A Stainless Protocol for High Quality RNA Isolation from Laser Capture Microdissected Purkinje Cells in the Human Post-Mortem Cerebellum
Summary
This protocol uses a stain-free approach to visualize and isolate Purkinje cells in fresh-frozen tissue from human post-mortem cerebellum via laser capture microdissection. The purpose of this protocol is to generate sufficient amounts of high-quality RNA for RNA-sequencing.
Abstract
Laser capture microdissection (LCM) is an advantageous tool that allows for the collection of cytologically and/or phenotypically relevant cells or regions from heterogenous tissues. Captured product can be used in a variety of molecular methods for protein, DNA or RNA isolation. However, preservation of RNA from postmortem human brain tissue is especially challenging. Standard visualization techniques for LCM require histologic or immunohistochemical staining procedures that can further degrade RNA. Therefore, we designed a stainless protocol for visualization in LCM with the intended purpose of preserving RNA integrity in post-mortem human brain tissue. The Purkinje cell of the cerebellum is a good candidate for stainless visualization, due to its size and characteristic location. The cerebellar cortex has distinct layers that differ in cell density, making them a good archetype to identify under high magnification microscopy. Purkinje cells are large neurons situated between the granule cell layer, which is a densely cellular network of small neurons, and the molecular layer, which is sparse in cell bodies. Because of this architecture, the use of stainless visualization is feasible. Other organ or cell systems that mimic this phenotype would also be suitable. The stainless protocol is designed to fix fresh-frozen tissue with ethanol and remove lipids with xylene for improved morphological visualization under high magnification light microscopy. This protocol does not account for other fixation methods and is specifically designed for fresh-frozen tissue samples captured using an ultraviolet (UV)-LCM system. Here, we present a full protocol for sectioning and fixing fresh frozen post-mortem human cerebellar tissue and purification of RNA from Purkinje cells isolated by UV-LCM, while preserving RNA quality for subsequent RNA-sequencing. In our hands, this protocol produces exceptional levels of cellular visualization without the need for staining reagents and yields RNA with high RNA integrity numbers (≥8) as needed for transcriptional profiling experiments.
Introduction
Laser capture microdissection (LCM) is a valuable research tool that allows the separation of pathologically relevant cells for subsequent molecularly driven evaluation. The use of molecular analyses in these heterogeneous tissue specimens and the correlation with pathological and clinical data is a necessary step in evaluating the translational significance of biological research1. When analyzing gene expression data from RNA, the use of frozen tissue sections is highly recommended as it allows for excellent quality of RNA as well as maximized quantity2. It has been well established that high quality and quantity of RNA are essential for meaningful data from RNA sequencing3. However, when using RNA from fresh frozen post-mortem tissue for LCM, RNA degradation is a major challenge, as it occurs immediately upon death and its extent is mediated by various factors associated with the tissue collection method4,5. Furthermore, RNA degradation is exacerbated when staining techniques are needed to recognize histologic details and cell identification. Specialized staining techniques, such as hematoxylin & eosin, Nissl stain, immunofluorescence and immunohistochemistry are helpful in differentiating cells from surrounding stroma but have been shown to degrade RNA and alter transcript expression profiles6. Therefore, our laboratory has created a stainless protocol specifically designed to preserve RNA in post-mortem human cerebellum for the purposes of RNA sequencing after LCM isolation of Purkinje neurons.
In processing fresh frozen tissue for LCM, the fixation method can variably affect both RNA and tissue integrity. Formalin fixation is standard for morphological preservation, but causes cross-linking that may fragment RNA and interfere with RNA amplification7. Ethanol fixation is a better alternative for RNA isolation, as it is a coagulative fixative that does not induce cross-linking1. To enhance the visualization of tissue morphology, xylene is the best choice, as it removes lipids from the tissue. However, there are known limitations when utilizing xylene in LCM, as the tissues can dry out and become brittle causing tissue fragmentation upon laser capture7. Xylene is also a volatile toxin and must be handled properly in a fume hood. Nevertheless, xylene has been shown to enhance tissue visualization while also preserving RNA integrity8. Therefore, our protocol centers around the use of 70% ethanol fixation and ethanol dehydration, followed by xylene incubation for morphological clarity.
It is important to note the different types of laser-based microdissection systems, as they have been shown to differ in speed, precision, and RNA quality. The infrared (IR) laser capture microdissection and the ultraviolet (UV) laser microbeam microdissection systems were both novel LCM platforms that emerged almost concurrently8. The IR-LCM system employs a "contact system" using a transparent thermoplastic film placed directly on the tissue section, and cells of interest selectively adhere to the film by focused pulses from an IR laser. Alternatively, the UV-LCM system is a "non-contact system" whereby a focused laser beam cuts away the cells or regions of interest in the tissue; depending on the configuration in two currently available commercial platforms that use either an inverted or upright microscope design, tissue is acquired into a collection device by a laser-induced pressure wave that catapults it against gravity or tissue is collected by gravity, respectively. Significant advantages of the UV-LCM system include faster cell acquisition, contamination-free collection with the non-contact approach and more precise dissection due to a much smaller laser beam diameter9. This protocol was specifically designed for a UV-LCM system and has not been tested in an IR-LCM system. In either UV-LCM system design, when accumulating cells into the collection cap, the use of a coverslip that enhances cellular clarity during microscopy is not suitable, as the cells would be unable to enter into the collection cap. Therefore, to enhance tissue visualization, we tested the use of Opaque collection caps, which are designed to act as a coverslip for microscopic visualization in UV-LCM systems, against liquid filled collection caps. Liquid filled collection caps can be challenging, as the liquid is subject to evaporation and must be replaced frequently while working at the microscope. Time becomes an important factor for RNA stability, as the captured tissue dissolves immediately7.
