Detection of Protease Activity by Fluorescent Peptide Zymography
Summary
Here, we present a detailed protocol for a modified zymographic technique in which fluorescent peptides are used as the degradable substrate in place of native proteins. Electrophoresis of biological samples in fluorescent peptide zymograms enables detection of a wider range of proteases than previous zymographic techniques.
Abstract
The purpose of this method is to measure the proteolytic activity of complex biological samples. The samples are separated by molecular weight using electrophoresis through a resolving gel embedded with a degradable substrate. This method differs from traditional gel zymography in that a quenched fluorogenic peptide is covalently incorporated into the resolving gel instead of full length proteins, such as gelatin or casein. Use of the fluorogenic peptides enables direct detection of proteolytic activity without additional staining steps. Enzymes within the biological samples cleave the quenched fluorogenic peptide, resulting in an increase in fluorescence. The fluorescent signal in the gels is then imaged with a standard fluorescent gel scanner and quantified using densitometry. The use of peptides as the degradable substrate greatly expands the possible proteases detectable with zymographic techniques.
Introduction
Gel zymography is a biological technique used to measure proteolytic activity within biological samples, such as body fluids or cell culture media1,2,3. The samples are separated by their molecular weights with electrophoresis through a polyacrylamide gel embedded with a degradable substrate. Common degradable substrates include gelatin, casein, collagen and elastin, which have been used to measure the activity of matrix metalloproteinases (MMPs) -1, -2, -3, -7, -8, -9, and -11, in addition to a variety of cathepsins1,2,4,5,6,7,8. After electrophoresis, the enzymes are renatured and allowed to degrade the protein within the gel. In traditional gel zymography, the gel is stained with a protein dye, such as Coomassie Blue, and protease activity is detected as a loss of signal, i.e., white bands (degradation of protein) on a dark blue background.
Here, we describe a protocol for an alternative method of gel zymography, in which the degradable substrate is a short, fluorogenic peptide covalently incorporated into the polyacrylamide gel (Figure 1). The substitution of synthetic peptides as the degradable substrates enables detection of a wider range of proteases as compared to traditional gel zymography with native proteins9. Covalent linkage of the fluorogenic peptide prevents peptide diffusion and migration during gel electrophoresis observed with previous methods9,10. Furthermore, the use of a fluorogenic substrate enables direct detection of protease activity without additional staining and de-staining steps. The overall goal of this method is the detection of protease activity in biological samples via the covalent incorporation of fluorogenic peptides in zymogram gels.
Protocol
1. Preparation of the Resolving Gel Layer
- Prepare a 10% polyacrylamide resolving gel solution as per Table 1. Add the Tetramethylethylenediamine (TEMED) and Ammonium Persulfate (APS) immediately prior to pouring the gel as their addition initiates the polymerization reaction.
- Fill an empty 1.5 mm mini-gel cassette half way (5 mL) with the 10% resolving gel solution.
- Add a thin layer of isopropanol (~500 µL) to the top of the polyacrylamide gel to produce a level gel and prevent bubbles. Use the leftover polyacrylamide solution to track the progress of the polymerization reaction. When the polyacrylamide in the tube has completely solidified, the reaction is complete (~40 min).
2. Preparation of the Azido-PEG3-maleimide Linker Molecule
- While the first resolving gel layer is polymerizing, retrieve the azido-PEG3-maleimide kit from the -20 °C storage and allow the components to reach room temperature. There are two components in each kit. Vial 1 contains a maleimide-NHS ester, an off-white to grey solid. Vial 2 contains azido-PEG3-amine, a slightly yellow oil.
NOTE: The authors suggest using the 25 mg azido-PEG3-maleimide kit as it can only be stored for short periods of time (1-2 hours) at -20 °C after being prepared before it begins to degrade. 25 mg is sufficient to produce 10 peptide gels. Use gels within 3 weeks of preparation. - Dissolve the components of Vial 2 in the manufacturer recommended volume of dimethyl sulfoxide (DMSO) and vortex for 30 s to ensure the liquids have been mixed well.
- Transfer the contents of Vial 1 into a clean, dry 100 mL round-bottom flask containing a stir bar.
