL tube, centrifuge at 1,500 x g at 4 °C, and spin the cells for 5 min.
- Check the pellet size (for estimating the volume of the buffer for the next step). Place the samples on ice. Discard the supernatant completely without disturbing the pellet.
- Add ice-cold buffer I (10 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, and protease inhibitor cocktail), mix the samples well by pipetting up and down, and then, proceed to sonication.
- Remove the cells from the 37 °C incubator. Look at the cells under a microscope to confirm the confluency.
- Sonication
- Set the sonicator for five cycles, 5 s each, and repeat 3x.
- Perform the sonication on ice. Proceed to the ultracentrifuge.
NOTE: Ice and cold condition preserve the proteins and make the results more reliable.
- Ultracentrifugation
- Use an ultracentrifuge to separate the cell homogenates into cytoplasmic and membrane fractions. Set the centrifuge to 100,000 x g for 35 min at 4 °C. As shown in Figure 1, check the pellet size.
NOTE: The membrane fraction is at the very bottom of the tube and the rest is other cytoplasmic components. - Collect the supernatant completely while being careful not to disturb the pellet. The supernatant is the cytosolic fraction. Place the supernatant in a newly labeled tube.
- Add 300 µL of dissociation buffer (buffer II) (50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM dithiothreitol, 1% SDS, 1 mM EDTA, 1 mM EGTA, and protease inhibitor cocktail) to the pellet (contains the membrane fraction). Mix well by pipetting up and down.
- Proceed to protein determination and western blot (immunoblot analysis) sample preparation.
- Use an ultracentrifuge to separate the cell homogenates into cytoplasmic and membrane fractions. Set the centrifuge to 100,000 x g for 35 min at 4 °C. As shown in Figure 1, check the pellet size.
- Immunoblotting
- Prepare the cell protein extracts from the separated fractions in lysis buffer (20 mM Tris-HCl [pH 7.5], 0.5 mM PMSF, 0.5% non-ionic detergent-40, 100 µM β-glycerol 3-phosphate, and 0.5% protease inhibitor cocktail).
- Measure the protein concentration using the Lowry method8 and calculate the volume of the lysis buffer (20 mM Tris-HCl [pH 7.5], 0.5 mM PMSF, 0.5% nondenaturing detergent, octylphenoxypolyethoxyethanol, 100 µM β-glycerol 3-phosphate, and 0.5% protease inhibitor cocktail) to normalize the concentration of protein between the samples.
- Heat the samples at 90 °C for 5 min and load 15 – 20 µL of the samples on a 15% SDS-PAGE gel to separate the proteins.
NOTE: Load 1 µg of protein for each sample. Calculate the volume that needs to be run accordingly. - Transfer the separated proteins to nylon membranes under reducing conditions (500 nM glycine, 50 mM Tris-HCl, and 20% methanol) for 2 h, at room temperature (RT) at 100 V.
NOTE: To confirm the successful protein transfer, use Ponceau stain or visualize the protein marker on the membrane. - Block the membranes with 5% nonfat dried milk and 1x Tris-buffered saline containing detergent (TBS/0.01% nonionic detergent; TBST) to block nonspecific antibody binding at 4 °C overnight or at RT for 1 h.
- Add primary antibodies for immunoblotting analysis and incubate overnight at 4 °C.
NOTE: In this experiment, Rac1/2/3, cdc42, RhoA, GAPDH, and pan-Cadherin were used at a 1:1,000 dilution in 1% milk in 1x TBST. Pan-Cadherin and GAPDH were used to confirm membrane and cytosolic fraction purity, respectively. - Wash the membranes 3x with a washing buffer with 1x TBST for 20 min.
- Incubate the membranes with anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody for the respective primary antibodies (for 1 h at RT).
- Wash the blots 3x for 20 min and develop them with enhanced chemiluminescence (ECL) detection.
2. Measurement of the RhoA GTP Load Using a Small G-protein Activation Assay
- Count 10,000 cells/mL and culture the U251 cells in a 100 mm dish.
- When they are 30% confluent, treat the cells with simvastatin as described in step 1.1.2.
