Mapping Metabolism: Monitoring Lactate Dehydrogenase Activity Directly in Tissue
Summary
We describe a protocol for mapping the spatial distribution of enzymatic activity for enzymes that generate nicotinatmide adenine dinucleotide phosphate (NAD(P)H) + H+ directly in tissue samples.
Abstract
Mapping enzymatic activity in space and time is critical for understanding the molecular basis of cell behavior in normal tissue and disease. In situ metabolic activity assays can provide information about the spatial distribution of metabolic activity within a tissue. We provide here a detailed protocol for monitoring the activity of the enzyme lactate dehydrogenase directly in tissue samples. Lactate dehydrogenase is an important determinant of whether consumed glucose will be converted to energy via aerobic or anaerobic glycolysis. A solution containing lactate and NAD is provided to a frozen tissue section. Cells with high lactate dehydrogenase activity will convert the provided lactate to pyruvate, while simultaneously converting provided nicotinamide adenine dinucleotide (NAD) to NADH and a proton, which can be detected based on the reduction of nitrotetrazolium blue to formazan, which is visualized as a blue precipitate. We describe a detailed protocol for monitoring lactate dehydrogenase activity in mouse skin. Applying this protocol, we found that lactate dehydrogenase activity is high in the quiescent hair follicle stem cells within the skin. Applying the protocol to cultured mouse embryonic stem cells revealed higher staining in cultured embryonic stem cells than mouse embryonic fibroblasts. Analysis of freshly isolated mouse aorta revealed staining in smooth muscle cells perpendicular to the aorta. The methodology provided can be used to spatially map the activity of enzymes that generate a proton in frozen or fresh tissue.
Introduction
Understanding the locations within tissues in which enzymes have high or low activity is essential for understanding development and physiology. Transcript or protein levels are often used as surrogates for enzymatic activity. While such studies can be informative, they do not provide information that can be critical for determining an enzyme's activity, such as post-translational modifications, the presence of protein complexes, or the enzyme's subcellular localization. When enzymatic activity is directly measured, it is often monitored in homogenized protein lysates that no longer contain information about individual cells within the mixture or the spatial distribution of the cells with high or low activity within a tissue.
We provide here a detailed protocol for mapping the spatial distribution of enzymatic activity within a tissue sample. The methodology is based on earlier studies demonstrating that tetrazolium salts can be used to localize the activity of dehydrogenases, reductases, and oxidases in frozen tissue1. With these methods, a water-insoluble formazan is formed when protons are transferred to a tetrazolium salt2,3. Glucose-6-phosphate dehydrogenase generates NADPH and a proton, and has been detected with tetrazolium activity. Glucose-6-phosphate dehydrogenase has been monitored in in European flounder hepatocytes4, in alveolar type 2 cells of the lungs5 and nephrons of the kidney5. Tetrazolium salts have also been used to monitor transketolase activity in frozen tissue6. A similar approach was recently used to monitor the activity of multiple dehydrogenases in the same tissue on adjacent slides7.
We describe here a method to use tetrazolium salts to monitor the spatial distribution of lactate dehydrogenase activity (Figure 1). Lactate dehydrogenase can convert pyruvate generated by glycolysis to lactate, and the reverse reaction. Lactate dehydrogenase activity is consequently an important determinant of pyruvate's entrance into the tricarboxylic acid cycle versus its secretion as lactate. Lactate levels in the blood are often used to diagnose a range of diseases, including cancer8,9,10, because it can signal that illness or injury has damaged cells and the enzyme has been released.
There are four lactate dehydrogenase genes: LDHA, LDHB, LDHC, and LDHD11. LDHA and LDHB are thought to have arisen from duplication of an early LDHA gene12. LDH is active as a tetramer and LDHA and LDHB can form homotetramers and heterotetramers with each other. LDHA is reported to have higher affinity for pyruvate, while LDHB is reported to have higher affinity for lactate, and to preferentially convert lactate to pyruvate13. The LDHA promoter contains binding sites for the HIF1α, cMYC and FOXM1 transcription factors11. In addition, like many other glycolytic enzymes14,15, LDH can be modified by post-translational modifications. Fibroblast growth factor receptor 1 can phosphorylate LDHA at Y10, which promotes tetramer formation, or Y83, which promotes NADH substrate binding16. LDHA can also be acetylated17. For these reasons, a complete understanding of LDH activity requires monitoring not only LDH protein levels, but the enzyme's activity as well.