NOTE: Rinse the flask with acetone and dry completely prior to usage to prevent moisture from interfering with the reaction. - Immediately insert a rubber septum stopper with a diaphragm that can be punctured with a syringe into the mouth of the flask. Work quickly to prevent moisture from entering the flask.
- Insert two 18 gauge syringe needles into the diaphragm and connect one to an inert gas tank (e.g. argon gas). Allow the inert gas to fill the flask for 3 min. Mix the components of Vials 1 and 2 under inert gas to prevent undesirable reaction products.
CAUTION: The second syringe needle is to provide a vent, thereby allowing the atmospheric air contained within the flask to flow out of the flask as it fills with inert gas. Do not forget to include a vent needle! - Shut off the inert gas and detach it from the needle. Using a syringe, inject the full contents of Vial 2 into the flask.
- Remove both needles and syringe and allow the components to mix for 30 min at room temperature while stirring.
- Remove the rubber septum stopper and transfer the contents to a clean 5 mL centrifuge tube. The azido-PEG3-maleimide solution must be used within 1 hour at room temperature.
3. Preparation of the Peptide Resolving Gel Layer
- Once the first resolving gel layer has polymerized, pour off the isopropanol layer. Rinse the top of the gel by pipetting 1 mL of deionized water on the top of the gel and then pour off the water.
- Retrieve the thiol-functionalized fluorescent peptide from -80 °C storage and allow it to thaw at room temperature.
NOTE: The thiol-functionalized peptide can be prepared as described previously11,12. Commercially available peptides can also be used but require the addition of a terminal cysteine residue to enable the maleimide-thiol click reaction. Dissolve the peptide to a stock concentration of 10 mM and store it at -80 °C in small (30 uL) aliquots to limit repeated freeze-thaw cycles. - Prepare a 10% resolving gel solution containing the azido-PEG3-maleimide linker molecule and the fluorescent peptide as per Table 1. Add the TEMED and APS immediately prior to pouring the gel as their addition initiates the polymerization reaction.
- Fill half of the remaining portion of the gel cassette (3 mL) with the peptide resolving gel solution.
NOTE: A multi-layer resolving gel approach reduces the amount of peptide and linker necessary for each gel. The size of the peptide resolving gel layer can be adjusted to accommodate a larger range of molecular weights as needed. - Pipette a thin layer of isopropanol (~500 µL) to the top of the polyacrylamide gel to produce a level gel and prevent bubbles. Use the leftover polyacrylamide solution to track the progress of the polymerization reaction. When the polyacrylamide in the tube has completely solidified, the reaction is complete (~40 min).
NOTE: The fluorescent peptide is light sensitive. Keep the gels covered with aluminum foil to prevent photobleaching during gel preparation, electrophoresis, washing and development. - Pour off the isopropanol layer and rinse the top of the peptide resolving gel with deionized water as in step 3.1.
- If using the gels immediately, proceed to step 4, otherwise, immerse the prepared gels in 100 mL of 1x phosphate buffered saline (PBS) at 4 °C in a plastic box to prevent the gels from drying out. Wrap the box in aluminum foil to prevent photobleaching. Gels can be stored in PBS for up to 3 weeks prior to usage.
4. Preparation of the Stacking Gel
- Prepare a 5% stacking gel solution as per Table 1. Add the TEMED and APS immediately prior to pouring the gel as their addition initiates the polymerization reaction.
- Fill the remaining empty portion of the gel cassette (~2 mL) with the stacking gel solution.
- Quickly insert a 1.5 mm gel comb into the stacking gel layer, making sure no bubbles remain trapped under the wells. Use the leftover polyacrylamide solution to track the progress of the polymerization reaction. When the polyacrylamide in the tube has completely solidified, the reaction is complete (~10 min).
- Gently remove the comb and the tape from the back of the gel cassette.
5. Preparation of Biological Samples for Electrophoresis
- Prepare conditioned cell media, cell lysates, tissue homogenates, and MMP standards as described elsewhere under non-reducing conditions2. Do not heat samples.