Our laboratory studies the postmortem neuropathologic changes in the cerebellum of patients with essential tremor (ET) and related neurodegenerative disorders of the cerebellum. We have demonstrated morphologic changes centered on Purkinje cells that distinguish ET cases versus controls, including a reduced number of Purkinje cells, increased dendritic regression and a variety of axonal changes, leading us to postulate that Purkinje cell degeneration is a core biologic feature in ET pathogenesis10,11,12,13. Transcriptional profiling has been used in many neurologic diseases to explore the underlying molecular basis of degenerative cellular changes. However, transcriptional profiles from heterogeneous samples, such as brain tissue regions, can effectually mask the expression of low abundance transcripts and/or diminish the detection of molecular changes that occur only in a small population of affected cells, such as Purkinje cells in the cerebellar cortex. For instance, Purkinje cells are vastly outnumbered by the abundant granule cells in the cerebellar cortex by approximately 1:3000; thus, to effectively target their transcriptome requires specific isolation of these neurons. The cerebellar cortex is delineated by distinct layers that differ in cell density and cell size. This cellular architecture is ideal to visualize in a fresh-frozen tissue sample without dye-containing staining reagents. In theory, this protocol could also be applied to other tissue types that have similar distinctive tissue organization.
This protocol was designed to work specifically for Purkinje cell visualization in the human post-mortem cerebellum. Numerous protocols exist for the fixation, staining visualization and RNA preservation of many types of tissues for the purposes of both IR- and UV-LCM. When contemplating an experimental design for UV-LCM, individuals should tailor their protocol to best fit the needs and requirements of starting and ending materials. Here, we combine many aspects of different LCM protocols to provide an enhanced method for visualizing Purkinje cells in the post-mortem human cerebellum without the need of dye containing staining reagents to prepare high-quality RNA for transcriptome sequencing.
Protocol
All human samples utilized in this protocol have been obtained with informed consent and have been approved by the Internal Review Board (IRB) at Columbia University and Yale University.
NOTE: The entirety of this protocol should follow strict RNA handling guidelines, whereby a gloved hand is always used, all surfaces are cleaned with an RNase decontaminator and all working materials are RNA/DNA/Nuclease free.
1. Test RNA Integrity of Tissue prior to Starting LCM
NOTE: Testing RNA quality can be done by many methods. Ensure that RNA from the entire section is tested to provide a representative RNA integrity number (RIN) for the sample.
- Carefully select tissue samples for inclusion that fit within the parameter of the experimental design. If cutting at the cryostat, move to Step 1.2. If not, process the tissue accordingly and move to Step 1.11.
- Get two containers of dry ice. The size will depend on the number of samples.
- In the first container of dry ice, place an empty, labeled 2 mL tube with the tissue code.
NOTE: It takes 10 min for the tubes to freeze. Tubes must be frozen prior to placing the tissue into them; otherwise, the tissue will melt on the tube surface. - In a second container of dry ice, place the tissue from a -80 °C freezer that requires RNA check.
- In the first container of dry ice, place an empty, labeled 2 mL tube with the tissue code.
- Clean the cryostat. See Step 4.7 for detailed cleaning procedure.
- Remove the tissue from dry ice and cut a small section of tissue with a RNase decontaminated razor blade. Tissue should be approximately 1 cm x 1 cm in size.
- Place an approximately 3 mm high mound of optimal cutting temperature (OCT) compound on a cryostat 'chuck' and allow it to partially freeze. Then, place the tissue on top of the OCT — do not push into OCT, simply allow it to rest on top.
- Once OCT hardens, place the chuck with the mounted tissue in the cryostat cutting arm and position in front of the cryostat blade.
- Trim at 30 μm sections until the tissue is even.
- Cut 300 μm of the tissue and place in a frozen 2 mL tube. Place the tube back into dry ice.
- Remove the tissue from the 'chuck' and place back into dry ice until ready to put away.
- Repeat Steps 1.4–1.9 for all tissues.
- Once all samples are complete, extract RNA using the RNA extraction kit provided (Table of Materials #15). A detailed protocol is provided with the RNA extraction kit. Follow all instructions including the optional step to dry the collection tube membrane and the optional step for DNase digestion (Table of Materials #16).
- Test the integrity of extracted RNA via a quality assessment bioanalyzer14.
2. Prepare prior to Starting Sectioning for LCM
- Clean the slides holders prior to each round of tissue sectioning for LCM. Perform the cleaning procedure the day before use to allow complete drying overnight.
NOTE: Slide holders are used for tissue section fixation in ethanol and clearing in xylene.- Slide holder cleaning procedure: Rinse with RNase decontaminator followed by diethyl pyrocarbonate treated (DEPC) water. Allow to dry overnight upside down on an RNA/DNA/Nuclease free surface, avoiding any dust or other airborne contamination.
CAUTION: DEPC is highly toxic and should be handled and disposed of using EH&S standard hazardous chemicals protocol.
- Slide holder cleaning procedure: Rinse with RNase decontaminator followed by diethyl pyrocarbonate treated (DEPC) water. Allow to dry overnight upside down on an RNA/DNA/Nuclease free surface, avoiding any dust or other airborne contamination.
