Intraventricular Transplantation of Engineered Neuronal Precursors for In Vivo Neuroarchitecture Studies
Summary
Based on in vitro lentiviral engineering of neuronal precursors, their co-transplantation into wild-type brains and paired morphometric evaluation of “test” and “control” derivatives, this method allows accurate modeling of in vivo gene control of neocortical neuron morphology in a simple and affordable way.
Abstract
Gene control of neuronal cytoarchitecture is currently the subject of intensive investigation. Described here is a simple method developed to study in vivo gene control of neocortical projection neuron morphology. This method is based on (1) in vitro lentiviral engineering of neuronal precursors as “test” and “control” cells, (2) their co-transplantation into wild-type brains, and (3) paired morphometric evaluation of their neuronal derivatives. Specifically, E12.5 pallial precursors from panneuronal, genetically labeled donors, are employed for this purpose. They are engineered to take advantage of selected promoters and tetON/OFF technology, and they are free-hand transplanted into neonatal lateral ventricles. Later, upon immunofluorescence profiling of recipient brains, silhouettes of transplanted neurons are fed into NeurphologyJ open source software, their morphometric parameters are extracted, and average length and branching index are calculated. Compared to other methods, this one offers three main advantages: it permits achieving of fine control of transgene expression at affordable costs, it only requires basic surgical skills, and it provides statistically reliable results upon analysis of a limited number of animals. Because of its design, however, it is not adequate to address non cell-autonomous control of neuroarchitecture. Moreover, it should be preferably used to investigate neurite morphology control after completion of neuronal migration. In its present formulation, this method is exquisitely tuned to investigate gene control of glutamatergic neocortical neuron architecture. Taking advantage of transgenic lines expressing EGFP in other specific neural cell types, it can be re-purposed to address gene control of their architecture.
Introduction
Here we describe a simple method we developed to dissect in vivo gene control of neuronal cytoarchitecture. Based on in vitro engineering of neuronal precursors, their transplantation into neonatal brain and paired morphometric evaluation of "test" and "control" cells, it allows to unveil functional implications of test genes in fine control of neuronal morphology in a fast and affordable way. To investigate in vivo gene control of neuronal architecture, three key technical issues must be addressed: (1) achieving an adequately patterned gene-of-interest (GOI) expression and an accurate quantitative control of it; (2) obtaining a properly segmented visualization of distinct neuronal silhouettes; (3) eliciting statistical significance of results while employing a limited number of animals.
When available, mouse mutant lines harboring tetracycline (tet)-controlled transgenes may be the best tool to address the first issue1. Alternatively, somatic transgenesis may be employed. In such cases, the transgene is delivered via electroporation2 or viral transduction3. Next, it is retained as an episome (e.g., upon standard electroporation4), or it is integrated into the genome (randomly, via retroviral integrase5; or in a defined location, via CRISPR-promoted homologous recombination (SLENDR)6).
Second, neuronal silhouette visualization may be achieved via (a) sparse uniform labeling or (b) dense differential labeling. As for sparse labeling, advanced Golgi-like methodologies may be employed7, selected neuronal minisets can be filled by biocytin8, and salt-and-pepper labeling can be obtained thanks to a sparsely expressed transgene. Such a transgene may display variegated transcription (Thy-EGFP)9 or may be activated by stochastic recombination (MORF)10. As for (b), state of art strategies include Cre-mediated stochastic recombination within a multi-floxed fluoroprotein transgene array (Brainbow)11, as well as piggyBac-transposase-driven genomic integration of fluoroprotein genes, previously delivered via somatic transgenesis (CLoNE)12.
As for the third issue, the morphometric outcome is often affected by a large random variability, originating from inter-animal differences and cell-injection contingencies. Because of this, a large number of animals is usually employed to achieve the statistical power necessary to assess GOI morphometric activity.
The approaches previously described often rely on advanced technical skills and require conspicuous financial resources, which may limit their diffusion within the scientific community. To circumvent these issues, we conceived an easy and straightforward pipeline to dissect gene control of neuroarchitecture in vivo in a fast and affordable way. This is inspired by a similar co-transplantation design previously developed for fast in vivo evaluation of antiblastic transgene activity13.
Specifically, it is thought that the co-transplantation of in vitro engineered "green" neural precursors ("test" and "control" cells) into a "black" recipient neonatal brain can simultaneously fix the three key issues listed above. In fact, in vitro lentiviral engineering of precursors, in well-controlled conditions, allows maintenance of variability of neuronal transgene expression at a minimum, far below that usually associated with in vivo somatic manipulations (performed previously14,15 and in our unpublished results). The resulting accurate control of gene expression is comparable with that achieved by tet-controlled transgenic models. However, costs of this procedure are far below those originating from maintenance of a transgenic mouse line. Next, free-hand cell injection is easy and requires minimal training. Moreover, the amount of labeled precursors injected into each brain can be easily tuned to achieve a sufficient cumulative number of sparsely distributed precursors, while also keeping the total number of transplanted animals at a minimum. Last but not least, co-injection of differently fluoro-labeled, "test" and "control" precursors and subsequent pairwise statistical analysis of the results counteract the effects of inter-animal experimental variability, allowing the reaching of statistical significance of results, even upon the analysis of a limited number of individuals13.
It should be emphasized that, albeit fast and cheap, this method has two main limitations. First, it is designed to investigate cell-autonomous gene control of neuronal architecture, and it is not appropriate to address environmental control. Second, as transplanted neuronal precursors reach their final location by a heterochronic schedule, this method is preferable to model neuroarchitectonics control occurring past migration completion.
Protocol
All methods and procedures described here have been approved by the SISSA Organismo preposto al Benessere Animale (SISSA IACUC).