- Bring the culture plates out of the incubator. Look at the cells under the microscope to confirm confluency. Ensure that the cells are 70% – 80% confluent. Place the Petri dish on ice, aspirate the media, and wash the cells 3x with ice-cold PBS (pH 7.2).
- Aspirate the PBS. Tilt the Petri dish on ice for an additional minute to remove all remnants of PBS.
NOTE: Residual PBS adversely affects this assay. - Lyse the cells in a 700 µL volume of ice-cold lysis buffer containing protease and phosphatase inhibitors.
NOTE: 700 µL is usually enough for a 100 mm Petri dish. See Table 1 to find the proper volume for each culture vessel. - Harvest the cell lysate with the cell scraper. Incline the culture plate for this technique.
- Transfer the lysate to a labeled ice-cold cryotube and keep it on ice.
- Mix thoroughly using a vortex. Keep 10 µL of the lysate for the protein assay, to measure the protein concentration in the sample.
- Snap-freeze the remaining cell lysate in liquid nitrogen.
NOTE: Prepare multiple aliquots of cell lysate before snap-freezing them, to avoid repeated freeze/thaw cycles which can lead to the loss of activity of RhoA GTPase. - Transfer the snap-frozen cryotubes to a -80 °C freezer and store the samples for the GTPase-linked immunosorbent assay.
NOTE: Do not store the samples for longer than 14 days. Work quickly and never leave the samples on ice for longer than 10 min. Never handle all Petri dishes simultaneously. - Measure the protein concentration using the Lowry method8 and calculate the volume of the lysis buffer to normalize the concentration of protein between the samples.
NOTE: The best concentration is usually 1 mg/mL; however, 0.3 – 2 mg/mL can be detectable. - Prepare a blank control by adding 60 µL of lysis buffer and 60 µL of binding buffer to a microtube.
NOTE: The blank control has all reagents except the antigen and is used for the subtraction of the background. - Prepare a positive control by adding 12 µL of Rho control protein, 48 µL of lysis buffer, and 60 µL of binding buffer.
NOTE: The positive control has all reagents plus a confirmed antigen for Rho-A-GTP. - Take the Rho affinity plate out of its bag and place it on ice.
- Dissolve the powder in the wells with 100 µL of ice-cold distilled water. Keep the plate on ice.
- Thaw the snap-frozen cell lysates in a water bath set to 25 °C.
- Add the calculated volume of ice-cold lysis buffer (from step 2.11) to each sample to normalize the protein concentration.
NOTE: Remove the PBS after washing the cells (using a vacuum tube aspirator) to avoid causing changes in the composition of the lysis buffer. Equalize the sample protein to a concentration between 0.8 and 2 mg/mL for an accurate comparison between samples in GTPase activation assays. Table 2 provides details about the buffer to be used for this assay. - Transfer 60 µL of the normalized ice-cold samples to microtubes and add 60 µL of binding buffer; mix the samples thoroughly and keep them on ice.
- Completely remove the water/solutions from the microplate by vigorous flicking, followed by five to seven hard taps on a lab mat.
- Add 50 µL of the normalized samples, a blank control, and a positive control to the wells in duplicates.
- Place the plate on an orbital shaker for 30 min at 4 °C at 300 rpm.
NOTE: The shaking step is very important, and it is recommended to use the orbital plate shaker at 300 rpm. - Clear the samples from the plate by flicking and wash them 2x with 200 µL of washing buffer at RT. Vigorously remove the washing buffer from the wells after each wash by flicking, followed by tapping, and keep the plate on the bench at RT.
- Add 200 µL of RT antigen-presenting buffer to each well and incubate at RT for 2 min.
- Flick out the solution from the wells and wash the wells 3x with 200 µL of washing buffer at RT.
- Add 50 µL of freshly prepared 1/250 anti-RhoA primary antibody to each well.
- Place the plate on an orbital shaker for 45 min at 300 rpm set to 25 °C. Flick out the solution from the well.
- Repeat the washing steps 2x (step 2.24).
- Add 50 µL of freshly prepared 1/250 anti-RhoA secondary antibody to each well.
- Place the plate on top of an orbital shaker for 45 min at 300 rpm set to 25 °C.