- Clean the brushes for sectioning with RNase decontaminator and allow to dry overnight. Avoid any dust and other contaminants.
- Place the membrane slides under UV light for 30 min at room temperature. Do not expose the slides to UV more than 1–2 days prior to use. This enhances the binding of the tissue to the membrane slide.
NOTE: Step 2.3 can be combined with Step 4, which allows for slide UV treatment to occur on the same day as tissue sectioning (see Step 4.4).
3. Prepare Fixation Solutions with RNase Free Water and High-Quality Ethanol
NOTE: All solutions are prepared fresh for every experiment. Ensure that the slide holder cleaning procedure from Step 1.1 is completed. All solutions are prepared in slide holders.
- Place 30 mL of 100% ethanol in one slide holder.
- Dilute ethanol with RNase/DNase/Nuclease free water. Place 30 mL of 95% ethanol in one slide holder and put on ice.
- Dilute ethanol with RNase/DNase/Nuclease free water. Place 30 mL of 70% ethanol in one slide holder and put on ice.
- Place 30 mL of 100% xylene in two separate slide holders and label them as #1 and #2.
4. Prepare Cryostat for Tissue Sectioning
- Get a bucket of ice for ethanol solutions and a container with dry ice for the tissue.
CAUTION: Dry ice is extremely cold, so use protective gloves. Do not place ice and dry ice in the same container. Differing temperatures in the ice and dry ice will cause them to form together at an indeterminate temperature. - Remove the tissue from the -80 °C freezer and place on dry ice for transport to the cryostat.
- Cryostat settings:
- Set the section thickness to 14 μm.
- Set the trim thickness to 30 μm to preserve tissue quantity.
- For cryostats with dual chamber and specimen holding temperatures, set the chamber temperature (CT) to -18 °C. For cryostats with one setting, set the temperature to -17 °C.
- For cryostats with dual chamber and specimen holding temperature, set the object temperature (OT) to -16 °C.
NOTE: The OT needs to be warmer than the CT to ensure even cutting. If the tissue is still shredding, the OT can be increased to -15 °C and the CT can be increased to -18 °C.
- Place the tissue in the cryostat for at least 20 min to allow to come to optimal temperature.
NOTE: If the tissue is not at optimal temperature, it will shred upon cutting. Do not place the tissue in -20 °C the night before to preserve RNA integrity and to prevent ice crystal formation. Allow the tissue as much time as necessary to equilibrate to optimal temperature to cut smoothly. Optional implementation of slide incubation under UV may occur here (see Step 1.3). - Place the 'chuck' in the cryostat for at least 20 min to allow to come down to the temperature of the cryostat.
- Place the clean brushes in the cryostat for at least 20 min to allow to come down to the temperature of the cryostat.
- Clean the cryostat
- Clean the stage with a 1:1 mixture of RNase decontaminator and 70% ethanol diluted with RNA/DNA/Nuclease free water to prevent freezing on stage.
- Clean a new disposable cryostat blade with RNase decontaminator and place in the blade holder on the stage. Allow at least 20 min for the cryostat blade to come to the temperature of the cryostat.
- Clean the anti-roll plate with RNase decontaminator and attach to the stage. Allow at least 20 min for the anti-roll plate to come to the temperature of the cryostat.
NOTE: An anti-roll plate is not required for cutting but is highly recommended. If an anti-roll plate is not available, a cleaned brush works as an alternative. If the anti-roll plate is not at the correct temperature, the tissue will melt or stick upon cutting.
5. Sectioning Tissue
- Cut a small piece of the tissue (~1 cm x 1 cm) for sectioning. Return the remaining tissue to dry ice/-80 °C freezer.
NOTE: Ensure that the tissue section is ~95% cerebellar cortex, where Purkinje cells are located. - Place an approximately 3 mm high mound of OCT to cover the 'chuck'. Start by building the layers on top in a slow, circular motion, until there is a small mound.
- Once OCT is partially frozen, but there is still some liquid in the center, place the tissue on top of OCT. Do not push the tissue deeply into OCT. Allow the tissue to sit on top of OCT until completely frozen. This will take 1–2 min.
- Place the 'chuck' with frozen OCT and the tissue into the cryostat cutting arm. Adjust the tissue so it is flush with cutting blade.
- Allow the tissue to sit in the cutting arm for 15–20 min to adjust to new temperature.
NOTE: Depending on the thickness of the tissue, time is needed for temperature acclimation. Allow the tissue to sit as long as required to cut cleanly. Adjust OT and CT to assist with tissue temperature acclimation. - Slowly move the tissue closer to the blade. Once the tissue has reached the blade, start the trim process. Trim thickness should be set to 30 μm.
- Trim 2–3x until the cortex layers are visible.
- Place and align the anti-roll plate just above the cryostat blade.
NOTE: A silver line will appear from the light in the cryostat at the interface of the blade and anti-roll plate. This indicates the anti-roll plate is directly over the edge of the blade. - Begin to cut the sections at 14 μm. Properly cut sections will be flat under the anti-roll plate.
NOTE: If the tissue is stuck, ensure that the anti-roll plate is cold enough. If necessary, placing a piece of dry ice on the outside (side not touching the stage) will rapidly chill the anti-roll plate. - Cut 4–6 sections and align them horizontally across the cryostat stage.
- Angling the slide, pick up all pieces of the tissue at one time.