1. Generation of engineered "green" progenitor pools
- Preparation of the "green" pool
- Mate a wild-type CD1 female with a MtaptEGFP/+ founder16. Euthanize the pregnant dam by cervical dislocation at 12.5 days post-coitum (day 0 is determined by vaginal plug inspection) and harvest the embryonic day 12.5 (E12.5) embryos. Set them in individual wells of a 24 multiwell plate in cold PBS solution.
- Quickly genotype the embryos by visual inspection under a blue light lamp, checking for green fluorescence emission in the brains.
- Place the "green" embryos into a 10 cm diameter Petri dish filled with cold 1x PBS with 0.6% glucose and transfer it to under a stereomicroscope.
- Cut the embryo heads using standard scissors and extract the telencephalon from them by means of #3 and #5 forceps.
- Separate the two telencephalic vesicles and dissect out the neocortices using forceps; make sure to remove the hippocampus and the basal ganglia17.
- Collect neocortices by transferring them with a P1000 pipette into a 1.5 mL tube kept on ice.
- Let the dissected neocortices settle to the bottom of the tube.
- Aspirate the supernatant as much as possible.
- Resuspend the neocortices in 400 µL of fresh proliferative medium [DMEM-F12, 1x Glutamax, 1X N2, 1 mg/mL bovine serum albumin (BSA), 0.6% glucose, 2 µg/mL heparin, 20 ng/mL basic fibroblast growth factor (bFGF), 20 ng/mL epidermal growth factor (EGF), 1X pen-strep, and 10 pg/mL amphotericin B].
- Gently pipette the neocortices up and down, sequentially with P1000, P200, and P20 tips, 4x per tip, to obtain a cloudy cell suspension.
NOTE: E12.5 neocortical tissue is very soft; dissociating it to single cells does not require enzymatic digestion at all; repeated pipetting is sufficient. - Let the meninges and residual neocortical clumps settle down for 2 min.
- Transfer 150 µL from the top of the cell suspension (mainly containing single cells) into a new 1.5 mL sterile tube.
- Add 150 µL of fresh proliferative medium and repeat steps 1.1.10-1.1.12 until no more tissue clumps are evident. While doing this, omit the P1000 pipetting step (in step 1.1.10).
NOTE: Three iteration of steps 1.1.10-1.1.12 are usually sufficient to achieve full tissue dissociation. - Count cells ("green" pool) with the trypan blue exclusion method in a Bürker chamber17 and add fresh proliferative medium to adjust the concentration to 103 cells/µL.
NOTE: Steps 1.1.7-1.1.14 must be performed under a sterile laminar flow hood disinfected by 70% ethanol and kept ventilated for at least 20 min.
- Engineering the "green" sub-pools
- Subdivide the "green" pool into two sub-pools in two distinct 1.5 mL tubes for lentiviral infection.
- Infect the sub-pools independently with the two dedicated lentiviral mixes. Each mix contains "red label" virus (Pgk1_mCherry) or "black" virus (pCAG_lacZ) as a control, as well as viruses for tetOFF driven transgene (Pgk1p_tTA and TRE_transgene) or control (Pgk1p_tTA and TRE_PLAP) expression.
CAUTION: Manipulate lentiviruses in a BLS-2 environment with all the protective measurements prescribed by applicable rules and laws.
NOTE: Each lentivirus must be administered at a multiplicity of infection (MOI) of 8, which (as previously shown14) is sufficient to transduce the vast majority of neural cells in such experimental conditions (Figure 1). The lentivirus harboring tet transactivators can be built by employing different neuronal promoters driving tTA/rtTA expression (e.g., pTα1, pSyn, pCaMKII, pGAD1, etc.) (Figure 2). The MOI is the ratio of (a) the number of infectious viral particles delivered to (b) the number of their target cells. Here, the former (a) is calculated according to the conventional procedure described elsewhere14, the latter (b) is simply evaluated using a Bürker chamber. - Plate 600,000 cells of each infected sub-pool per well of a 12 multiwell plate, in 600 µL of proliferative medium with 2 µg/mL of doxycycline.
NOTE: Here, wells are not pretreated with poly-lysine, which prevents neural cell attachment to their bottoms. - Transfer cells to the incubator in 5% CO2 at 37 °C.
- After 2 days, to assess the efficiency of infection and the differential marking of the engineered pools, inspect the cells under a conventional fluorescent microscope. The two sub-pools must appear as suspensions of small neurospheres, homogenously expressing the red fluorescent protein ("control" sample) or not ("test" sample). Within these neurospheres, the MtaptEGFP transgene is active limited to a minority of cells (post-mitotic neurons) and silent in the majority of them (proliferating precursors) (Figure 3).
2. Setting up of the surgical instruments and the operating area
- Preparing borosilicate needles
- Use the borosilicate glass capillaries with external and internal diameter equaling 1.5 mm and 1.12 mm, respectively.
- Pull the capillary with a P 1000 puller.
- Set up the pulling program. Typical program parameters are: heat = 614; vel = 60; time = 1; pressure = 600. They must be adjusted according to the puller model and the heating resistor employed.
- Place the capillary into the puller holders and tighten them.
- Start the program and take the two pulled microcapillaries.
- Cut the capillary tip by hand, with a scalpel, under the stereomicroscope to obtain a tip with an external diameter of ~200-250 µm.
- Place the capillaries on a modelling clay (e.g., plasticine) support in a closed Petri dish and transfer it to under the hood.
- Setting up the hood and preparing the cell tracer solution
- Clean up the horizontal flow hood carefully and disinfect it using 70% ethanol.
- Disinfect optical fibers using 70% ethanol and place them under the hood.
- Mix 10 µL of 125 mM EGTA solution and 10 µL of Fast Green to prepare the "cell tracer solution" and place it under the hood.
- Cut small pieces (4 cm x 2 cm of size) of laboratory sealing film for cell suspension spotting.