- Prepare the HRP detection reagent by mixing equal volumes of reagent A and reagent B.
- Flick out the solution from each well and wash the wells 3x with 200 µL of washing buffer at RT.
- Add 50 µL of freshly prepared HRP detection reagent to each well.
- Read the luminescent signal within 3 – 5 min to obtain the maximum signal and analyze the results using an appropriate software package.
NOTE: Readings must be taken within 3 – 5 min to obtain the maximum signal. Run a "test plate" to confirm the proper lysis buffer volume is being used for the cell lysates so that the protein concentration is high enough to detect RhoA GTPase activity. (A test plate is a plate of cells used to determine if the protein concentration falls within the acceptable range and also to determine if the volume of the lysis buffer being used is appropriate.) The positive control should read 4- to 10-fold higher than the blank wells if it is in the linear range. If not, then adjust the luminometer by consulting the manufacturer. The settings for the luminometer are given in Table 3. - Enter raw data in the columns where the headings read Sample, Mean, Standard Deviation, rep1, rep2, rep3, and rep4, which is to show the number of replicates being done on each sample.
- Under Mean, enter the formula =average(Xn:Yn) where X = the column designator for rep1, Y = the column designator for rep4, and n = the row designator of the row being worked on.
- Under Standard Deviation, enter the formula =stdev (Xn:Yn) where X = the column designator for rep1, Y = the column designator for rep4, and n = the row designator of the row being worked on.
- Enter the replicate data into rep1, rep2, etc.
- After entering the data, use the click-and-drag method to select the Sample, Mean, and Standard Deviation.
- Then, in data analysis software, select the function for chart making which looks like a square with a mini bar chart inside.
NOTE: This brings up the chart making process where it is possible to design charts based on the data entered. - Choose column chart and, for input values, designate the Mean numbers.
NOTE: The chart for the Mean numbers is first made, and then, the Standard Deviation column for the y-axis error bars is designated. To do this, double-click on the graph bars, select the Y-axis error tab, click the Custom option, and select the area in the worksheet to enter the location of the Standard Deviation data. The difference between the groups that need to be compared can be seen after the creation of the desired charts.
Representative Results
Membrane Fractionation:
Ultracentrifugation was used for the fractionation of membrane and cytosol components. As shown in Figure 1, the supernatant contains the cytosolic fraction and the pellet contains the membrane fraction. The abundance of RhoA in cytosolic andmembrane fractions obtained from U251 cells was examined after the treatment with simvastatin using immunoblotting. The purity and loading control of the membrane and cytosol fraction were confirmed by pan-Cadherin and GAPDH respectively. As shown in Figure 2, the simvastatin treatment reduced the amount of membrane-bound RhoA GTPase, while it increased its cytosolic content. This is consistent with the known effects of statins on the translocation of GTPases based on the inhibition of RhoA prenylation. Simvastatin inhibits the prenylation of RhoA GTPase, and therefore, unprenylated RhoA is unable to anchor in cell plasma membranes, which results in its higher cytosolic concentrations.
RhoA-GTP Bound:
We measured GTP-bound RhoA protein using a GTPase-linked immunosorbent assay and showed that simvastatin significantly (P < 0.05) increased GTP-bound RhoA in U251 cells (Figure 3). Therefore, while simvastatin inhibited RhoA GTPase protein prenylation (Figure 2), it also increased its GTP loading as compared to the control cells. This points to the fact that prenylation and GTP binding both play a role in the activity and regulation of RhoA GTPase. For additional details regarding this phenomenon, please refer to the original publication using this protocol8.
Figure 1: Schematic view of the cytosolic and membrane fractions after ultracentrifugation. The pellet contains the membrane fraction and the supernatant contains the cytosolic fraction.