In addition to the method we present here, other approaches have been used to monitor lactate dehydrogenase activity. Lactate dehydrogenase activity can be monitored spectrophotometrically in homogenized protein lysate. The generation of NADH as lactate is converted to pyruvate can be measured based on absorbance at 340 nm, while the disappearance of NADH can be monitored as pyruvate is converted to lactate18. Lactate dehydrogenase activity has also been monitored with magnetic resonance imaging (MRI). 13C- pyruvate can be administered and the conversion of pyruvate to lactate can be monitored as the ratio of [1-13C]lactate/[1-13C]pyruvate. Elevated ratios of [1-13C]lactate/[1-13C]pyruvate have been observed in cancer tissue19. While MRI-based approaches can provide information on lactate dehydrogenase activity in normal and disease tissues, the methodologies do not have the resolution needed to determine the activity level in specific cells. The methodology provided here can provide information on lactate dehydrogenase activity in tissue areas and even in single cells.
Using in situ activity assays, we found that the activity of lactate dehydrogenase is high in the hair follicle stem cells of mouse skin20. We also used the method to monitor the activity of lactate dehydrogenase in cultured embryonic stem cells and found the activity is higher in the stem cells than the feeder layer. Finally, we monitored the activity of lactate dehydrogenase in fresh mouse aorta and observed staining in smooth muscle cells. We describe here a detailed protocol for monitoring lactate dehydrogenase activity in frozen mouse skin.
Protocol
All experiments described were approved by the Animal Care Committee at the University of California, Los Angeles.
1. Generate Slides of Frozen Mouse Skin
- Euthanize mice in accordance with institution policy.
Note: Follow institutional policy for protective clothing. All protocols involving animals must be approved by the Institutional Animal Care Committee. - Remove the mouse’s hair with an animal hair trimmer.
- Make incisions in the skin using scissors. Using forceps, lift the skin away from the mouse. Using scissors, cut the skin section away.
- Fill the cryomold with freezing reagent compound (see Table of Materials).
- Place skin sections into filled cryomolds using needle-nosed forceps. Orient skin so that tissue slices will generate a cross section of the skin from the epidermis to the dermis, hypodermis and muscle (Figure 2).
Note: If possible, mount experimental and control mice together in the same cryomold so they can be sectioned onto the same slide and processed together. Take care not to create bubbles. - Place cryomold on the flat surface of a block of dry ice in an ice bucket and let freeze. Continue to observe the orientation of the skin. Adjust if necessary.
Note: Wear cryogenic gloves when handling dry ice. - Transfer cryomolds into a -80 °C freezer for storage. Samples can be maintained in the freezer for approximately 3 months without significant loss of enzymatic activity. Do not let frozen sections dry out.
- Using a cryostat at -20 °C, slice tissue to create sections 7 – 10 µm thick for the best skin morphology21.
2. Preparing Slides for Staining
- Establish the set of slides to be tested. Include a control slide on which NAD will be withheld and another control slide on which the substrate, lactate, will be withheld. Include a positive control slide from a frozen tissue block previously investigated.
- Briefly fix slides containing skin sections with 4% formalin for 5 min either by pipetting 1 mL of 4% formalin onto the slide, or when processing multiple slides, by dipping slides into a container containing 4% formalin.
Note: The fixation will ensure the skin does not peel off from the slides during the following procedures and preserve the skin morphology.
Caution: Formalin is a carcinogen and hazardous; wear gloves, do not let it touch your skin and perform this step in a chemical fume hood. - Wash slides with at least 1 mL phosphate buffered saline, pH 7.4, per slide either by pipetting or dipping.
3. Preparing Staining Solution
- Vortex together reagents for lactate dehydrogenase activity staining (50 mM Tris pH 7.4, 750 µM NADP, 80 µM phenazine methosulfate (PMS), 600 µM nitrotetrazolium blue chloride, and 30 mM lactate) in an appropriately sized tube depending on the amount of staining solution required.