- For example, prepare conditioned cell media as follows:
- Plate 40,000 cells/cm2 cells in a 6-well plate in 10% fetal bovine serum (FBS) culture media. Incubate cells in a humidified chamber (5% CO2 at 37 °C) for 24 hours and allow them to reach 70-80% confluence.
NOTE: If the cells have not reached the desired confluency after 24 hours, allow them to grow in 10% FBS culture media for an additional 24 hours. - Wash the cells twice with PBS and add 2 mL of serum-free culture media. Incubate the cells in a humidified chamber (5% CO2 at 37 °C) for an additional 24 hours.
- Using a serological pipette, collect the conditioned media from each well. Centrifuge the media at 1200 rpm for 3 minutes to remove any cell debris. Take the supernatant and concentrate using a 15 mL, 10 kDa molecular weight cutoff centrifugal filter unit. Centrifuge the filter units at 4,000 x g for 15 min in a swinging bucket rotor or 5,000 x g for 15 min in a fixed angle rotor.
NOTE: This step is optional but can enhance the intensity of the proteolytic bands in the peptide zymography gels. - Transfer the concentrated filtrate to a fresh 1.5 mL centrifuge tube. Aliquot and store samples at -80 °C for up to three freeze-thaw cycles.
- Plate 40,000 cells/cm2 cells in a 6-well plate in 10% fetal bovine serum (FBS) culture media. Incubate cells in a humidified chamber (5% CO2 at 37 °C) for 24 hours and allow them to reach 70-80% confluence.
- Quantify protein content using a standard protein quantification assay (e.g. BCA, Bradford Assay, etc.).
6. Electrophoresis of Biological Samples in Peptide Zymography Gels
- Dissolve samples in conventional zymography sample buffer (62.5 mM Tris-HCl, pH 6.8, 25% glycerol, 4% SDS, 0.01% bromophenol blue). For cell and tissue samples, ~30 µg of total protein per well is recommended, and 50-100 ng of protein for MMP standards.
- Add 400 mL of 1x Tris-Glycine SDS Running Buffer to the gel apparatus. Load up to 35 µL of sample per well. Run the samples at 120 V at 4 °C for 1.5 hours or until the molecular weight standards indicate that the proteases of interest are within the peptide resolving gel layer (which has a visible orange color).
NOTE: Most MMPs and their variants fall within the range of 35-100 kDa. When the molecular weight standards indicate that those weights are within the peptide resolving gel layer, electrophoresis can be stopped. The same principle can be applied to other classes of proteases with known molecular weights. If there is an interest in detecting multiple proteases over a larger range of molecular weights, reduce the size of the resolving gel layer and increase the size of the peptide resolving gel layer. - Following electrophoresis, remove the gels from the plastic cassette and wash gels three times for 10 min each at room temperature under gentle agitation in renaturing buffer containing 2.5% Triton X-100, 1 µM ZnCl2, and 5 mM CaCl2 in 50 mM Tris-HCl, pH 7.5.
- Transfer gels to a developing buffer solution containing 1% Triton X-100, 1 µM ZnCl2 and 5 mM CaCl2 in 50 mM Tris-HCl, pH 7.5 for 15 min. Replace with fresh developing buffer solution and incubate gels at 37 °C under gentle agitation for 24 hours, making sure the gels are fully submersed in the solution.
7. Imaging of Peptide Zymography Gels
- After 24 hours, image gels using a fluorescent gel scanner/imager using the appropriate excitation and emission filters. For example, the peptide gels shown in the representative results are conjugated with Fluorescein and were imaged using an excitation filter of 488 nm and an emission filter of 521 nm. Using the appropriate filters for your fluorophore will maximize the detection of proteolytic activity.
NOTE: Images can also be taken with a gel imager equipped with a UV transilluminator, often used for the imaging of DNA gels stained with ethidium bromide. Image the gels using the UV transilluminator (365 nm) setting and an emission filter of 590 nm. - Conduct densitometric evaluation of band intensities using ImageJ as described elsewhere13.