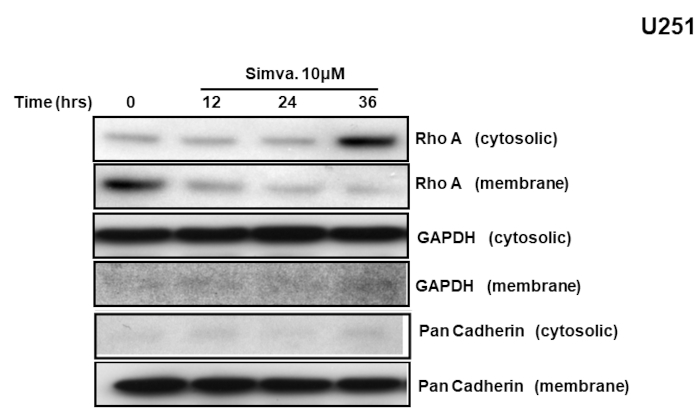
Figure 2: Simvastatin changes the localization of RhoA. U251 cells were treated with simvastatin (10 µM; 12, 24, and 36 h) and the abundance of RhoA in the membrane and cytosolic fractions was determined by immunoblotting. The GAPDH and pan-Cadherin abundance was also assessed to control for loading in the cytosolic and membrane fractions and to confirm the lack of cytosolic contamination in the membrane fractions. The data are typically from three independent experiments using different primary cultures. This figure has been modified from Alizadeh et al.8. Please click here to view a larger version of this figure.
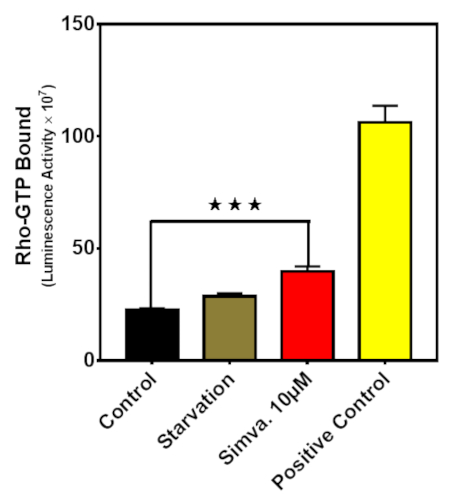
Figure 3: Simvastatin modulates RhoA GTPase activity. The GTPase-linked immunosorbent assay was used to measure GTP-bound Rho protein in U251 cells. Different conditions were tested for 36 h, including starvation and simvastatin (10 µM). For each experiment, a constitutively active RhoA protein provided in the kit was used as a positive control. The results are expressed as the mean ± SD of two replicates in independent experiments (*** P < 0.001). This figure has been modified from Alizadeh et al.8.
| Cell Culture Vessel | Surface area (cm2) | Lysis Buffer (µl) | |
| Dishes | 35 mm | 9 | 100 |
| 60 mm | 21 | 300 | |
| 100 mm | 55 | 700 | |
| 150 mm | 145 | 1500 | |
| Plates | 6-well | 9.4 / well | 100 |
| 12-well | 3.8 / well | 70 | |
| 24-well | 1.9/ well | 40 | |
| Flasks | T-25 | 25 | 250 |
| T-75 | 75 | 1000 | |
| T-150 | 150 | 1500 | |
Table 1: Recommended volumes of lysis buffer for U251 Cells. The volume is adjustable for different cell types.
| Lysis Buffer Name | Lysis Buffer Composition | Small G-Protein Target | Notes |
| GL35 | Tris (1X GL36), MgCl2 (8X GL36), NaCl (2X GL36), IGEPAL (1X GL36), SDS (5X GL36) | Cdc42 | Identical components to GL36, composition varies as follows; |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2, NaCl, IGEPAL and SDS | RalA | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2, NaCl, IGEPAL and SDS | Rac1 | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2, NaCl, IGEPAL and SDS | Rac1,2,3 | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2, NaCl, IGEPAL and SDS | Ras | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2, NaCl, IGEPAL and SDS | RhoA | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
| GL36 | a proprietary formulation of Tris pH 7.5, MgCl2 | Arf1 | Standard buffer, compatible with most GTPase-linked immunosorbent assays. |
Table 2: List of GTPase-linked immunosorbent assay lysis buffers and recommended lysis composition for small GTPases.