- Prepare a solution of 1 mg/mL sterile doxycycline and aspirate it into a 0.3 mL syringe.
- Switch on the hood and keep the laminar flow on for 20 min before the operation.
- Assembling the injection tube
- Take a hard-plastic mouthpiece, two latex tubes (each about 30 cm long) and a capillary holder from an aspirator tube assembly kit for calibrated microcapillary pipettes.
- Fix the hard-plastic mouthpiece to one end of a latex tube.
- Fix the capillary holder to one end of another latex tube.
- Connect the free ends of the two latex tubes with a 0.45 µm sterile filter, as a barrier against operator germs.
- Place the resulting aspirator tube assembly on a modelling clay holder to be kept under the hood.
3. Cell mix and intraventricular transplantation
- Mixing "test" and "control" green cell sub-pools
- Collect "test" and "control" neurospheres (see step 1.2.5) in separate 1.5 mL tubes.
- Centrifuge neurospheres suspension at 200 g for 5 min.
- Collect and discard the supernatant, avoiding disturbance of the cell pellet.
- Gently resuspend neurospheres in 500 µL of 1x PBS.
- Centrifuge them at 200 g for 1 min.
- Remove the supernatant.
- Repeat steps 3.1.4-3.1.6 two more times.
- Gently resuspend spheres in 200 µL of 2x Trypsin solution (2x Trypsin, 1x PBS) and pipette cell pellet up and down 4x-5x.
NOTE: Pay attention not to make air bubbles. - Leave cells in the incubator for 5 min at 37 °C to achieve a single-cell suspension.
- Block Trypsin with 200 µL of Trypsin inhibitor solution (DMEM-F12, 10 µg/mL DNAse I, 0.28 mg/mL Trypsin inhibitor) by pipetting up and down 4x-5x.
- Centrifuge cells at 200 x g for 5 min and resuspend them in 1 mL of fresh proliferative medium (DMEM-F12, 1x Glutamax, 1x N2, 1 mg/mL BSA, 0.6% glucose, 2 µg/mL heparin, 20 ng/mL bFGF, 20 ng/mL EGF, 1x pen-strep, 10 pg/mL amphotericin B).
- Count cells of the two pools with trypan blue exclusion method in a Bürker chamber.
- Adjust their concentration to 100,000 cells/µL in proliferative medium.
- Mix 1:1 "test" cells with "control" ones.
- Check the resulting mix under a fluorescence microscope (objective magnification: 10x) to ensure that the mix is a single-cell suspension of the two pools (Figure 3C).
- Place the mix on ice under the hood.
- Intraventricular transplantation
- Prepare the cage with the mother and the P0 pups on a table far away from the surgical operatory area.
- Place a recovery cage on a cart close to the hood.
- Put a mixture of sawdust taken from the mother's cage on the bottom of the recovery cage and place it under a lamp.
- Aside, set an ice box covered by an aluminum foil for the anesthesia of P0 pups.
- Just prior to transplantation, add 1/10 volume of cell tracer solution (from step 2.2.3) to 1 volume of cell suspension (from step 3.1.16) and spot 3 µL of the resulting "injection mix" on a piece of laboratory sealing film.
- Place the pup on the cool aluminum foil for 1 min and check that it is fully anesthetized.
NOTE: To confirm anesthesia, gently squeeze a paw and monitor the pup for lack of movements, while still breathing. - Meanwhile, aspirate the 3 µL of injection mix (from step 3.2.5) into the glass capillary.
- Gently wipe the head of the anesthetized pup with 70% ethanol.
- Place the pup's chin on the tip of the optical fiber to clearly identify the cortical hemispheres.
NOTE: At P0-P1, the head skin is very thin, allowing for straightforward visual identification of the hemispheres just above the eyeballs and behind the olfactory bulbs. - Keep the capillary (loaded as described in step 3.2.7) in either one of the two mid-parasagittal planes (left or right), above the olfactory bulb, and puncture the forehead skin at its intersection with the frontal plane containing the eyeballs centers. Pay attention not to damage blood vessels. Enter the capillary into the frontal cortex and access the ventricular cavity (Figure 4A).
- To prevent accidental damage of the ganglionic eminence, rotate the needle laterally by 30 degrees, pointing its tip caudal-medial-ward (Figure 4B).
- Gently inject the cells into the ventricular cavity and monitor their diffusion by means of Fast Green (Figure 4C).
- Wait 5-10 s.
- Remove the glass needle taking care not to aspirate the cell suspension and discard it in a suitable disposal bin.
- Before pup recovery from anesthesia inject 150 µL of doxycycline solution intraperitoneally to keep transgene expression still off after the transplantation.
- After the injection, place the pup immediately under the lamp for recovery.
- Leave the pups under the lamp for 5-10 min and, once fully awake, place them in the cage with the mother.
- Observe the operated pups for 2-3 h, to ensure that the mother accepts them.
- Supply the cage with drinking water containing 0.5 mg/mL doxycycline and 50 g/L sucrose to maintain transgene expression off.
- Replace the doxycycline-containing water by doxy-free water 4 days after transplantation, thus activating the transgene in post-mitotic neurons. Keep mice in a doxy-free regime for 6 days and euthanize them at P10 by carbon dioxide inhalation followed by decapitation.
4. Analysis of transplanted brains
- Histology and immunofluorescence
- Euthanize the transplanted pups 10 days after cell transplantation by carbon dioxide inhalation followed by decapitation. Fix brains of euthanized pups in 4% paraformaldehyde (PFA) at 4 °C overnight.
CAUTION: PFA is highly toxic; handle it with care, strictly complying with manufacturer's prescriptions, under a chemical fume hood; discard PFA solution residual in a labeled waste container. - Remove the PFA solution and replace it with 30% sucrose solution.
- Leave the brains at 4 °C or until they sink to the vial bottom.