Representative Results
Using the method described here, two fluorescent protease-degradable peptides were incorporated into polyacrylamide gels: GGPQG↓IWGQK(PEG)2C (abbreviated as QGIW throughout the text and figures) and GPLA↓CpMeOBzlWARK(PEG)2C (abbreviated as LACW throughout the text and figures). ↓ indicates the site of cleavage. QGIW is a collagen-I derived sequence designed to detect cellular collagenases14. LACW is a sequence that has been optimized for the detection of MMP-14 and MMP-1115. The peptides are labeled with dabcyl (quencher) and fluorescein (fluorophore) using N-hydroxysuccinimide (NHS)-ester-amine chemistry11. It can be difficult to develop new fluorogenic peptides that have adequate fluorescence quenching and are soluble in standard buffers. Therefore, adapting peptide sequences from commercially available fluorescent protease substrates to include a C terminal cysteine is often a successful strategy to develop new fluorogenic sensors. To demonstrate the ability of peptide zymography to separate complex protease mixtures, conditioned media was collected from two different cancer cell lines. HT1080 fibrosarcoma cells and MDA-MB-231 breast adenocarcinoma cells were plated in 10% FBS media for 24 hours, after which the media was replaced with serum-free media for an additional 24 hours. Conditioned media samples were collected and concentrated using 10 kDa molecular weight cutoff centrifugal filter units. The protein content of the media was measured using a standard µBCA assay. 30 µg of protein from conditioned media were electrophoresed. As positive controls, wells with type I bacterial collagenase (100 µg) or purified, activated MMP-9 (125 ng) were also included. The gels were incubated for 24 h in developing buffer to allow MMP cleavage of the degradable substrates within the gels (Figure 2) and then imaged. Fluorescent imaging revealed numerous bands were visible within the LACW peptide gels (Figure 2A14), while only a single band was apparent within QGIW gels (Figure 2D14). In comparison to gelatin zymography (Figure 2G14), LACW gels were able to detect more proteolytic bands, demonstrating the ability of peptide zymography to detect a wider range of proteases present within biological samples than traditional methods using native substrates.
To verify the identity of the visualized bands as MMPs, peptide zymography gels were incubated in development buffer containing either 20 µM GM6001, a broad-spectrum MMP inhibitor, or 10 µM E-64, a general cathepsin inhibitor. Treatment of the LACW peptide gels with GM6001 (Figure 2B14) decreased the intensity of the bands, while treatment with E-64 (Figure 2C14) had no discernable effect. Treatment of the QGIW peptide gels with GM6001 resulted in complete ablation of the previously seen bands (Figure 2E14). As expected, E-64 did not have any effect (Figure 2F14). In both peptide gels, GM6001 inhibited purified MMP-9 activity but did not affect bacterial collagenase activity, further verifying that the visualized increase in fluorescence was a result of proteolytic activity by MMPs present within the tested biological samples.
To compare the sensitivity of peptide zymography to the current gold standard, gelatin zymography, a sensitivity analysis was conducted using purified, activated MMP-9. Serial dilutions of MMP-9 (1-250 ng) were electrophoresed in LACW, QGIW and gelatin zymography gels (Figure 3A14). Following development and fluorescent imaging, band intensities were quantified with ImageJ and plots of normalized band intensity were generated to calculate EC50 values-the concentration that produces 50% of the maximum signal (Figure 3B14). LACW peptide gels were able to detect the smallest concentrations of MMP-9, with an EC50 value of 17.28 ng, as compared to QGIW and gelatin zymograms with values of 59.50 ng and 31.85 ng, respectively. These data indicate that the use of peptide zymography can match or exceed the sensitivity limits of native substrates like gelatin.