| Parameters | Notes | ||
| GAIN | GAIN option adjusts the machine sensitivity. Most luminometers use automatic calibration or limited calibration function. User should read the GAIN at low, medium, and high in order to see if the reading is within the linear range which varies in different instruments. | ||
| INTEGRATION TIME | It is recommended to have this option at its lowest as the very high times might read out of the linear range. This might require unnecessary work like using lower dilution or primary and secondary antibodies. INTEGRATION TIME also varies in different instruments. | ||
| SHAKING | It is recommended to use the 5s orbital SHAKING. It is not as critical as other parameters for the accuracy of the assay. | ||
| TEMPERATURE | Room temperature is recommended | ||
| PLATE TYPE | Use according to the plate you are using. Usually the plate is 96-well, flat and white. | ||
| FILTERS | Excitation or emission filters are not required for Luminescence. Excitation can be set at any desired value and optimal range for emission is 430-445 nm. Leave the spaces of the filter blank. If this is not an option, | ||
Table 3: A detailed description of luminometer settings.
Discussion
Here we describe an accurate method to measure small GTPase prenylation and GTP binding shown as small GTPase subcellular localization (membrane versus cytosol) and Rho GTP loading. Small GTPases are expressed in eukaryotic cells and play essential roles in cellular proliferation, motility, and structure. Both prenylation and GTP binding are involved in the regulation of GTPase activity; therefore, assays to evaluate the prenylation and GTP binding of these proteins are important tools for cell biologists1,8.
Based on the results of a recent study, Rho protein GTP loading is cell-type specific8, and therefore, differs among different cell types. Also, the subcellular localization and geranylgeranylation (GGT) of Rho proteins are the determining steps in the regulation of their function. This is also further regulated by the interplay of the effector proteins GEF, GAP, and GDI for the major GTPases.
Simvastatin is known to increase Rac GTP loading in THP-1 monocytes, decrease the prenylation of Rac in the presence of amyloid β stimulation, and reduce inflammatory responses from these cells17. We also know that T-cell function is not affected by Rho GTP loading but, rather, the GGT of Rho determines function18,19. Therefore, we recommend that, in order to detect Rho GTPase activity, both prenylation and GTP loading must be simultaneously measured in cells.
Of note, to achieve high-purity cytosolic and membrane fractions, the most important steps of the protocol are the sonication and the ultracentrifugation. For the Rho GTP-binding assay, the most critical step is snap-freezing the cell lysates before sequential processing. Following these important steps will produce consistent and reproducible results in the study of GTPases in living systems.
A key step to examine a particular intracellular structure or membrane protein is the separation of cellular compartments from one another. Fractionation takes advantage of the properties of each cellular compartment, like size and shape, surface charge density, and buoyant density20. It is mainly based on differential centrifugation in media of high viscosity at 4 °C. The membrane fractionation method which was used here is mainly based on the size of each compartment and on high gravitational speed where, after ultracentrifugation, membrane proteins go to the bottom of the tube and the cytosolic proteins remain in the supernatant.
It is important to note that samples containing high protein concentrations and, possibly, high levels of GAPs can potentially inactivate the target GTPase. This issue can occur even in lysis buffer and may lead to false negative results. One of the key parameters that determine the success and reproducibility of GTPase activity assay results is the health and responsiveness of the cells being used in the experiment8. It is strongly recommended that investigators identify the optimal/appropriate growth conditions and doubling time for the cells under study to determine the GTPase activation/inhibition. Additionally, the GTPase activity of all small GTPases is tightly regulated and is, therefore, susceptible to rapid decreases via hydrolysis of the GTP molecule bound to the enzyme via the action of GAPs (during and after the cell lysis procedure). This action results in the rapid inactivation of the GTPase of interest. Therefore, it is strongly recommended that cell lysis is rapidly performed at 4 °C, to achieve accurate and reproducible results.
There are several factors depending on experimental conditions that determine the final cell lysate. First, the total amount of RhoA GTPase in the cell line or specific tissue: the amount of endogenous RhoA GTPase is variable in different types of cells and tissues; therefore, this can result in a more vigorous response to an activator or deactivator. Second, the amount of activation/deactivation achieved under the experimental conditions: it is important to consider that approximately 2% to 10% of the total cellular small GTPase is possibly activated in response to a specific stimulus8. The amount of deactivation also solely depends on the type of stimuli and it is variable in different cells and tissues. Therefore, for each type of small Rho GTPase activity assay, the composition of the lysis buffer and cellular compartment is crucial. Table 3 shows the recommended composition of the lysis buffer for each specific small GTPase protein.