NOTE: Using the brushes, lift up the ends of the tissue to be more readily available for the slide upon pick up. Do not pick up one section at a time. Exposure to room temperature air will degrade RNA integrity.
CAUTION: Do not let the membrane touch the stage of the cryostat. If the membrane touches the stage, it will begin to detach from the glass slide and will cause errors when attempting LCM. - Immediately move to Step 6. Do not delay fixation.
6. Fixing Tissue/Stain-less Visualization
- Immediately move to the ethanol fixation protocol
- Place the slide in the slide holder with 70% ethanol for 2 min — on ice.
- Place the slide in the slide holder with 95% ethanol for 45 s — on ice.
- Place the slide in the slide holder with 100% ethanol for 2 min — at RT.
- Dip the slide 3 times in the slide holder with xylene 1 — at RT.
- Place the slide in the slide holder with xylene 2 for 5 min — at RT.
- Allow to dry in a clean area for at least 30 min. Longer dry times are optimal, up to 60 min.
CAUTION: Stand up slides for drying in a fume hood. Xylene is volatile. Laying slides flat will cause xylene to pool in the tissue section, which will cause tissue to be too dark.
7. Optional — Slide Storage
- Place dried slides in individual 50 mL tubes (one slide per tube) and place in the -80 °C freezer for up to 7 days.
NOTE: Slides must be dry prior to placing in the -80 °C freezer. Do not use desiccant inside the tube as particulates will embed in the tissue and membrane. Ensure that the tube cap is tight; if not, condensation will occur on the slide when thawing for use.
CAUTION: RNA integrity decreases after 7 days, as does the quality of tissue visualization.
8. Thawing Stored Slides for Use
- Remove the tube with a single slide from the -80 °C freezer 1 h prior to intended use.
- Do not immediately open the tube. Allow the tube to come to room temperature. Condensation will occur on the outside of the tube only.
- Once the tube has come to room temperature and all condensation is gone (approximately 30–40 min), remove the cap and expose the slide to the room temperature air.
- Allow the slide to acclimate to room temperature for 15–20 min. Leave the slide inside the tube with the cap removed.
NOTE: Slide is now ready for LCM.
9. Laser Capture Microdissection of Purkinje Cells:
NOTE: This section of the protocol will only discuss specifics related to capturing Purkinje cells for subsequent RNA sequencing. This section assumes that the user is familiar with the UV laser capture microscope and the affiliated software.
- Clean the microscope stage and the cap collection arm with RNase decontaminator.
- With gloved hands, place the slide on the microscope and 500 μL opaque cap in the collection arm.
NOTE: Always ensure proper RNA technique by cleaning the work area with RNase decontaminator and use gloved hands while touching the slide or opaque cap. Gloves can be removed once the slide and cap are in place and laser capture has commenced. - At low magnification, align the opaque cap over the cerebellar tissue, ensuring that the cap covers the entire area visualized in the eye piece.
NOTE: Only the eye piece of the microscope will show if the opaque cap is centered. The camera will not show if the cap is centered over the tissue. Depending on the laser capture scope design, the visualization through the opaque cap will either be in the eye piece only or visualized in both the eye piece and camera. - Visualize the cerebellar layers with 5x – 10x objective lens and place the cursor over the section where molecular layer and granule cell layer intersect (the Purkinje cell layer).
- Move to 40X and visualize Purkinje cells.
NOTE: See Figure 1 and Figure 2 for example visualizations. - Begin capturing Purkinje cells.
NOTE: To perform RNA-sequencing, at least 5 ng of RNA is required. Approximately 1600–2000 Purkinje cells result in enough RNA material for sequencing. RNA will be stable for up to 8 h at the microscope. Test UV energy and UV focus prior to collection to ensure levels are correct. Over time, the tissue will continue to dry out and the UV energy and UV focus may need to be changed.
10. RNA Collection post LCM
- Prepare the cell lysis buffer (prepare fresh each time)
- Dilute 2-mercaptoethanol in lysis buffer at 1:100 (10 μL:1 mL, respectively). Lysis (RLT) buffer is provided in Table of Materials (#14 and #15).
- Add 50 μL of cell lysis buffer to the opaque cap, with the cap facing up. Carefully close the tube over the cap.
- Leave the tube upside down for 1 min.
- Vortex the tube for 30 s and quickly spin the lysis buffer to the bottom of the tube.
- Reopen the tube and repeat Steps 10.2–10.4.
- Place the tube containing 100 μL of lysis buffer and RNA from the -80 °C freezer until all samples are finished and ready for RNA extraction (Table of Materials #14).
Representative Results
This protocol details the steps for preparing fresh frozen post-mortem human brain tissue for UV-LCM. Thorough specifications and annotations are given for cryostat cutting, as it can be difficult to cut fresh frozen brain tissue with a high degree of precision. The most important point to consider is that fresh frozen tissue is extremely cold and requires a significant amount of time to acclimate to the warmer temperature of a cryostat. This step cannot be rushed, and it cannot be overstated how central this step is in the success or failure of this protocol. If the tissue is not prepared properly at the cryostat, all subsequent attempts at identifying cells or regions of interest will be very difficult, if not impossible.