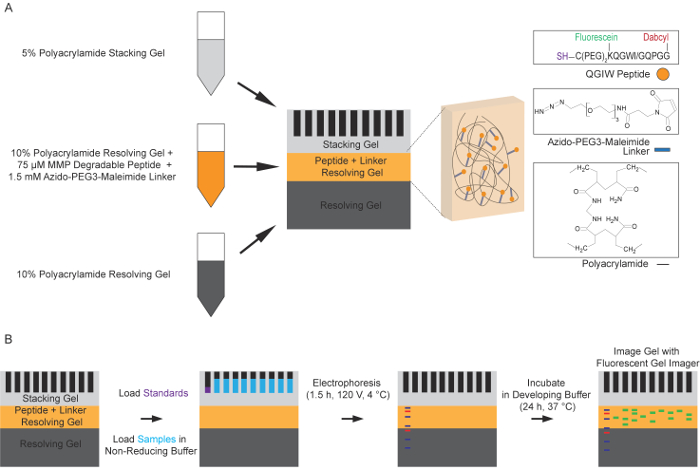
Figure 1: Schematic of the fluorescent peptide zymography process. (A) Preparation of the multi-layer polyacrylamide resolving gel. A standard 10% resolving gel solution is used to form the first layer of the peptide zymography gel. A second 10% resolving gel layer containing a quenched, fluorescent peptide and an azido-PEG3-maleimide linker molecule is then polymerized on top of the first layer. The final top layer is a 5% stacking gel. (B) Standard electrophoresis under non-reducing conditions is used to separate protease-containing samples in the functionalized polyacrylamide gels. The gels are washed to remove SDS and to allow the proteins to renature. The gels are then incubated in a development buffer for 24 h at 37 °C, allowing the proteases to cleave the fluorogenic peptides, resulting in increased fluorescence. This fluorescence, corresponding with protease activity, is then captured using a fluorescent gel imager at an excitation of 488 nm and emission of 521 nm (Adapted with permission from Biotechniques and Future Science14). Please click here to view a larger version of this figure.
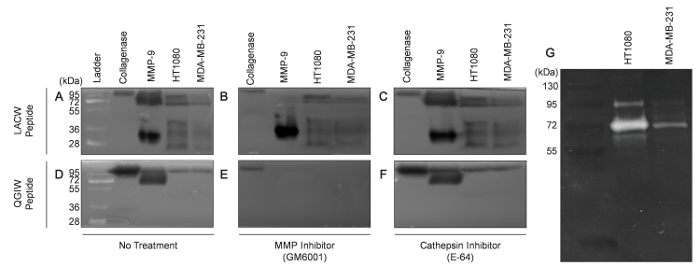
Figure 2: Detection of cell-secreted proteases in human cancer cell lines. Analysis of collagenase enzyme (100 µg), MMP-9 (125 ng) and conditioned cell media from HT1080 fibrosarcoma (30 µg) and MDA-MB-231 adenocarcinoma breast cancer (30 µg) cell lines in LACW and QGIW peptide gels. Gels were treated with DMSO (vehicle control) (A & D), treated with GM6001 (B & E) or treated with E-64 (C & F). (G) Gelatin zymogram of HT1080 and MDA-MB-231 conditioned cell media (Adapted with permission from Biotechniques and Future Science14). Please click here to view a larger version of this figure.
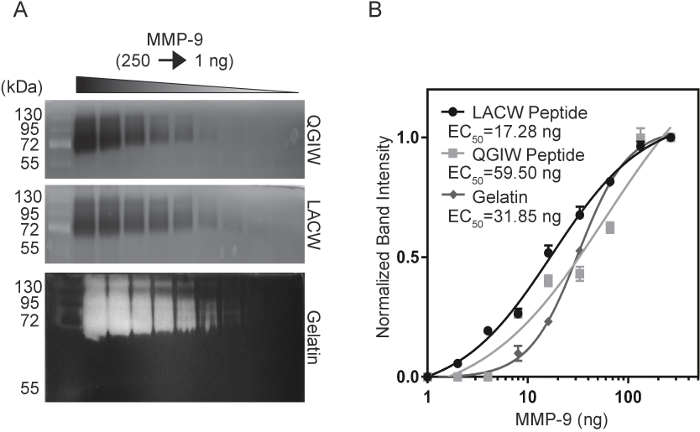
Figure 3: Comparison of MMP-9 Sensitivity of Peptide and Gelatin Zymograms. (A) QGIW (top), LACW (middle) and gelatin (bottom) zymography gels were subjected to serial dilutions of MMP-9. (B) Normalized band intensities were plotted against MMP-9 concentration and fit to a four parameter variable slope curve. EC50 values indicate concentration at half the maximum signal. Results are represented as n=3, mean SD (Adapted with permission from Biotechniques and Future Science14). Please click here to view a larger version of this figure.