- Transfer the brains into a disposable embedding mold approximately half-filled with cryo-inclusion medium.
- Freeze the included brains at -80 °C.
- Cut 60 µm thick coronal sections using a cryostat.
NOTE: Avoid employing thinner sections, as this may result in unacceptable loss of information regarding the whole architecture of single neurons. - Process sections for immunofluorescence18,19, using anti-GFP [1:400] and anti-RFP (mCherry) [1:500] antibodies. Counterstain nuclei with 1 mg/mL DAPI solution [1:200].
- Euthanize the transplanted pups 10 days after cell transplantation by carbon dioxide inhalation followed by decapitation. Fix brains of euthanized pups in 4% paraformaldehyde (PFA) at 4 °C overnight.
- Morphometry
- Set working parameters of the confocal microscope as follows: z-stack height = 40 µm; step = 2 µm.
- Collect pictures of immunoassayed slices selecting GFP positive neuron-rich photographic fields, blind of RFP signal distribution.
- Export the images as .nd2 files.
- Generate maximum Z-projections of them as .tiff files (Figure 5A).
- To allow for sub-sequential neuronal skeletonization blind to cells genotype, hide the red signal. To do so, open each .tiff file by a suitable software and add an adjustment layer. Select "levels", "red", then set "output" to zero. Save the file as such.
NOTE: Skeletonization is subsequently performed by a new operator who had no previous access to original plain red files. - For each hidden red file, add a drawing layer to the primary image and select the pencil tool (white color). Trace the soma on the basis of GFP signal, then trace the neurites.
NOTE: the pencil tool may be set at 40 pixels for the soma and at three pixels for neurites. - Save the multilayer file including neuronal silhouettes (Figure 5B).
NOTE: Subsequent analysis of neuronal skeleton can be performed by either operator. - Create a new 1024 x 1024 16-bit grayscale file with black background, one per neuron.
- Copy and paste single neuronal silhouettes from the primary image to the new grayscale file. Get back to multilayer file and switch off the adjustment layer to unveil neuronal genotypes. Save the 16-bit grayscale file annotating the corresponding neuronal genotype.
- Import grayscale images in ImageJ one by one and analyze skeletons by NeurphologyJ plugin of ImageJ software20 (Figure 5C).
- Copy NeurphologyJ primary data (neurite_length, attachment_points and endpoints; for each, take "Total Area" values), paste them into a spreadsheet and employ them to compute secondary morphometric parameters (average neurite length, branching index) (Figure 5D).
Representative Results
There are five primary datasets providing useful information about key aspects of the procedure, the first being (1) efficiency of neural precursors transduction and co-transduction by lentiviral vectors. (2) An example of key features of the promoters employed to drive the "test gene". (3) An example of engineered cells ready for transplantation. (4) A cartoon including key procedural details of cell microinjection into the neonatal brain. (5) A synopsis of the whole morphometric pipeline.
As for (1), the capability of the prototypical lentiviral vectors delivered at different MOIs (2,4,8) to effectively transduce neocortical precursors was evaluated. In the first assay, neural precursors originating from early wild-type neocortices were infected by a lentiviral reporter, harboring the mCherry coding sequence (cds) under the control of the neuronogenic-lineage-specific pTα1 promoter, at MOI = 2, 4, 8, then allowed to differentiate. Co-immunoprofiling of their progenies for mCherry and the pan-neuronal marker Tubb3 showed that neurons were effectively transduced at frequencies of 64%, 69%, and 78%, respectively (Figure 1A). In a second assay, neocortical precursors were acutely coinfected by two lentiviral reporters, expressing EGFP and mCherry, under the control of the constitutive pPgk1 promoter, at MOIs = (2,2), (4,4), and (8,8). A few days later, co-immunoprofiling of their derivatives showed that the ratio between EGFP+mCherry+ and EGFP+mCherry± cells was 80%, 88%, and 93%, respectively (Figure 1B). These data suggest that, upon delivery of the engineering protocol envisaged for the present study, (a) the vast majority of neurons are transduced, and (b) almost all transduced neurons received the full lentiviral set needed for their characterization.
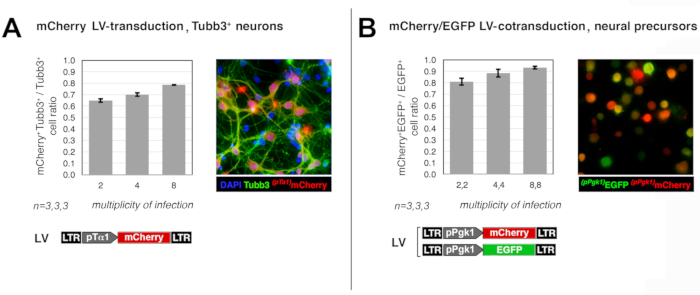
Figure 1: Efficiency of neural precursor cells transduction by lentiviral vectors. Panels report an evaluation of the efficiency by which E12.5 neocortical precursor cells are transduced (or co-transduced) upon delivery of dedicated lentiviral reporters at different multiplicities of infection (MOIs). In the former case (A), cells were acutely infected with the lentivirus (LV) pTα1-mCherry at MOI = 2, 4, 8, cultured in proliferative medium for 2 days, transferred to differentiative medium, and allowed to differentiate for 1 week. Upon fixation at day in vitro (DIV) 9, cultures were co-immunoprofiled for mCherry and the pan-neuronal marker Tubb3. mCherry and EGFP signals were detected by anti-RFP and anti-EGFP primary antibodies and revealed by Alexa-594- and Alexa-488-conjugated secondary antibodies, respectively. Finally, mCherry+Tubb3+/ mCherryTubb3+ ratios were calculated, averaged and plotted against the corresponding MOIs. In the latter case (B), cells were acutely infected with a 1:1 mix of two lentiviruses, encoding for the constitutively active pPgk-mCherry and pPgk-EGFP transgenes, at different MOIs (2, 4, 8 for each virus). Cells were cultured in proliferative medium for 4 days, trypsinized and, upon fixation, profiled for mCherry and EGFP immunofluorescence as above. Finally, mCherry+EGFP+/ mCherryEGFP+ ratios were calculated, averaged and plotted against the corresponding MOIs. Error bars represent S.E.M. Please click here to view a larger version of this figure.
