VirWaTest, A Point-of-Use Method for the Detection of Viruses in Water Samples
Summary
Here we present VirWaTest, which is a simple, affordable and portable method for the concentration and detection of viruses from water samples at the point of use.
Abstract
Viruses excreted by humans and animals may contaminate water sources and pose a risk to human health when this water is used for drinking, food irrigation, washing, etc. The classical fecal bacteria indicator does not always check for the presence of viral pathogens so the detection of viral pathogens and viral indicators is relevant in order to adopt measures of risk mitigation, especially in humanitarian scenarios and in areas where water-borne viral outbreaks are frequent.
At present, several commercial tests allowing the quantification of fecal indicator bacteria (FIB) are available for testing at the point of use. However, such commercial tests are not available for the detection of viruses. The detection of viruses in environmental water samples requires concentrating several liters into smaller volumes. Moreover, once concentrated, the detection of viruses relies on methods such as nucleic acid extraction and molecular detection (e.g., polymerase chain reaction [PCR]-based assays) of the viral genomes.
The method described here allows the concentration of viruses from 10 L water samples, as well as the extraction of viral nucleic acids at the point of use, with simple and portable equipment. This allows the testing of water samples at the point of use for several viruses and is useful in humanitarian scenarios, as well as at any context where an equipped laboratory is not available. Alternatively, the method allows concentrating viruses present in water samples and the shipping of the concentrate to a laboratory at room temperature for further analysis.
Introduction
During the first phases of any humanitarian emergency, access to clean water supplies, sanitation, and hygiene are critical for the survival of those affected. Therefore, monitoring water quality is a priority to prevent waterborne outbreaks. It is well-known that contaminated water is frequently the origin of diseases, but it is often difficult to determine the sources of viral outbreaks such as Hepatitis E virus (HEV), even with the availability of conventional laboratory methods. The control of water quality is based on the quantification of FIB1,2,3,4. However, it has been extensively documented that there is no correlation between the absence of FIB and the presence of viral waterborne pathogens such as rotavirus (RoV), norovirus (NoV), or HEV5,6. Thus, using the water quality criteria based on FIB might result in an underestimation of risks associated with the presence of waterborne viral pathogens. The surveillance of indicator viruses, such as human adenoviruses (HAdV), or specific pathogens would be helpful in defining the exposure to viral pathogens and identifying the potential source of human infection7,8,9,10 and in validating the efficacy of sanitation measures11.
Until now, the detection of viruses in these scenarios relied on skilled staff and complex logistics. VirWaTest (virwatest.org) is aimed at the development of a simple, affordable, and portable method for the concentration and subsequent detection of viruses from water samples at the point of use.
The virus concentration is based on the principle of organic flocculation of 10 L water samples, by which viruses are recovered in smaller volumes12,13. The flocs are collected and added to a buffer that lyses the viruses and prevents the nucleic acids from degradation when they stored at room temperature for not more than 2 weeks.
The nucleic acid extraction method is based on the use of magnetic particles to which the nucleic acids get adsorbed. They can be transferred from one washing buffer to another and finally into the elution buffer by using a magnetic pipette to which the particles attach. Viral nucleic acid suspensions obtained can be shipped to a reference laboratory where the detection can be performed using molecular methods based on PCR. For each nucleic acid extraction, two different quantities are tested to rule out enzymatic inhibition originated by the sample. Alternatively, with minimum equipment availability, PCR tests can be run at the point of use. The entire process is designed to be performed independently of a power supply (Figure 1).
A quantitative PCR assay to detect HAdV, excreted by humans and found in wastewater samples in high concentrations, has been adapted to be run at the point of use. HAdV are used as human fecal viral indicators. A PCR for the quantification of MS2 bacteriophage has been also adapted since MS2 is used in VirWaTest as process control. The method can be customized for the detection of any virus of interest.
After development, the VirWaTest method has been applied by the users in two different settings in the Republic of Central Africa (RCA) and Ecuador, providing feedback on the application of the protocol in real situations.
To our knowledge, this is the first procedure that allows the concentration and detection of viruses at the point of use, independent of any power supply, large equipment, and freezing/cooling conditions. It is recommended to collect two replicates of each water sample in order to obtain robust results.