| Resolving Gel | ||||
| Stock Conc. | Final Conc. | 5 Gels | 10 Gels | |
| Acrylamide/Bis-Acrylamide (19:1) | 40% | 10% | 10 mL | 20 mL |
| Tris-HCl pH 8.7 | 1 M | 0.375 M | 15 mL | 30 mL |
| Sodium Dodecyl Sulfate (SDS) | 20% | 0.10% | 200 µL | 400 µL |
| Deionized H2O | — | — | 14.4 mL | 28.7 mL |
| TEMED | — | — | 40 µL | 80 µL |
| APS | 10% | 0.10% | 400 µL | 800 µL |
| Total Volume | 40 mL | 80 mL | ||
| Peptide Resolving Gel | ||||
| Stock Conc. | Final Conc. | 5 Gels | 10 Gels | |
| Acrylamide/Bis-Acrylamide (19:1) | 40% | 10% | 5 mL | 10 mL |
| Tris-HCl pH 8.7 | 1 M | 0.375 M | 7.5 mL | 15 mL |
| Sodium Dodecyl Sulfate (SDS) | 20% | 0.10% | 100 µL | 200 µL |
| Deionized H2O | — | — | 6.5 µL | 13 mL |
| Fluorescent Peptide | 10 mM | 75 µM | 150 µL | 300 µL |
| Azido-PEG3-Maleimide CrossLinker | 75 mM | 1.5 mM | 400 µL | 800 µL |
| TEMED | — | — | 20 µL | 40 uL |
| APS | 10 | 0.10% | 200 µL | 400 µL |
| Total Volume | 20 mL | 40 mL | ||
| Stacking Gel | ||||
| Stock Conc. | Final Conc. | 5 Gels | 10 Gels | |
| Acrylamide/Bis-Acrylamide (19:1) | 40% | 5% | 2.5 mL | 5 mL |
| Tris-HCl pH 6.9 | 1 M | 0.125 M | 2.5 mL | 5 mL |
| Sodium Dodecyl Sulfate (SDS) | 20% | 0.10% | 100 µL | 200 µL |
| Deionized H2O | — | — | 14.75 mL | 29.5 mL |
| TEMED | — | — | 50 µL | 100 µL |
| APS | 10% | 0.10% | 100 µL | 200 µL |
| Total Volume | 20 mL | 40 mL | ||
Table 1: Reagent table for preparing fluorescent peptide zymography gels. Concentrations and volumes for the preparation of the multi-layer peptide zymography gel.
Discussion
Current zymographic techniques rely on the incorporation of native substrates into polyacrylamide gels for the detection of proteolysis. While these techniques have garnered widespread use, they are still limited in the number of proteases they can detect. Here, a protocol was described in which fluorescent, protease-degradable peptides are incorporated into the polyacrylamide resolving gel. Covalent coupling using an azido-PEG3-maleimide linker molecule enables the separation and detection of a wider variety of proteases than is currently attainable with native substrates. The highly tunable nature of fluorescent peptides affords researchers the ability to design substrates that can target their proteases of interest. Numerous peptide substrates have been identified for a wide variety of proteases using peptide libraries, and there are a growing number of commercial sources manufacturing custom peptides. It can be difficult to develop new fluorogenic peptides that have adequate fluorescence quenching and are soluble in standard buffers. Therefore, adapting peptide sequences from commercially available fluorescent protease substrates to include a C-terminal cysteine is often a successful strategy to develop new fluorogenic sensors.
While completing this protocol, care should be taken while handling fluorescent peptide gels to prevent excessive exposure to light as this can significantly reduce the detected fluorescent signal. Additionally, the current concentration of peptide used in each gel is 75 µM. This can be adjusted to lower concentrations to conserve peptide, keeping in mind that the azido-PEG3-maleimide solution must be added to the solution in at least a 20 molar excess to the peptide. The azido-PEG3-maleimide kit can be purchased in 3 sizes (25, 100 and 1000 mg). The authors recommend purchasing the 25 mg kit as the prepared solution can only be stored at short periods of time at -20 °C. Furthermore, a 25 mg kit is sufficient to prepare 10 peptide gels, which must be used within 3 weeks of preparation.