Note: 1 mL is sufficient to completely cover one slide. - Prepare a second solution in which all reagents are present except NAD. Prepare a third solution in which all reagents are present except lactate.
4. Incubate Slides in Staining Solution
- Gently pipet the staining solution or control solutions onto the prepared slides covering the section in its entirety (1 mL is sufficient for one slide). If multiple slides are being processed together, dip all slides into the staining solution at the same time (make sure the container has enough solution to completely cover the tissue section).
- Incubate the slides in a humidified environment at 37 °C in the dark. If staining solution is on top of the slide, place the slide in a humidified environment to prevent evaporation.
5. Monitor the Slides
- Monitor the conversion of the staining solution from clear to blue by visual inspection. When the samples have reached the desired level of blueness, stop the reaction by removing the staining solution.
Note: For skin, 10 min is sufficient. The time will depend on the organ and the enzymatic activity. - Rinse slides with phosphate buffered saline by pipetting (1 mL is sufficient for each slide) or dipping.
Note: Any samples that will be compared to each other must be maintained in staining solution for the same amount of time.
6. Counterstain and mount
- Pipet a counterstain onto the slides when the slides have reached an appropriate level of blueness.
Note: Counterstains that turn the nuclei red or green will provide good contrast with the blue created by the formazan precipitant. - Mount with aqueous or non-aqueous mounting medium22. One to three drops (40 – 50 µL) of mounting medium is sufficient depending on the size of the cover slip.
7. Image slides and quantify
- Image slides under a light microscope. Take photographs of experimental and control samples and all negative controls at 10X, 20X, and 40X magnification.
- Determine the intensity of blue stain in different regions with image analysis software.
Representative Results
We have previously reported results for in situ activity assays in mouse skin20. As shown in Figure 3, we observed high levels of lactate dehydrogenase activity in the hair follicle stem cells at the base of the hair follicle when the procedures described above were followed. These findings were corroborated by fluorescence activated cell sorting of skin for hair follicle stem cells and confirming high lactate dehydrogenase activity in the stem cell compartment with activity assays on sorted cells.
Based on our findings that the hair follicle stem cells have high lactate dehydrogenase activity, we extended our studies of cultured embryonic stem cells. Mouse embryonic fibroblasts serving as a feeder layer were analyzed alone or in the presence of embryonic stem cells. To analyze cultured cells, the medium was aspirated, and the cells were briefly fixed and incubated with lactate dehydrogenase staining solution as described above. We observed high lactate dehydrogenase activity in the embryonic stem cells as compared with the mouse embryonic fibroblasts (Figure 4).
We have also applied the staining protocol to freshly isolated tissue. Mouse aortas were dissected and sliced open. The vessels were opened and immobilized so that the inside lumen of the aortas was accessible. Samples were incubated with Hank's Balanced Salt Solution containing 0.5% Triton-X for 10 min, then with the lactate dehydrogenase staining solution for 10 min. Lactate dehydrogenase staining solution was applied to the fresh tissue. After incubation, images were collected, showing staining in the smooth muscle cells (Figure 5).
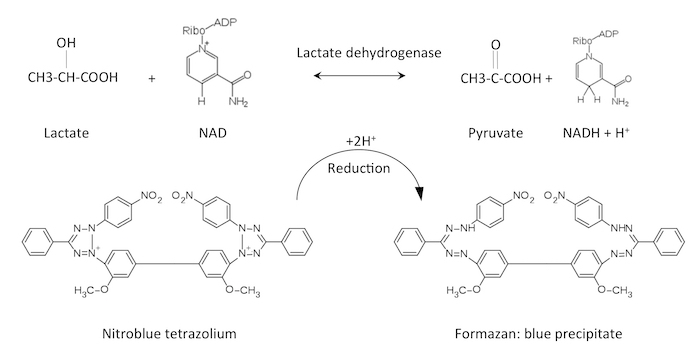
Figure 1: Chemical activity of lactate dehydrogenase and its detection. Lactate dehydrogenase converts lactate + NAD to pyruvate + NADH + H+. The proton that is generated will reduce nitroblue tetrazolium to formazan, which will form a blue precipitate. Please click here to view a larger version of this figure.