The normalized cell lysate protein concentration is a major requirement because it enables investigators to compare the GTPase activity of different samples. Therefore, washing the cells from all samples in all conditions with cold PBS is mandatory to remove protein from the tissue culture media. It is also essential that all reagents and buffers are used at cold temperatures (4 °C) in all steps of the experiment. This cold temperature will minimize the hydrolysis of GTPases, including Rho GTPase, during sample preparation. It is critical that this processing of cell lysates is conducted rapidly (in 10 – 15 min in total) in order to avoid the loss of RhoA GTPase activity. Furthermore, the most important step of the cell lysate preparation is to snap-freeze aliquots of the lysate in liquid nitrogen to maintain the RhoA small GTPase enzymatic activity. This is especially important if there are different timepoints or multiple samples in the experiment. After the preparation of snap-frozen lysates, the samples can be kept in -80 °C without losing their Rho GTPase activity.
The provided protocol for the analysis of the membrane anchoring of RhoA GTPase represents only an indirect tool to measure the prenylation of small GTPases and is not able to directly detect or quantify the binding of isoprenoid residues to the target protein. This is one of the very few limitations of this assay. Therefore, it gives an estimation of the prenylation of proteins. Some approaches have been defined that are able to directly measure FT (farnesylation) and/or GGT by farnesyl transferase and/or geranylgeranyl transferase, respectively, both in cultured cells and in animals and human-derived tumors. The assays use electrophoretic mobility shift, [3H]farnesyl diphosphate and [3H]geranylgeranyl diphosphate, and [3H]mevalonic acid labeling, followed by immunoprecipitation and SDS-PAGE21.
We used the RhoA GTPase-linked immunosorbent assay to detect any membrane anchoring and activity of RhoA GTPase. It consists of a Rho-GTP-binding protein which is linked to the wells of a 96-well plate. So, the GTP-bound active Rho in cell or tissue lysates binds to the wells, while GDP-bound inactive Rho is washed away during the washing steps. Then, the bound, active RhoA in the wells will be detected using a RhoA-specific antibody and chemiluminescence. It is possible to determine the degree of RhoA activation by comparing readings from activated to nonactivated cell lysates. Serum starvation (the use of serum-free medium on cultured cells) is usually used to inactivate RhoA in tissue culture. It should also be mentioned that the GTPase-linked immunosorbent assay's range of activation requires 10 – 50 µg of protein for the detection of RhoA GTPase activity.
It is strongly recommended that untreated samples have low basal cellular levels of GTPase activity (control state). As an example, proper cell starvation conditions can downregulate GTPase activity and provide ideal conditions to show their activation under experimental conditions. Also, both activation and inhibition assays are performed in a time- and dose-response manner to get the best GTPase activation/inhibition responses. More importantly, during cellular preparation, it is very important to use cells which are not overconfluent (>70%), to avoid any nonresponsiveness of the cells to activation/inhibition stimuli.
Luminometers differ greatly in terms of sensitivity and absolute readings. Therefore, in order to determine that it is in the linear range, we suggest running a GTPase-linked immunosorbent assay with a blank and a positive control. If the assay is out of the linear range (the positive control should be 4x – 10x higher than the buffer-only reading) or the blank reading is higher than 9 – 10 million, then it is recommended to use further antibody dilutions. Moreover, we highly recommend calibrating the luminometer to read within the linear range of the assay before beginning the assay.