One of the limitations of zymography is the difficulty in discerning the exact identity of visualized protease bands due to significant overlap in molecular weights. In future studies, it will be critical to conduct secondary analysis to determine their identity using techniques such as mass spectrometry16,17. Another limitation of zymography is the refolding of proteins to their active conformation following partial denaturing by SDS and electrophoresis. These processes can cause a change in the active conformation of the protease, rendering proteolytically inactive proteins, active. For example, pro-MMP-2 can be detected in gelatin zymograms despite having the inhibitory pro-domain intact due to its renaturation to an intermediate active form. Supplementary methods like enzyme-linked immunosorbent assay (ELISA) or Western Blots can be used to determine the identity and total presence of a protease of interest.
This article demonstrates the use of fluorescent peptide substrates for enhancing the sensitivity of current zymographic techniques. Using purified MMP-9, a concentration gradient analysis was conducted comparing LACW, QGIW and gelatin zymography gels. Currently, gelatin zymography is the gold standard technique by which the gelatinases (MMP-2 and -9) are detected in biological samples. Comparing the EC50 values of the three substrates, LACW peptide gels had the lowest values, indicating the highest sensitivity. Utilizing different peptide sequences designed for detection of specific proteases can potentially enhance these sensitivities even further. Treatment of the gels with an MMP activating agent such as 4-aminophenylmercuric acetate (APMA) or heparin can also be used to boost a weak signal as previously described18.
In addition to the measurement of protease activity for biological studies, protease-degradable peptides are also often used for crosslinking synthetic hydrogels for tissue engineering and drug delivery applications. Controlled degradation is critical for these applications. Currently, the degradation kinetics of these peptides are characterized using single, purified enzymes. However, determining which enzymes cells actually produce and are responsible for cleavage of these peptides has been difficult to determine. The use of peptide zymography to quantify cell and tissue-specific enzyme release will greatly aid in the rational design of these peptide crosslinking sequences.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Funding provided by The Ohio State University College of Engineering, Biomedical Engineering Department, and the Comprehensive Cancer Center – Arthur G. James Cancer Hospital and Richard J. Solove Research Institute.
Materials
| 1.5 mm Empty Gel Cassettes | ThermoFisher Scientific | NC2015 | |
| 1.5 mm, 10 well Empty Gel Cassette Combs | ThermoFisher Scientific | NC3510 | |
| 1x Phosphate Buffered Saline | Fisher Scientific | 10-010-049 | |
| 20% SDS Solution | Ambion | AM9820 | |
| 3x Zymography Sample Buffer | Bio-Rad | 1610764 | |
| 40% (w/v) Acrylamide/Bis (19:1) | Ambion | AM9022 | |
| 6 Well Tissue Culture Plates | ThermoFisher Scientific | 087721B | |
| Amicon Ultra-2 Centrifugal Filter Unit (10 kDa MWCO) | Sigma-Aldrich | UFC201024 | |
| Ammounium Persulfate | Sigma-Aldrich | A3678 | |
| Azido-PEG3-Maleimide Kit | Click Chemistry Tools | AZ107 | |
| Calcium Chloride | ThermoFisher Scientific | BP510100 | |
| Dimethyl Sulfoxide | Fisher Scientific | BP231 | |
| Isopropanol | Fisher Scientific | A416P | |
| Micro BCA Protein Assay Kit | ThermoFisher Scientific | 23235 | |
| N N N' N'-Tetramethylethylenediamine (TEMED) | Sigma-Aldrich | T9281 | |
| PowerPac Basic Power Supply | Bio-Rad | 1645050 | |
| Precision Plus Protein Dual Color Standard | Bio-Rad | 161-0374 | |
| PrecisionGlide Hypodermic Needles | Fisher Scientific | 14-826 | |
| Round Bottom Flask (100 mL) | Fisher Scientific | 50-873-144 | |
| Septum Rubber Stopper | Fisher Scientific | 50-872-546 | |
| Sterile Slip Tip Syringe (1 mL) | Fisher Scientific | 14-823-434 | |
| Triton X-100 | Sigma-Aldrich | X100 | |
| Trizma hydrochlroide | Sigma-Aldrich | T5941 | |
| Typhoon 9410 Molecular Imager | GE Amersham | 8149-30-9410 | |
| Zinc Chloride | Sigma-Aldrich | 208086 |
References
- Vandooren, J., Geurts, N., Martens, E., Vanden Steen, P. E., Opdenakker, G. Zymography methods for visualizing hydrolytic enzymes. Nature Methods. 10 (3), 211-220 (2013).