Following cryostat sectioning and allotted drying time, the tissue is ready for LCM. When visualizing the tissue under the microscope, the cellular layers of the cerebellum are easily visible with 5X and 10X objective lenses. Figure 1 shows representative images of tissue fixed in ethanol only (Figure 1A) and tissue incubated in xylene following ethanol fixation (Figure 1B). We rigorously tested various ethanol and xylene incubation times/temperatures on the ability to both fix and visualize the tissue. If the tissue is not incubated long enough in xylene, the resulting image will more closely resemble Figure 1A, rather than Figure 1B. Xylene incubation causes the tissue to become darker and better delineates cellular layers than does ethanol alone. When cutting at the laser capture microscope, the 40X objective lens is required to ensure capturing only Purkinje cells, and not the surrounding tissue. Incubation in xylene produces a high quality, morphologically intact image (Figure 1D) compared to ethanol fixation alone (Figure 1C).
To further enhance the visualization of our tissue, we tested the coverslip ability of an Opaque cap. Other UV-LCM protocols utilize a liquid filled cap of a 200–500 μL tube that dissolves the tissue on contact. Liquid filled caps can reduce tissue visualization, causing the resulting image to be granular and iridescent under the microscope (Figure 2A). Tissue visualization through the Opaque cap (Figure 2B) results in a smoothened tissue appearance that is softer and sharper in form. Notably, the Opaque cap allows for collection up to 8 h without the need for replacement. Representative images of excised Purkinje cells at low power (Figure 2C) and at high power (Figure 2D, E) show precise removal of just the Purkinje cell body. For reference, Figure 2F shows a Luxol Fast Blue/ Hematoxylin and Eosin (LH&E) stained cerebellar cortex, which specifically highlights the different layers of the cerebellum and the placement of the Purkinje cells at the molecular layer and granule cell layer juxtaposition.
The purpose of this protocol is to obtain high quality RNA for subsequent RNA sequencing. We tested our protocol on six different post-mortem human brain samples, which each underwent three days of LCM to collect 1500–2000 cells for RNA extraction. Approximately 6–8 h at the microscope produced 500–700 cells, which were then combined with the previous days LCM products prior to RNA extraction. Samples underwent RNA extraction and were resuspended in 14 μL of RNAse-free water (Table of Materials #14). All six samples produced high quality RNA, with RNA integrity numbers (RINs) ≥8 (Figure 3A-F). This procedure resulted in RNA quantities of 11.5 ng (Figure 3A), 8.3 ng (Figure 3B), 13.6 ng (Figure 3C), 11.5 ng (Figure 3D), 14.9 ng (Figure 3E) and 26.9 ng (Figure 3F). Of note is that all post-mortem human brain samples were tested for RNA quality prior to LCM (Table 1). If the sample of origin has RNA degradation, LCM will only further degrade its integrity.
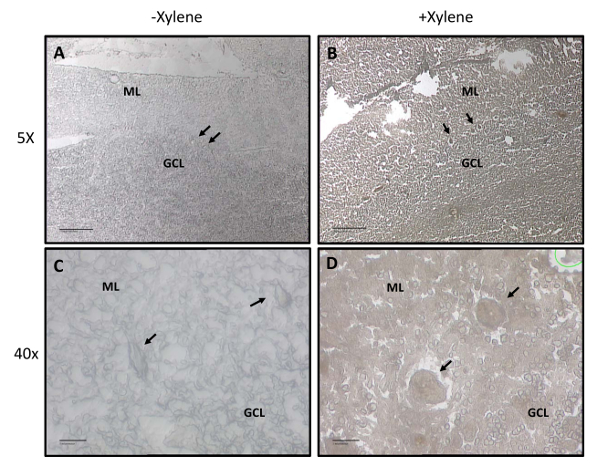
Figure 1: Cerebellar layers and Purkinje Cell visualization with and without xylene treatment. Representative images with and without xylene following ethanol fixation and dehydration. The molecular layer (ML) and granule cell layer (GCL) are labeled. Purkinje cells are marked with arrows. (A) 5X representation of cerebellar layer visualization without xylene. Scale bar: 10 µm. (B) 5X representation of cerebellar layer visualization with xylene. Scale bar: 10 µm. (C) 40X representation of cerebellar layer visualization without xylene. Scale bar: 1 µm. (D) 40X representation of cerebellar layer visualization with xylene. Scale bar: 1 µm. Please click here to view a larger version of this figure.
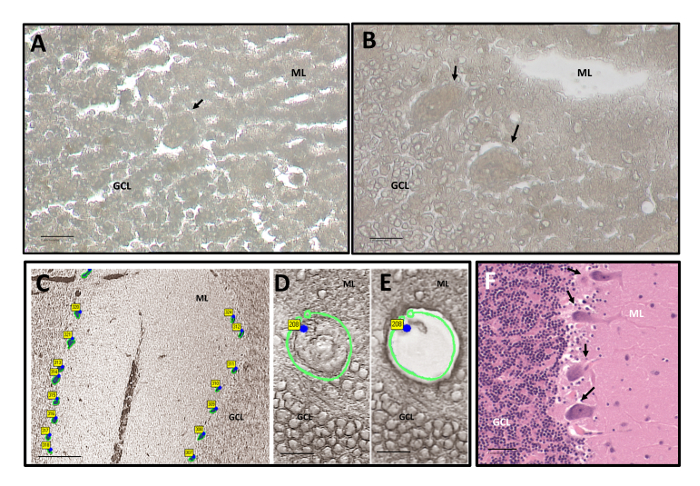
Figure 2: Purkinje Cell Visualization before and after LCM. Representative images at 40X with xylene following ethanol fixation and dehydration. The ML and GCL are labeled. Purkinje cells are marked with arrows. (A) Visualization at the microscope under liquid filled cap at 40x. Scale bar: 1 µm. (B) Visualization at the microscope under Opaque collection cap at 40X. Scale bar: 1 µm. (C) 5X visualization of the cerebellar cortex showing the excised Purkinje cells between the ML and GCL. Scale bar: 10 µm. (D) 40X visualization of a Purkinje cell prior to excision. Scale bar: 1 µm. (E) 40X visualization of successful Purkinje cell capture. Scale bar: 1 µm. (F) 20X representative LH&E stained cerebellum showing Purkinje cell bodies with marked arrows. Scale bar: 5 µm. Please click here to view a larger version of this figure.