As for (2), pTα1 and pSyn activity patterns can be inferred on the basis of expression profiles of fluoroprotein genes under their control. In this example, such control is direct in case of mCherry (Figure 2A), and indirect [i.e. mediated by a tetON second generation interface (Figure 2E)], in the case of EGFP (Figure 2B,C,D). Both promoters are specifically active within Tubb3+ postmitotic neurons (Figure 2B,C,D), but pTα1 also fires in a subset of Ki67+ intermitotic (neuronogenic) precursors (Figure 2A). Here, to provide an idea of limited variability of promoter activity when cells are engineered according to these standard conditions (Figure 2E), pTα1- and pSyn-driven EGFP expression levels within Tubb3+ neurons were quantified by Photoshop CS6 Histogram plugin. Then, raw data were normalized against medians and plotted against promoters (Figure 2F). Remarkably, single-cell fluorescence levels were densely clustered around medians: first and third quartiles equaled 0.86 and 1.16, as well as 0.89 and 1.12 in cases of pTα1 and pSyn, respectively.
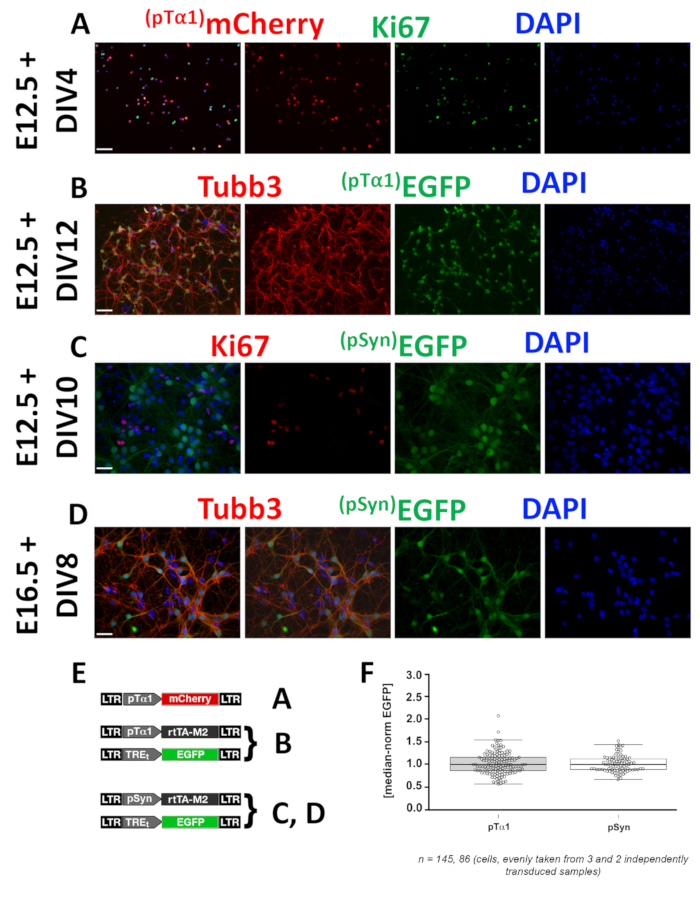
Figure 2: Profiling of neural precursor type-specific promoters suitable to drive GOI overexpression. Panels refer to characterization of Tubulin alpha 1 (pTα1) and Synapsin (pSyn) promoters, specifically active within the whole neuronal lineage and postmitotic neurons, respectively. Test were run in neocortical precursors harvested at different embryonic (En) ages, acutely infected with dedicated lentiviral mixes [pTα1-mCherry in (A); pTα1-rtTA, TREt-EGFP in (B); pSyn-rtTA, TREt-EGFP in (C,D); 2µg/ml doxy ab initio], and cultured in proliferative medium (A), proliferative (2 days) followed by differentiative medium (B,C), or differentiative (D) medium. Upon fixation at day in vitro (DIV) n, postmitotic neurons were identified by αTubb3 and intermitotic precursors by αKi67 immunofluorescence; mCherry and EGFP were detected as shown in Figure 1. Signals were revealed as shown in Figure 1. Scale bars: 100 µm in (A,B) and 50 µm in (C,D). Shown are (E) dedicated lentiviral mixes used in this study with references to the figure panels. Shown is a (F) a scatter plot of pTα1- and pSyn-driven EGFP signal in Tubb3+ cells referred to in (B) and (D), respectively. Boxed are cells falling between 25th and 75th percentiles; whiskers refer to 10th and 90th percentiles. Please click here to view a larger version of this figure.
As for (3), 3 days after lentiviral transduction, MtaptEGFP/+ "green" neurospheres express the Pgk1p promoter-driven, mCherry red fluorescent protein ("control" spheres) or not ("test" spheres) (Figure 3A,B). All spheres are dissociated to single cells, ensuring there are no clumps lefts, and the corresponding suspensions are mixed 1:1. The resulting mix (Figure 3C) is placed on ice and used within 20 min for transplantation.