- Toth, M., Fridman, R. Assessment of Gelatinases (MMP-2 and MMP-9) by Gelatin Zymography. Methods in Molecular Medicine. 57, (2001).
- Heussen, C., Dowdle, E. B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Analytical Biochemistry. 102 (1), 196-202 (1980).
- Gogly, B., Groult, N., Hornebeck, W., Godeau, G., Pellat, B. Collagen zymography as a sensitive and specific technique for the determination of subpicogram levels of interstitial collagenase. Analytical Biochemistry. 255 (2), 211-216 (1998).
- Inanc, S., Keles, D., Oktay, G. An improved collagen zymography approach for evaluating the collagenases MMP-1, MMP-8, and MMP-13. BioTechniques. 63 (4), 174-180 (2017).
- Perera, H. K. I. Detection of Aspartic Proteinase Activities Using Gel Zymography. Zymography. , 43-52 (2017).
- van Beurden, P. A. M. S. n. o. e. k. -., Vonden Hoff, J. W. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. BioTechniques. 38 (1), 73-83 (2005).
- Oliver, G. W., Stetler-Stevenson, W. G., Kleiner, D. E., Zymography, Zymography, Casein Zymography, and Reverse Zymography: Activity Assays for Proteases and their Inhibitors. Proteolytic Enzymes. Springer Lab Manual. , 63-76 (1999).
- Deshmukh, A. A., Weist, J. L., Leight, J. L. Detection of proteolytic activity by covalent tethering of fluorogenic substrates in zymogram gels. BioTechniques. 64 (5), 203-210 (2018).
- Yasothornsrikul, S., Hook, V. Y. Detection of proteolytic activity by fluorescent zymogram in-gel assays. BioTechniques. 28 (6), 1172-1173 (2000).
- Leight, J. L., Alge, D. L., Maier, A. J., Anseth, K. S. Direct measurement of matrix metalloproteinase activity in 3D cellular microenvironments using a fluorogenic peptide substrate. Biomaterials. 34 (30), 7344-7352 (2013).
- Leight, J. L., Tokuda, E. Y., Jones, C. E., Lin, A. J., Anseth, K. S. Multifunctional bioscaffolds for 3D culture of melanoma cells reveal increased MMP activity and migration with BRAF kinase inhibition. Proceedings of the National Academy of Sciences of the United States of America. 112 (17), 5366-5371 (2015).
- Ren, Z., Chen, J., Khalil, R. A. Zymography as a Research Tool in the Study of Matrix Metalloproteinase Inhibitors. Methods in Molecular Biology. 1626, 79-102 (2017).
- Nagase, H., Fields, G. B. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptide. Biopolymers. 40 (4), 399-416 (1996).
- Mucha, A., et al. Membrane Type-1 Matrix Metalloprotease and Stromelysin-3 Cleave More Efficiently Synthetic Substrates Containing Unusual Amino Acids in Their P1′ Positions. Journal of Biological Chemistry. 273 (5), 2763-2768 (1998).
- Thimon, V., Belghazi, M., Labas, V., Dacheux, J. -. L., Gatti, J. -. L. One- and two-dimensional SDS-PAGE zymography with quenched fluorogenic substrates provides identification of biological fluid proteases by direct mass spectrometry. Analytical Biochemistry. 375 (2), 382-384 (2008).
- Sun, X., Salih, E., Oppenheim, F. G., Helmerhorst, E. J. Activity-based mass spectrometric characterization of proteases and inhibitors in human saliva. Proteomics. Clinical Applications. 3 (7), 810-820 (2009).
- Yu, W., Woessner, J. F. Heparin-Enhanced Zymographic Detection of Matrilysin and Collagenases. Analytical Biochemistry. 293 (1), 38-42 (2001).