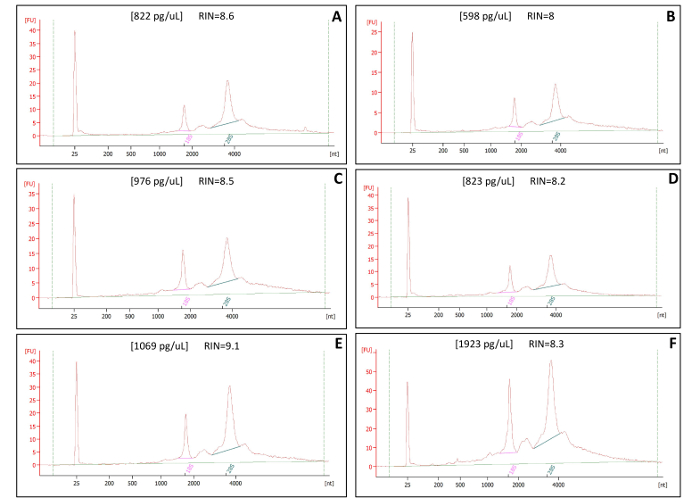
Figure 3: RNA Quality Control Bioanalyzer Results of LCM samples. Panels A-F show representative RNA quality control readouts. Each panel is a different sample that underwent the same LCM process of fixation and visualization with ethanol and xylene only. RNA integrity numbers (RINs) are all >8.0. Representative concentrations [ ] result in a yield of at least 5 ng of total RNA. All samples were eluted in 14 µL of RNAse-free water. Please click here to view a larger version of this figure.
| Sample | RIN – Section Prep | RIN – LCM | [RNA] – LCM | PMI-Frozen |
| A | 9.2 | 8.6 | 822 pg/µL | 450 min |
| B | 9.8 | 8 | 598 pg/µL | 550 min |
| C | 9.1 | 8.5 | 976 pg/µL | 455 min |
| D | 9.8 | 8.2 | 823 pg/µL | 463 min |
| E | 9.8 | 9.1 | 1069 pg/µL | 1139 min |
| F | 9.6 | 8.3 | 1923 pg/µL | 1080 min |
Table 1: Summary of RNA Integrity. Sample numbers correspond to the bioanalyzer results presented in Figure 3. Section preparations are performed prior to LCM to ensure that starting tissue is of good quality. Shown are the original tissue RINs from section preps, LCM RINs and concentration of RNA from LCM, as well as the post-mortem intervals to frozen (PMI-Frozen) for the tissues.
Discussion
The protocol presented here is specifically modified to be a stainless approach in visualizing morphologically distinct tissues for UV-LCM. This method is designed to maximize RNA integrity for subsequent direct RNA sequencing, while maintaining an enhanced level of tissue visualization. The ability to distinguish different cell types for capture, to create the purest population of cells possible, is essential for understanding different molecular profiles in human tissues15. Within the context of this protocol, tissue selection is paramount, as the use of antigen or dye specific reagents are not utilized and therefore is not suitable for studies that require such differentiation. While this is a limitation of this protocol, the resulting tissue visualization and RNA integrity are quite superior and maintain relevance in other experimental designs. Many other protocols exist that also utilize alternative staining methods specifically for UV- and IR-LCM with the intention of preserving RNA. However, most other methods contain at least one staining or antigen specific reagent6,16,17, are designed to one cell or organ type (that are not human autopsy tissue)18,19,20, or require specialized RNA-seq kits to enhance integrity21. Cresyl violet (Nissl) staining is a popular dye containing reagent used in many protocols, as it stains nuclei in neurons in the brain and causes the least amount of RNA degradation6. However, the larger nucleus of the Purkinje cell is not well stained by cresyl violet, providing no significant benefit for visualizing Purkinje cells. Importantly, we identified only one LCM study in the literature that described the collection of human Purkinje cells, which are of considerable interest to the study of cerebellar degenerative and developmental disorders. This study stained frozen tissues with cresyl violet and isolated Purkinje cells with an IR-LCM system; sample RINs as low as 5 were used but deemed acceptable for microarray analysis22. Therefore, this is the first protocol study that is designed specifically for high-quality RNA from Purkinje cells in the post-mortem human cerebellum excised via UV-LCM.