There are also advantages to the GTPase-linked immunosorbent assay that are worth mentioning. GTPase-linked immunosorbent assays improve the current experimental design and enable technology to facilitate experiments that were not possible with old pull-downs techniques22. The GTPase-linked immunosorbent assays also provide detection accuracy and sensitivity that allows analyses of GTPase activity in preparations previously off-limits to pull-down assays23. A couple of recent studies compared GTPase-linked immunosorbent activation assays with pull-downs and concluded that GTPase-linked immunosorbent assay has some clear advantages, namely that GTPase-linked immunosorbent assays are superior due to their ability to use small amounts of protein22,24, their greater sensitivity24, and their quantitative measurements23. The GTPase-linked immunosorbent assay kit is available in either luminometric or colorimetric detection versions, where the luminometric assays are more sensitive. This GTPase-linked immunosorbent assay is based on a rather simple and fast protocol, requires only small amounts of sample, and yields quantitative and accurate results. Therefore, it might be a good idea to further develop this assay to detect other types of GTPase-based proteins in different cell lines and tissue culture cells with a much higher specificity and accuracy.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Saeid Ghavami was supported by a Health Science Centre Operating Grant, CHRIM operating grant and Research Manitoba New Investigator Operating Grant. Javad Alizadeh was supported by Research Manitoba studentship. Shahla Shojaei was supported by a Health Science Foundation Operating grant and the MITACS Accelerate postdoctoral fellowship. Adel Rezaei Moghadam was supported by an NSERC operating grant which was held by Joseph W. Gordon. Amir A. Zeki was supported by the NIH/NHLBI K08 award (1K08HL114882-01A1). Marek J. Los kindly acknowledges the support from LE STUDIUM Institute for Advanced Studies (region Centre-Val de Loire, France) through its Smart Loire Valley General Program and co-funded by the Marie Sklodowska-Curie Actions, grant #665790. Simone da Silva Rosa was supported by UMGF studentship.
Materials
| DMEM high Glucose | VWR (Canada) | VWRL0101-0500 | |
| Fetal Bovine Serum | VWR (Canada) | CA45001-106 | |
| Penicillin/Streptomycin | VWR (Canada) | 97062-806 | |
| EDTA (Ethylenediamine tetraacetic acid) | VWR (Canada) | CA71007-118 | |
| EGTA (Ethylene glycol bis(2-aminoethyl ether)-N,N,N',N'-tetraacetic acid) | VWR (Canada) | CAAAJ60767-AE | |
| DTT (DL-Dithiothreitol) | VWR (Canada) | CA97061-340 | |
| Ammonium Persulfate | VWR (Canada) | CABDH9214-500G | |
| Tris-Hydroxymethylaminomethane | VWR (Canada) | CA71009-186 | |
| 30% Acrylamide/Bis Solution | Biorad (Canada) | 1610158 | |
| TEMED | Biorad (Canada) | 1610801 | |
| Protease Inhibitor cocktail | Sigma/Aldrich (Canada) | P8340-5ML | 1:75 dilution |
| Rho-GTPase Antibody Sampler Kit | Cell Signaling (Canada) | 9968 | 1:1000 dilution |
| Pan-Cadherin antibody | Cell Signaling (Canada) | 4068 | 1:1000 dilution |
| GAPDH antibody | Santa Cruz Biotechnology (USA) | sc-69778 | 1:3000 dilution |
| RhoA G-LISA Activation Assay (Luminescence format) | Cytoskeleton Inc. (USA) | BK121 | Cytoskeleton I. G-LISA Activation Assays Technical Guide. 2016. |
| RhoA Antibody | Cell Signaling | 2117 | |
| ECL | Amersham-Pharmacia Biotech | RPN2209 | |
| Anti-Rabbit IgG (whole molecule) Peroxidase antibody | Sigma | A6154-1ML | |
| SpectraMax iD5 Multi-Mode Microplate Reader | Molecular Devices | 1612071A | Spectrophotometer |
| Nonidet P-40 | Sigma | 11332473001 | non-denaturing detergent, octylphenoxypolyethoxyethanol |
| DMSO | Sigma | D8418-50ML | |
| PBS | Sigma | P5493-1L | |
| Phophatase Inhibitor cocktail | Sigma | P5726-5ML | 1:75 Dilution |
References
- Yeganeh, B., et al. Targeting the mevalonate cascade as a new therapeutic approach in heart disease, cancer and pulmonary disease. Pharmacology & Therapeutics. 143 (1), 87-110 (2014).