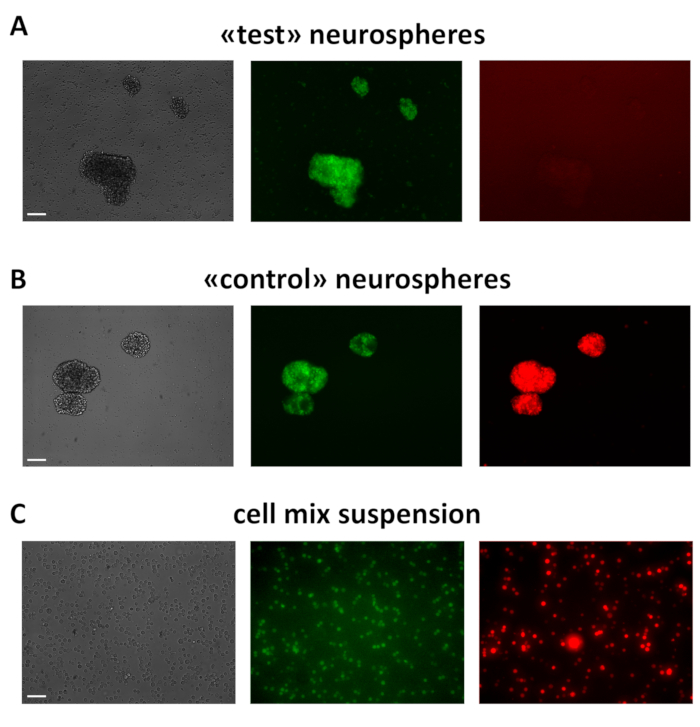
Figure 3: Example of test ("only green") and control ("green-red") DIV2 neurospheres and cell suspension ready for transplantation. (A,B) These spheres were obtained from MtaptEGFP/+E 12.5 neocortical precursors, acutely infected by lentiviral mixes "pCAG_lacZ, Pgk1p_tTA, TREt_GOI" (A) and "Pgk1p_mCherry, Pgk1p_tTA, TREt_PLAP" (B), respectively, each LV at MOI = 8, and kept in proliferative medium. (C) Cell suspension was obtained by dissociating "test" and "control" neurospheres (A,B) to single cells and mixing them 1:1. Scale bars: 200 µm in (A,B) and 100 µm in (C). Please click here to view a larger version of this figure.
As for (4), the procedure for cell injection into the neonatal brain includes three key steps. The capillary, placed on the mid-parasagittal plane, is entered into the lateral ventricular cavity via the frontal cortical wall (Figure 4A). Next, to prevent damage of the ganglionic eminence, the capillary is rotated 30° within the horizontal plane, so that the tip points medially/caudally (Figure 4B). Last, the cell suspension is delicately ejected into the lateral ventricular cavity, where it forms an easily distinguishable, light-blue "cloud" (Figure 4C).
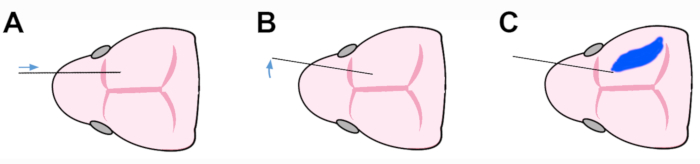
Figure 4: Schematics of the three steps procedure employed for cell injection into the neonatal brain. (A) The capillary is entered into the lateral ventricle through the frontal neocortical wall. Then, (B) it is rotated medialward, to prevent damage of ganglionic eminence. Finally (C), the cell suspension is gently injected into the lateral ventricle, where it forms a light blue cloud. Please click here to view a larger version of this figure.
As for (5), the analytical procedure includes four main steps. First, optical confocal sections of transplanted neuron-rich fields are flattened, according to MAX Z-projection modality so generating RGB .tiff files. Here, as an example, only "green test" neurons and "yellow control" neurons, originating from co-transplanted precursors, can be easily distinguished (it should be noted that mCherry may alternatively be used to label "test" neurons) (Figure 5A). Then, an idealized, skeletonized camera lucida version of the picture is generated by a suitable graphic software (see step 4.2.6) (Figure 5B) by an operator blind of red channel. Blindness of red channel is of paramount importance in order to prevent any unintentional bias in data evaluation. Next, single-neuronal silhouettes are fed into NeurphologyJ software as black-and-white pictures and values of the three primary parameters "total number of exitpoints" (attachment_points), "total number of endpoints" (endpoints) and "total neurite length" (neurite_length) are collected (Figure 5C). Finally, secondary parameters of each neuron are calculated, averaged and statistically evaluated by Excel software (Figure 5D).
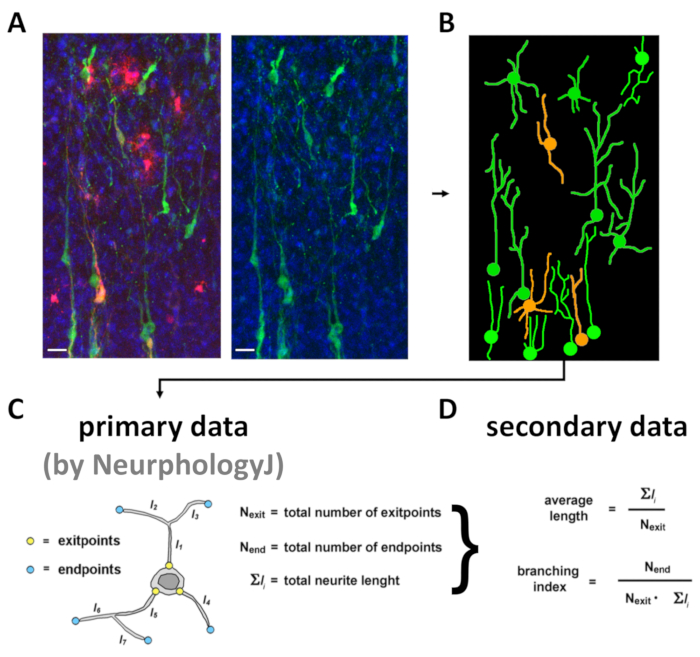
Figure 5: Representative flowchart of morphometric analysis of transplanted brains. Upper row panels show: (A) a MAX z-projection .tiff image of medial neocortex, fixed 10 days after engineered cell transplantation (green = Mtapt-driven, neuron-restricted EGFP; red = mCherry, constitutively labeling "control" cells; blue = DAPI), the green channel signal, and (B) its skeletonized e-camera lucida rendering. Scale bars: 50 µm. The bottom row includes: (C) primary morphometric parameters extracted from neuronal skeletons by NeurphologyJ software analysis (neurite_length as Σli, attachment_points as Nexit and endpoints as Nend) and (D) secondary parameters calculated on their basis. This figure has been modified from Chiola et al.21. Please click here to view a larger version of this figure.