The stability of RNA in post-mortem human tissue is a well-known obstacle, as RNA molecules within the cells are quite subject to natural decay after death. Specifically, mRNA has been shown to be the most susceptible to nucleolytic degradation5. Monitoring of the post-mortem interval (PMI) time to freezing (PMI-frozen) is one metric that has shown some correlation to RNA degradation in some studies23,24,25. However, Table 1 shows our PMI-frozen intervals for the six samples shown in Figure 3, which indicates no relative correlation with PMI values and RNA quality. Therefore, it is necessary to perform due diligence in checking RNA integrity prior to starting a laser capture project. If the RNA integrity of the starting sample is of low quality, the resulting RNA from an LCM product will be of even lower quality. When low RNA integrity cannot be avoided, other methods for enhancing RNA for sequencing could be employed in addition to this protocol21. Notably, low-input or degraded RNA can lead to muted complexity and suboptimal results that often necessitate additional amplification steps. The addition of PCR cycles for amplification has been shown to amplify sequences unequally, as well as create read duplicates upon sequencing26,27. Therefore, utilization of high-quality RNA from LCM samples for sequencing is highly advantageous.
The visualization method employed in this protocol heavily relies on the user's expertise when cutting fresh frozen tissue on a cryostat. The protocol goes into extensive detail on how to best prepare the tissue for sectioning and place the tissue on the slide. These are undoubtedly the two most critical steps of this protocol. If the tissue is not acclimated to the cryostat temperature, shredding will occur, which significantly hinders the morphological quality of the tissue. Shredding is the result of a few potential issues; troubleshooting possibilities include waiting longer for the tissue to come to temperature or altering the cryostat OT and/or CT to a warmer range. Once the tissue has been successfully cut and placed on the stage, it is important to orient the tissue and the slide properly to ensure all tissue fits within the membrane and can be accurately picked up from the stage. When attempting to pick up the tissue from the cryostat stage, the tissue must be cold, and the slide should be warm. Alternative protocols exist that recommend to chill the slide prior to picking up the tissue and warmed with a finger over the area of tissue placement28; however, in our hands, this does not provide any benefit, and often causes difficulty in picking up the tissue and excessive tissue folds. With a slide at room temperature, the tissue melts immediately upon contact with the slide. To ensure a clean pickup, it is necessary to angle the slide so that it comes into contact with the tissue at the membrane area but does not go so far as to touch the stage itself. The membrane will begin to detach from the glass slide if it touches the stage, which damages the membrane and makes laser capture difficult. If this occurs, it will be noticeable once LCM has begun and cannot be undone. Troubleshooting options are limited; if at any time it is thought the membrane or slide have been compromised, it is wise to discard. It is recommended to practice fresh frozen tissue sectioning prior to starting a project or using a pathology core that can produce high quality tissue sectioning. However, if using a core service, ensure that proper RNA technique is followed to prevent RNase contamination. RNA degradation can readily occur when improper technique is used.
We have presented here a complete method for stainless visualization of Purkinje cells in the post-mortem human cerebellum for the purposes of UV-LCM. We have also included proper RNA preservation methods and techniques to ensure high levels of RNA integrity. This protocol is not necessarily specific to any one cell or tissue type but does maintain the requirement that the region/cell of interest is morphologically distinguishable from the surrounding stroma without the need for antigen or dye specific recognition.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge The New York Brain Bank, Dr. Jean Paul Vonsattel and Dr. Etty Cortés, for their assistance in cutting and preserving the frozen human brain tissue samples for these experiments. The authors would like to acknowledge the individuals who generously donated their brain for the continuing research into Essential Tremor. Dr. Faust and Dr. Martuscello would like to acknowledge Columbia University Department of Pathology and Cell Biology for their continued support and core research space. The authors would like to acknowledge NIH R01 NS088257 Louis/Faust) for research funding for this project. LCM images were performed in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant #P30 CA013696 (National Cancer Institute). The authors would like to acknowledge Theresa Swayne and Laura Munteanu for their continued assistance with specialized microscopy.
Materials
| MembraneSlide NF 1.0 PEN | Zeiss | 415190-9081-000 | Membrane slides for tissue. We do not recommend using glass slides. |
| AdhesiveCap 500 Opaque | Zeiss | 415190-9201-000 | Opaque caps are used to enhance visualization on inverted scopes only |
| RNase Away | Molecular BioProducts | 7005-11 | Other RNase decontamination products are also suitable. Use to clean all surfaces prior to work. |
| 200 Proof Ethanol | Decon Laboratories | 2701 | If using alternative Ethanol, ensure high quality. Essential for tissue fixation. |
| Xylenes (Certified ACS) | Fisher Scientific | X5P-1GAL | If using alternative xylene, ensure high quality. Essential for tissue visualization. |
| UltraPure Distilled Water | Invitrogen | 10977-015 | Other RNA/DNA/Nuclease free water also suitable. Utilized in ethanol dilution only. |
| Slide-Fix Slide Jars | Evergreen | 240-5440-G8K | Any slide holder/jar is also suitable. Must be cleaned prior to use. Used for fixation following sectioning. |
| Anti-Roll Plate, Assy. 70mm 100um | Leica Biosystems | 14041933980 | Anti-roll plate type is determined by type of cryostat and attachment location on stage. All cryostats have differing requirements. |
| Diethyl pyrocarbonate (DEPC) | Sigma Aldrich | D5758-50mL | DEPC from any vendor will do. Follow directions for water treatment. |
| Kimwipes | Fisher Scientific | 06-666A | Kimwipes can be ordered from any vendor and used at any size. Kimwipes are to be used to clean microscope and cryostat. |
| Tissue-Plus O.C.T. Compound | Fisher Scientific | 23-730-571 | Any brand of OCT is acceptable. No tissue will be embedded in the OCT, it is strickly to attach tissue to 'chuck'. |
| Edge-Rite Low-Profile Microtome Blades | Thermo Scientific | 4280L | Blade type is determined by type of cryostat. All cryostats have differing requirements. |
| Leica Microsystems 3P 25 + 30MM CRYOSTAT CHUCKS | Fisher Scientific | NC0558768 | Chuck type is determined by type of cryostat. All cryostats have different requirements. Either size is suitable. |
| RNeasy Micro Kit (50) | Qiagen | 74004 | Required for RNA extraction post LCM. RLT Lysis buffer essential to gather captured cells from opaque cap. |
| RNeasy Mini Kit (50) | Qiagen | 74104 | Required for RNA extraction of section preps, which are performed prior to LCM to test RNA integrity of starting tissues. |
| RNase-Free DNase Set | Qiagen | 79254 | Dnase will remove any DNA to allow for a pure RNA population. Ensure Dnase is made fresh. |
| 2-Mercaptoethanol | Sigma Aldrich | M6250-100ML | Purchased volume at user discretion. Necessary for addition to RLT buffer for cell lysis. Prevents RNase activity. |
| Globe Scientific Lot Certified Graduated Microcentrifuge Tube (2 mL) | Fisher Scientific | 22-010-092 | Any 2 mL tube that is RNase free, DNase free, human DNA free, and Pyrogen free will do. |
References
- Liu, H., et al. Laser capture microdissection for the investigative pathologist. Veterinary Pathology. 51 (1), 257-269 (2014).