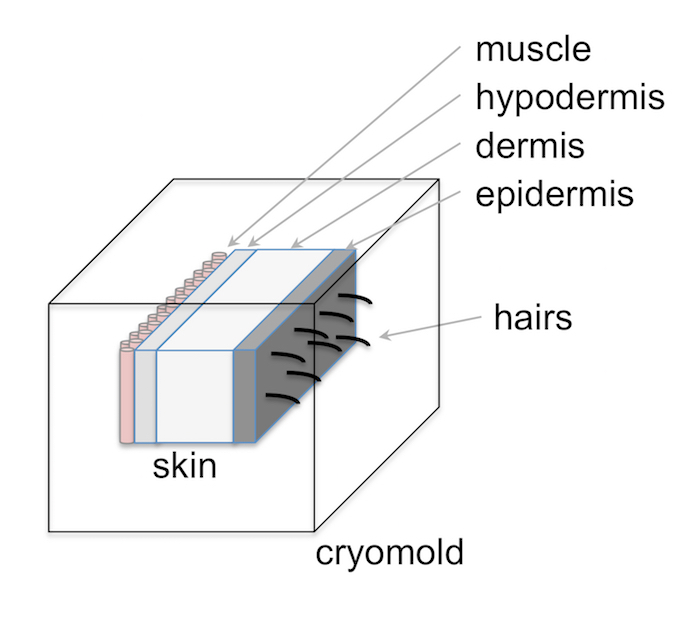
Figure 2: Schematic of orienting mouse skin in cryomolds. Mouse skin should be oriented within cryomolds so that each cut section will contain a cross section of the entire skin from the epidermis, dermis, hypodermis, and muscle. Please click here to view a larger version of this figure.
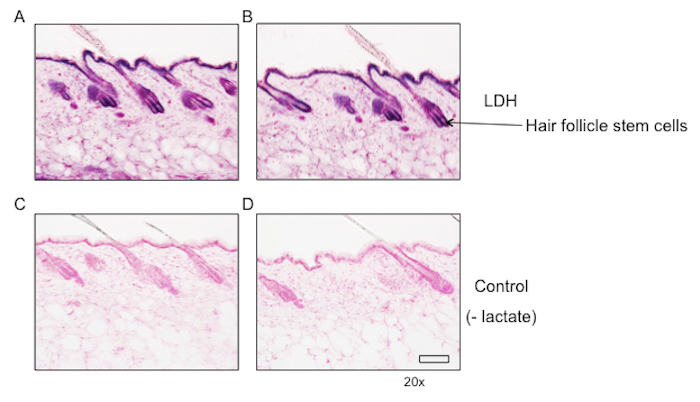
Figure 3: Lactate dehydrogenase staining of mouse skin reveals high activity in hair follicle stem cells. Mouse skin was frozen in cryomolds and incubated with lactate dehydrogenase staining solution contain lactate. Lactate dehydrogenase activity was prominent in hair follicle stem cells. (A, B) Two images of lactate dehydrogenase activity assay of mouse skin. (C, D) Two images of control staining with the substrate lactate withheld. Scale bar = 50 µm. Please click here to view a larger version of this figure.
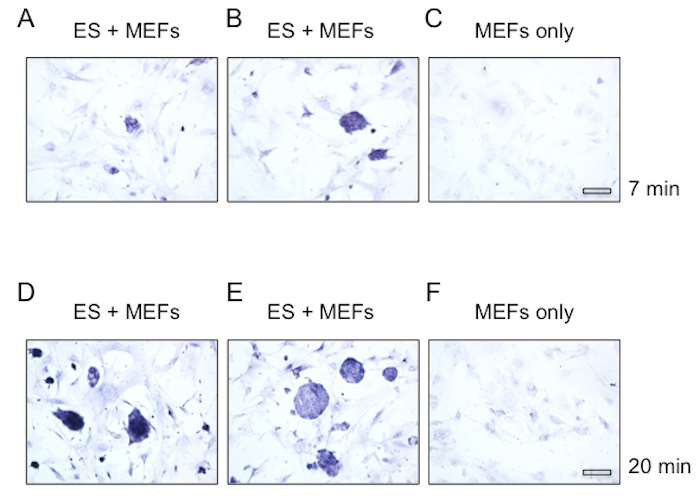
Figure 4: High lactate dehydrogenase activity in cultured mouse embryonic stem cells. Cultured mouse embryonic stem cells grown on an irradiated mouse embryonic fibroblast feeder layer were analyzed for lactate dehydrogenase activity. High activity was observed in the stem cells compared with the fibroblasts. Mouse embryonic fibroblasts showed similar levels of staining with and without irradiation. Images were taken with a 20X objective. Scale bar = 50 µm. (A, B, D, E) Mouse embryonic fibroblasts and stem cells. (C, F) Mouse embryonic fibroblasts alone. (A-C) Images were taken after 7 minutes of incubation with staining solution. (D-F) Images were taken after 20 minutes of incubation with staining solution. Please click here to view a larger version of this figure.