Discussion
Specific aspects/steps of this procedure are critical and require special attention. First, (a) operators must be adequately pretrained to safely manipulate lentiviruses in a BSL-2 compliant lab environment. Second, (b) prior to mix "test" and "control" neural preparations, it is mandatory to carefully wash the two corresponding neurosphere suspensions as described, in order to prevent any delayed cross-infection of the two preparations due to unwanted lentiviral carry over. Third, (c) while transplanting cells, care must be placed while targeting the ventricular cavity; in this respect, the Fast Green tracer included into the cell suspension is of substantial help. Additionally, (d) to prevent cannibalism, transplanted pups must be re-transferred to the mother only after their full recovery from anesthesia (usually 10 min is sufficient). (e) Cryo-sectioning of the transplanted tissue should be performed at 60 µm; in fact, this thickness simultaneously allows an easy antibody penetration into the tissue and limits loss of information originating from artifactual neuron fragmentation. Finally, (f) to prevent any unintentional bias in data evaluation, we emphasize that neuron skeletonization must be performed by an operator blind of red channel.
The protocol described here refers to GOF gene manipulations. Alternatively, "test" neurons may be engineered to downregulate GOI function by means of lentiviruses encoding for RNAi effectors. Next, we usually employ mCherry to label "control" neural cells; however, the mCherry/"control" and LacZ/"test" scheme may be obviously inverted. Last, the protocol described here refers to intraventricular cell injections; "test" and "control" neurons may be alternatively co-injected into the neural parenchima21.
Despite its advantages, this method has two main limits. While allowing to investigate cell–autonomous gene control of neuroarchitecture, it does not apply to environmental control of it. Moreover, as transplanted neuronal precursors migrate from the ventricular aspect of the cortical wall to their final laminar location after birth (i.e. within a non-physiological timeframe), this method should be preferably employed to model neuroarchitectonics control after migration completion.
Last but not least, special attention should be paid to critical interpretation of results. In particular, if the X gene subject of investigation reduces the morphological complexity of "test" neurons, this might not specifically reflect its genuine impact on neuron architecture. Rather, such phenomenon might be a non-specific index of neuronal damage elicited by X overexpression. To address this issue, it may be useful to compare local densities of transplanted, "test" and "control" neurons, then look for possible numerical shrinkage of the "test" population, as an index of neuronal suffering induced by X [this shrinkage should be even more pronounced upon further delayed analysis of transplanted brains]. In such cases, lowering the overepression level of the X gene or moving to a loss-of-function design might fix the issue.
Our method adds to a wide array of technologies employed to investigate gene control of neuronal morphology in vivo1,2,3,4,7,8,9,10,11,12. As recalled in the introduction, these technologies rely on a variety of ad hoc genetic manipulations aimed to perturb GOI expression levels and ease the extraction of morphological information from the biological samples. These technologies usually allow accurate dissection of this control; however, they require not valuable experimental skills and financial resources. In this respect, the method offers three key advantages. It allows an accurate control of GOI expression levels comparable to those peculiar to transgenic models; however, in the absence of mutant colony maintenance costs. It does not require sophisticated experimental (surgical) skills. It often achieves statistical significance of results upon analysis of a relatively limited number of animals.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Mihn Duc Do for his contribution to early setting up of this procedure.
Materials
| 0.3 mL syringe | BD | 320840 | store at RT |
| 0.45 μm sterile filter | Millex-HV | SLHU033RS | store at RT |
| 12 multiwell plate | Falcon | 353043 | store at RT |
| 1X PBS | Gibco | 14190-094 | store at RT |
| 24 multiwell plate | Falcon | 351147 | store at RT |
| anti-EGFP antibody, chicken polyclonal, RRID:AB_371416 | Tebubio | GTX13970 | store at -20 ℃ |
| anti-RFP antibody, rat monoclonal, RRID:AB_10795839 | Antibodies Online | ABIN334653 | store at +4 ℃ |
| anti-Tubb3 antibody, mouse monoclonal, RRID:AB_2313773 | Covance | MMS-435P | store at -20 ℃ |
| Aspirator tube assemblies kit for calibrated microcapillary pipettes | Sigma | A5177-5EA | store at RT |
| Blue light lamp | Nightsea | BLS2 | store at RT |
| Borosilicate capillaries | Kwik-Fil | TW150-4 | store at RT |
| BSA | Sigma | A9647 | store at +4 ℃ |
| Bürker chamber | Sigma | BR719520 | 0.0025 mm2, 0.100 mm |
| Cryo-inclusion medium (Killik) | Bio-Optica | 05-9801 | store at RT |
| DAPI | Sigma | D9542 | store at +4 ℃ |