- Datta, S., et al. Laser capture microdissection: Big data from small samples. Histology and Histopathology. 30 (11), 1255-1269 (2015).
- Romero, I. G., Pai, A. A., Tung, J., Gilad, Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biology. 12 (42), 1-13 (2014).
- Bevilacqua, C., Ducos, B. Laser microdissection: A powerful tool for genomics at cell level. Molecular Aspects Medicine. 59, 5-27 (2018).
- Sidova, M., Tomankova, S., Abaffy, P., Kubista, M., Sindelka, R. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomolecular Detection and Quantification. 5, 3-9 (2015).
- Wang, H., et al. Histological staining methods preparatory to laser capture microdissection significantly affect the integrity of the cellular RNA. BMC Genomics. 7, 97 (2006).
- Mahalingam, M., Murray, G. I. . Laser Capture Microdissection: Methods and Protocols. , (2018).
- Vandewoestyne, M., et al. Laser capture microdissection: should an ultraviolet or infrared laser be used. Analytical Biochemistry. 439 (2), 88-98 (2013).
- Graeme, M. I. . Laser Capture Microdissection. Third edn. , (2018).
- Louis, E. D. Re-thinking the biology of essential tremor: From models to morphology. Parkinsonism & Related Disorders. 20, 88-93 (2014).
- Choe, M., et al. Purkinje cell loss in essential tremor: Random sampling quantification and nearest neighbor analysis. Movement Disorders. 31 (3), 393-401 (2016).
- Louis, E. D., et al. Neuropathological changes in essential tremor: 33 cases compared with 21 controls. Brain. , 3297-3307 (2007).
- Louis, E. D., et al. Neuropathologic findings in essential tremor. Neurology. 13 (66), 1756-1759 (2006).
- Schroeder, A., et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 7, 3 (2006).
- Jensen, E. Laser Capture Microdissection. The Anatomical Record. (296), 1683-1687 (2013).
- Waller, R., et al. Isolation of enriched glial populations from post-mortem human CNS material by immuno-laser capture microdissection. Journal of Neuroscience Methods. 208 (2), 108-113 (2012).
- Lee, S., et al. Laser-capture microdissection and transcriptional profiling of the dorsomedial nucleus of the hypothalamus. Journal of Comparative Neurology. 520 (16), 3617-3632 (2012).
- Cummings, M., et al. A robust RNA integrity-preserving staining protocol for laser capture microdissection of endometrial cancer tissue. Analytical Biochemistry. 416 (1), 123-125 (2011).
- Sonne, S. B., et al. Optimizing staining protocols for laser microdissection of specific cell types from the testis including carcinoma in situ. PLoS One. 4 (5), 5536 (2009).
- Butler, A. E., et al. Recovery of high-quality RNA from laser capture microdissected human and rodent pancreas. Journal of Histotechnology. 39 (2), 59-65 (2016).
- Schuierer, S., et al. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics. 18 (1), 442 (2017).
- Kumar, A., et al. Age-associated changes in gene expression in human brain and isolated neurons. Neurobiology of Aging. 34 (4), 1199-1209 (2013).
- Sampaio-Silva, F., Magalhaes, T., Carvalho, F., Dinis-Oliveira, R. J., Silvestre, R. Profiling of RNA degradation for estimation of post mortem [corrected] interval. PLoS One. 8 (2), 56507 (2013).
- Walker, D. G., et al. Characterization of RNA isolated from eighteen different human tissues: results from a rapid human autopsy program. Cell Tissue Bank. 17 (3), 361-375 (2016).
- Birdsill, A. C., Walker, D. G., Lue, L., Sue, L. I., Beach, T. G. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 12 (4), 311-318 (2011).
- Adiconis, X., et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nature Methods. 10 (7), 623-629 (2013).
- Parekh, S., Ziegenhain, C., Vieth, B., Enard, W., Hellmann, I. The impact of amplification on differential expression analyses by RNA-seq. Scientific Reports. 6, 25533 (2016).
- . . Carl Zeiss Microscopy. , (2012).