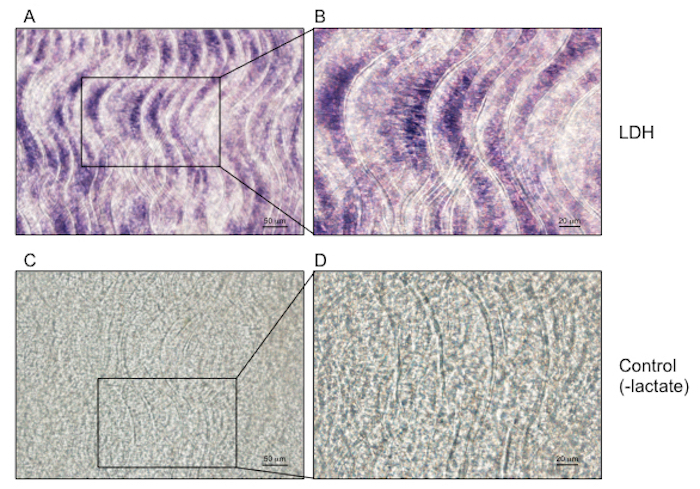
Figure 5: High lactate dehydrogenase activity in the smooth muscle cells surrounding the mouse aorta. Aortas were isolated from euthanized mice. Vessels were fixed for 2 minutes in 2% paraformaldehyde and washed 3 x 10 minutes in 1x phosphate buffered saline. Vessels were incubated with Hank's balanced salt solution with 0.5% Triton-X and washed 3 x 5 minutes in phosphate buffered saline. Lactate dehydrogenase staining solution was added and samples were incubated for 10 min at 37°C and washed 3 x 5 min with 1x phosphate-buffered saline. Samples were then fixed for 1 hour in 2% paraformaldehyde, washed 3 x 5 min with 1x phosphate-buffered saline and imaged. (A) Lactate dehydrogenase solution containing lactate. (B) Lactate dehydrogenase solution without lactate as a negative control. For each section, there is a lower magnification image on the left and a designated area is shown with higher magnification on the right. (A, B) Complete lactate dehydrogenase solution containing lactate was used for staining. B is a magnified image of the indicated area in image A. (C, D) Lactate dehydrogenase solution without lactate was used for staining as a negative control. D is a magnified image of the indicated area in C. A and C were taken with 20X objectives (scale bar = 50 µm) and B and D were taken with a 40X objective (scale bar = 20 µm). Please click here to view a larger version of this figure.
Discussion
The method described here can be used to monitor the activity of lactate dehydrogenase or other metabolic enzymes that generate NADH or NADPH, in different cell types within a tissue or within different portions of a tissue over time. Lactate dehydrogenase is an important enzyme for understanding the biology of stem cells and tumors, and the ability to monitor lactate dehydrogenase activity in individual cells is likely to provide important insights into the function of this enzyme.
One important advantage of this protocol compared with other methodologies is that the ability to monitor enzymatic activity, rather than just protein abundance, can result in more conclusive information about not just where lactate dehydrogenase enzymes are present, but where they are actually functional. The approach described here can be combined with immunohistochemistry so that the level of a selected protein, for instance, a cell lineage marker, is determined in the same tissue samples as lactate dehydrogenase activity. As compared with MRI-based approaches for monitoring lactate dehydrogenase activity, the protocol provided has the advantage that it can provide greater spatial resolution and help to determine the specific cells within a tissue that have high enzymatic activity. As compared with approaches to mapping metabolism based on fluorogenic substrates, detecting signal with the protocol described is not limited by autofluorescence1. Miller and co-authors performed a similar analysis of the time dependence and substrate dependence of five different enzymes by monitoring conversion to formazan7. The reactions were discovered to follow stoichiometric principles of enzyme kinetics. Miller and co-authors distinguished mitochondrial enzymes from non-mitochondrial enzymes by co-localization with mitochondrial staining7. They used 5-Cyano-2,3-di-(p-tolyl)tetrazolium chloride (CTC) as a detection reagent followed by confocal microscopy for subcellular co-localization analysis7.