| Disposable embedding mold | Bio-Optica | 07MP7070 | DispoMold07 |
| DMEM/F-12 | Gibco | 31331-028 | store at +4 ℃ |
| Dnase I | Roche | 10104159001 | store at +4 ℃ |
| Doxycicline | Sigma | D1822 | store at +4 ℃ |
| Dumont forceps #3c | Fine Science Tools | 11231-20 | store at RT |
| Dumont forceps #5 | Fine Science Tools | 11251-20 | store at RT |
| EGF | Gibco | PHG0311 | store at -20 ℃ |
| EGTA | Sigma | E3889 | store at RT |
| FGF | Gibco | PHG0261 | store at -20 ℃ |
| Fine scissors – Sharp | Fine Science Tools | 14060-09 | store at RT |
| Fungizone (Amphotericin B) | Gibco | 15290018 | store at -20 ℃ |
| Glucose | Sigma | G8270 | store at RT |
| GlutaMAX Supplement | Gibco | 35050061 | store at RT |
| Goat anti-chicken Alexa 488 | Invitrogen | A11039 | store at -20 ℃ |
| Goat anti-rat Alexa 594 | Invitrogen | A11007 | store at -20 ℃ |
| Heparin Solution | Stem Cell Technologies | 07980 | store at +4 ℃ |
| N2 Supplement | Gibco | 17502048 | store at -20 ℃ |
| Optical fibers | Leica | CLS150X | store at RT |
| P1000 puller | Sutter Instruments | P-1000 model | Flaming/Brown Micropipette Puller |
| Parafilm | Bemis | PM-996 | store at RT |
| Pen Strep | Sigma | P0781 | store at -20 ℃ |
| Petri dish | Falcon | 353003 | store at RT |
| PFA | Sigma | 158127 | store at RT |
| Plasmid #363 [LV_TREt_(IRES)PLAP] | built in house | ||
| Plasmid #386 [LV_pTa1_mCherry] | built in house | ||
| Plasmid #401 [LV_pTa1_rtTA(M2)] | built in house | ||
| Plasmid #408 [LV_Ppgk1p_rtTA(M2)] | built in house | ||
| Plasmid #484 [LV_lacZ] | Addgene | 12108 | |
| Plasmid #529 [LV_Pgk1p_mCherry] | built in house | ||
| Plasmid #730 [LV_pSyn_rtTA(M2)] | built in house | ||
| Scalpel | Braun | BB515 | store at RT |
| Steromicroscope | Leica | MZ6 | store at RT |
| Trypan blue | Gibco | 15250-061 | store at RT |
| Trypsin | Gibco | 15400-054 | store at -20 ℃ |
| Trypsin inhibitor | Sigma | T6522 | store at +4 ℃ |
References
- Kistner, A., et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. , (1996).
- Matsui, A., Yoshida, A. C., Kubota, M., Ogawa, M., Shimogori, T. . Mouse in Utero Electroporation: Controlled Spatiotemporal Gene Transfection. Journal of visualized experiments: JoVE. , (2011).
- Kim, J. Y., Grunke, S. D., Levites, Y., Golde, T. E., Jankowsky, J. L. Intracerebroventricular Viral Injection of the Neonatal Mouse Brain for Persistent and Widespread Neuronal Transduction. Journal of Visualized Experiments. , (2014).
- Puram, S. V., et al. A CaMKIIβ 2 signaling pathway at the centrosome regulates dendrite patterning in the brain. Nature Neuroscience. , (2011).
- Zuccotti, A., Le Magueresse, C., Chen, M., Neitz, A., Monyer, H. The transcription factor Fezf2 directs the differentiation of neural stem cells in the subventricular zone toward a cortical phenotype. Proceedings of the National Academy of Sciences. , (2014).
- Mikuni, T., Nishiyama, J., Sun, Y., Kamasawa, N., Yasuda, R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by in Vivo Genome Editing. Cell. , (2016).
- Spiga, S., Acquas, E., Puddu, M. C., Mulas, G., Lintas, A., Diana, M. Simultaneous Golgi-Cox and immunofluorescence using confocal microscopy. Brain Structure and Function. , (2011).
- Chen, B., et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences. , (2008).
- Feng, G., et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. , (2000).
- Lu, X. H., Yang, X. W. Genetically-directed Sparse Neuronal Labeling in BAC Transgenic Mice through Mononucleotide Repeat Frameshift. Scientific Reports. , (2017).
- Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W., Sanes, J. R. Improved tools for the Brainbow toolbox. Nature Methods. , (2013).
- Garcia-Moreno, F., Vasistha, N. A., Begbie, J., Molnar, Z. CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. Development. , (2014).
- Falcone, C., Daga, A., Leanza, G., Mallamaci, A. Emx2 as a novel tool to suppress glioblastoma. Oncotarget. , (2016).
- Brancaccio, M., Pivetta, C., Granzotto, M., Filippis, C., Mallamaci, A. Emx2 and Foxg1 inhibit gliogenesis and promote neuronogenesis. Stem Cells. , (2010).
- Fimiani, C., Goina, E., Su, Q., Gao, G., Mallamaci, A. RNA activation of haploinsufficient Foxg1 gene in murine neocortex. Scientific Reports. , (2016).
- Tucker, K. L., Meyer, M., Barde, Y. A. Neurotrophins are required for nerve growth during development. Nature Neuroscience. , (2001).
- Currle, D. S., Hu, J. S., Kolski-Andreaco, A., Monuki, E. S. Culture of mouse neural stem cell precursors. Journal of Visualized Experiments. , (2007).
- Park, J. J., Cunningham, M. G. Thin Sectioning of Slice Preparations for Immunohistochemistry. Journal of Visualized Experiments. , (2007).
- Puzzolo, E., Mallamaci, A. Cortico-cerebral histogenesis in the opossum Monodelphis domestica: Generation of a hexalaminar neocortex in the absence of a basal proliferative compartment. Neural Development. , (2010).
- . . NeurphologyJ Manual. , (2012).
- Chiola, S., Do, M. D., Centrone, L., Mallamaci, A. Foxg1 Overexpression in Neocortical Pyramids Stimulates Dendrite Elongation Via Hes1 and pCreb1 Upregulation. Cerebral Cortex. , (2018).