- Valencia, A., Chardin, P., Wittinghofer, A., Sander, C. The ras protein family: evolutionary tree and role of conserved amino acids. 생화학. 30 (19), 4637-4648 (1991).
- Hall, A. Rho family GTPases. Biochemical Society Transactions. 40 (6), 1378-1382 (2012).
- Rojas, A. M., Fuentes, G., Rausell, A., Valencia, A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. The Journal of Cell Biology. 196 (2), 189-201 (2012).
- Cherfils, J., Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiological Reviews. 93 (1), 269-309 (2013).
- Shojaei, S., et al. Perillyl Alcohol (Monoterpene Alcohol), Limonene. Enzymes. 36, 7-32 (2014).
- Ghavami, S., et al. Airway mesenchymal cell death by mevalonate cascade inhibition: integration of autophagy, unfolded protein response and apoptosis focusing on Bcl2 family proteins. Biochimica et Biophysica Acta. 1843 (7), 1259-1271 (2014).
- Alizadeh, J., et al. Mevalonate Cascade Inhibition by Simvastatin Induces the Intrinsic Apoptosis Pathway via Depletion of Isoprenoids in Tumor Cells. Scientific Reports. 7, 44841 (2017).
- Ghavami, S., et al. Mevalonate cascade regulation of airway mesenchymal cell autophagy and apoptosis: a dual role for p53. PLoS One. 6 (1), e16523 (2011).
- Tang, Y., Olufemi, L., Wang, M. T., Nie, D. Role of Rho GTPases in breast cancer. Frontiers in Bioscience: A Journal and Virtual Library. 13, 759-776 (2008).
- DerMardirossian, C., Bokoch, G. M. GDIs: central regulatory molecules in Rho GTPase activation. Trends in Cell Biology. 15 (7), 356-363 (2005).
- Garcia-Mata, R., Boulter, E., Burridge, K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nature Reviews Molecular Cell Biology. 12 (8), 493-504 (2011).
- Etienne-Manneville, S., Hall, A. Rho GTPases in cell biology. Nature. 420 (6916), 629-635 (2002).
- Ghavami, S., et al. Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. American Journal of Physiology – Lung Cellular and Molecular Physiology. 302 (4), L420-L428 (2012).
- Clark, E. A., Golub, T. R., Lander, E. S., Hynes, R. O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 406 (6795), 532-535 (2000).
- Ghavami, S., et al. Statin-triggered cell death in primary human lung mesenchymal cells involves p53-PUMA and release of Smac and Omi but not cytochrome c. Biochimica et Biophysica Acta. 1803 (4), 452-467 (2010).
- Cordle, A., Koenigsknecht-Talboo, J., Wilkinson, B., Limpert, A., Landreth, G. Mechanisms of statin-mediated inhibition of small G-protein function. Journal of Biological Chemistry. 280 (40), 34202-34209 (2005).
- Waiczies, S., Bendix, I., Zipp, F. Geranylgeranylation but not GTP-loading of Rho GTPases determines T cell function. Science Signaling. 1 (12), pt3 (2008).
- Waiczies, S., et al. Geranylgeranylation but not GTP loading determines rho migratory function in T cells. Journal of Immunology. 179 (9), 6024-6032 (2007).
- Satori, C. P., Kostal, V., Arriaga, E. A. Review on Recent Advances in the Analysis of Isolated Organelles. Analytica Chimica Acta. 753, 8-18 (2012).
- Berndt, N., Sebti, S. M. Measurement of protein farnesylation and geranylgeranylation in vitro, in cultured cells and in biopsies, and the effects of prenyl transferase inhibitors. Nature Protocols. 6 (11), 1775-1791 (2011).
- Keely, P. J., Conklin, M. W., Gehler, S., Ponik, S. M., Provenzano, P. P. Investigating integrin regulation and signaling events in three-dimensional systems. Methods in Enzymology. 426, 27-45 (2007).
- Oliver, A. W., et al. The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. British Journal of Cancer. 104 (2), 324-331 (2011).
- Moniz, S., Matos, P., Jordan, P. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cellular Signalling. 20 (10), 1762-1768 (2008).