The method described has disadvantages as well. While MRI-based approaches can be used in living organisms, the approach described requires frozen or fresh tissue. Frozen tissues are only suitable for a few months because the activity of the enzymes degrade over time, even when frozen. Further, because the substrate is provided in excess, the approach provides information on "activity potential" assuming substrate is present. If the levels of the substrate are limiting, then the activity distribution observed with this assay may deviate from the amount of activity that occurs physiologically. In addition, if there are different isoforms of the same enzyme that perform the same enzymatic activity, the method presented here is not able to distinguish between them. For instance, the method presented cannot distinguish between the activity of LDHA versus LDHB. One approach to address this issue would be to use genetically engineered mice in which a single enzyme isoform is genetically inactivated.
We have experimented with PMS, a photochemically stable electron carrier that mediates electron transfer between NADH and tetrazolium dyes23. While some scientists have reported that the PMS inhibits glucose 6-phosphate dehydrogenase activity and found PMS unhelpful5, we found the PMS to be valuable for enzyme localization and include it in our protocol. Some other authors have reported using polyvinyl alcohol to limit diffusion of glucose 6-phosphate dehydrogenase enzyme1. In our experiments, we did not find the polyvinyl alcohol to be helpful and eliminated it.
A few steps are critical for the success of the staining. One important issue is ensuring that the tissue samples are placed properly in the cyromolds. Skin sections should be laid flat so that slices cut across the skin. In addition, if multiple sections are to be prepared, it is ideal if they are embedded within the same cryomold so they can be sliced together on the same slide. This will help to eliminate variation due to the thickness of the slide that is cut with the cryostat. It is also important to ensure that the tissue still retains enzymatic activity.
The best time to end the reaction should be determined empirically. It is necessary to monitor the slides closely during the incubation period and determine the appropriate time to add counterstain and mount the slides based on the intensity of the blue stain. If the samples are too lightly stained, it will not be possible to determine where the activity is. If the samples are overstained, the formazan precipitate can spread, making it difficult to ascertain the specific areas that were originally high in activity.
We anticipate this protocol will be valuable for scientists with a wide range of questions about the role of metabolism. Applications include co-staining for metabolic activity and cell lineage markers or proliferation markers, assessing the contributions of different enzymes with genetic models, assessing subcellular localization of enzymatic activity, monitoring enzymatic activity in diseased tissue or during development.
Disclosures
The authors have nothing to disclose.
Acknowledgements
HAC was the Milton E. Cassel scholar of the Rita Allen Foundation (http://www.ritaallenfoundation.org). This work was funded by grants to HAC from National Institute of General Medical Sciences R01 GM081686, R01 AR070245, National Institute of General Medical Sciences R01 GM0866465, the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research (Rose Hills and Hal Gaba awards), the Iris Cantor Women’s Health Center/UCLA CTSI NIH Grant UL1TR000124, the Leukemia Lymphoma Society, Impact awards from the Jonsson Comprehensive Cancer Center to WEL and HAC. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number P50CA092131. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. HAC is a member of the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research, the UCLA Molecular Biology Institute, and the UCLA Bioinformatics Interdepartmental Program.
Materials
| Surgical instruments | For collecting skin from euthanized mice | ||
| Tissue-tek cryomold 25 mm x 20 mm x 5 mm | Fisher Scientific | NC9511236 | For freezing mouse skin |
| Tissue-Tek O.C.T. compound | Fisher Scientific | NC9638938 | For mounting cryomolds |
| Ice bucket | Fisher Scientific | 07-210-106 | |
| Dry Ice | |||
| Polysine Adhesion Slide | Fisher Scientific | 12-545-78 | |
| 4% formalin | Fisher Scientific | 23-245-684 | Dilluted in water |
| phosphate buffered saline, pH 7.4 | |||
| vortex | |||
| Tris base | Fisher Scientific | 23-245-684 | |
| NAD | Sigma-Aldrich | N7004 | |
| Phenazine methosulfate | Sigma-Aldrich | P9625 | |
| Nitrotetrazolium blue chloride | Sigma-Aldrich | N6876 | |
| Lithium L-lactate | Sigma-Aldrich | L2250 | Substrate |
| 37°C incubator (or tissue culture incubator) | |||
| Braziliant! Counter stain | Anatech | 861 | Counter stain |
| Mounting medium | Vector Laboratories | H-5000 | |
| Cover slips for slides | Fisher Scientific | 12-544D | |
| Light microscope |
References
- Boonacker, E., Van Noorden, C. J. Enzyme cytochemical techniques for metabolic mapping in living cells, with special reference to proteolysis. J Histochem Cytochem. 49, 1473-1486 (2001).
- Seidler, E. The tetrazolium-formazan system: Design and histochemistry. Prog Histochem Cytochem. 24, 1-86 (1991).
- Van Noorden, C. J. F., Frederiks, W. M. . Enzyme Histochemistry: A Laboratory Manual of Current Methods. , (1992).
- Winzer, K., Van Noorden, C. J., Kohler, A. Quantitative cytochemical analysis of glucose-6-phosphate dehydrogenase activity in living isolated hepatocytes of European flounder for rapid analysis of xenobiotic effects. J Histochem Cytochem. 49, 1025-1032 (2001).
- Negi, D. S., Stephens, R. J. An improved method for the histochemical localization of glucose-6-phoshate dehydrogenase in animal and plant tissues. J Histochem Cytochem. 25, 149-154 (1977).
- Boren, J., et al. In situ localization of transketolase activity in epithelial cells of different rat tissues and subcellularly in liver parenchymal cells. J Histochem Cytochem. 54, 191-199 (2006).
- Miller, A., et al. Exploring metabolic configurations of single cells within complex tissue microenvironments. Cell Metab. , (2017).
- Shen, J., et al. Prognostic value of serum lactate dehydrogenase in renal cell carcinoma: a systematic review and meta-analysis. PLoS One. 11, e0166482 (2016).
- Zhang, X., et al. Prognostic significance of serum LDH in small cell lung cancer: A systematic review with meta-analysis. Cancer Biomark. 16, 415-423 (2016).
- Petrelli, F., et al. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta Oncol. 54, 961-970 (2015).
- Valvona, C. J., Fillmore, H. L., Nunn, P. B., Pilkington, G. J. The regulation and function of lactate dehydrogenase A: Therapeutic potential in brain tumor. Brain Pathol. 26, 3-17 (2016).
- Markert, C. L., Shaklee, J. B., Whitt, G. S. Evolution of a gene: Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science. 189, 102-114 (1975).
- Read, J. A., Winter, V. J., Eszes, C. M., Sessions, R. B., Brady, R. L. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins. 43, 175-185 (2001).
- Holness, M. J., Sugden, M. C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 31, 1143-1151 (2003).
- Yi, W., et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 337, 975-980 (2012).
- Fan, J., et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol. 31, 4938-4950 (2011).
- Zhao, D., et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 23, 464-476 (2013).
- Vanderlinde, R. E. Measurement of total lactate dehydrogenase activity. Ann Clin Lab Sci. 15, 13-31 (1985).
- Nelson, S. J., et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Sci Transl Med. 5, 198ra108 (2013).
- Flores, A., et al. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat Cell Biol. , (2017).
- Fischer, A. H., Jacobson, K. A., Rose, J., Zeller, R. Cryosectioning tissues. CSH Protoc. 2008, (2008).
- Espada, J., et al. Non-aqueous permanent mounting for immunofluorescence microscopy. Histochem Cell Biol. 123, 329-334 (2005).
- Hisada, R., Yagi, T. 1-Methoxy-5-methylphenazinium methyl sulfate. A photochemically stable electron mediator between NADH and various electron acceptors. J Biochem. 82, 1469-1473 (1977).