Protocol
1. Preparation and packaging
NOTE: The materials/equipment to be packed is listed in Table 1. Use gloves to handle the reagents required for the process control, the concentration reagents, the nucleic acid extraction reagents and the detection reagents. Wear protective glasses to handle the reagents required for nucleic acid extraction.
- Process control
- Prepare MS2 bacteriophage stock culture (American Type Culture Collection [ATCC] 15597-B1) containing 1 x 1010 PFU/mL in the laboratory by following the ISO procedure 10705-1:199514.
- Then, aliquot 10 µL of the suspension per tube containing 1 x 108 PFU into 10 mL tubes and let the water evaporate at 37 °C. Use one tube per water sample.
- Additionally, prepare one tube containing 10 mL of sterile distilled water per collected sample.
- Concentration reagents
- For the preflocculated skimmed milk solution (PSM), use the following amounts to concentrate five 10 L water samples.
- Prepare a plastic tube containing 5 g of skimmed milk.
- Prepare a zipper plastic bag containing 16.66 g of sea salts.
- Prepare a plastic bottle containing 500 mL of distilled water.
- Prepare the reagents required for pH adjustment. Use the following amounts to adjust the pH of the PSM and of five 10 L water samples.
- Add 5 g of citric acid 1-hydrate to a 10 mL plastic tube.
CAUTION: Citric acid causes serious irritation when it comes in contact with eyes. If this occurs, rinse the eye cautiously with water for several minutes. If irritation persists, seek medical advice. - Put 2 g of sodium hydroxide into a 10 mL plastic tube.
CAUTION: Sodium hydroxide can cause severe skin burns and eye damage. If swallowed, rinse the mouth. Do not induce vomiting. If it comes in contact with the eyes, rinse cautiously with water for several minutes. Then, seek medical advice. - Put 25 mL of sterile distilled water into a plastic container.
- Put 50 mL of sterile distilled water into a plastic container.
- Add 5 g of citric acid 1-hydrate to a 10 mL plastic tube.
- Prepare the sample conditioning sachet used for the flocculation, the preservative solution used to preserve the nucleic acids, and the neutralizing sachet to neutralize the discarded water and materials. Use the following amounts for each 10 L water sample to be tested.
- Put 15 g of sea salts and 8 g of citric acid 1-hydrate into a zipper plastic bag for the conditioning sachet.
- Add 2 mL of lysis buffer (Table of Materials) and 0.3 mL of nucleic acid preserving agent (Table of Materials) to a 5 mL tube.
CAUTION: Lysis buffer contains guanidine thiocyanate and Triton X-100. Contact with acid liberates toxic gas, and it is harmful if swallowed, inhaled, or comes in contact with the skin. If swallowed, rinse the mouth. Do not induce vomiting. If it comes in contact with the skin, wash with plenty of water. Then, seek medical advice. Nucleic acid preserving agent causes skin and eye irritation and is harmful if swallowed. If it comes in contact with the skin or the eyes, rinse with abundant water for several minutes. In any case, especially if swallowed and feeling unwell, seek medical advice. - Put 40 g of detergent powder into four zipper plastic bags.
CAUTION: Powder detergent causes eye irritation. If it comes in contact with the eyes, rinse with abundant water for several minutes. If irritation persists or if swallowed, seek medical advice.
- For the preflocculated skimmed milk solution (PSM), use the following amounts to concentrate five 10 L water samples.
- Nucleic acid extraction reagents
- Put 50 µL of magnetic particles (Table of Materials) and 1 mL of ethanol 96% into a 5 mL tube which constitute the Binding Buffer.
CAUTION: The magnetic particles buffer contains sodium azide, which forms a toxic gas when it comes in contact with acids or heavy metal ions and is toxic if ingested or when it comes in contact with skin. If ingested or in contact with skin, rinse the mouth or the skin with abundant water and seek medical advice. Ethanol is a highly flammable liquid and vapor and causes serious eye irritation. Keep away from heat, hot surfaces, sparks, and any other ignition source. If it comes in contact with the skin or the eyes, rinse with abundant water. In case of ingestion of large amounts of water and/or if inhaled, go outside into the fresh air. In any case, seek medical advice. - Add 500 µL, 600 µL, and 200 µL of washing buffers 1, 2, and 3, respectively (Table of Materials), to three 2 mL tubes.
CAUTION: Washing buffers contain guanidinium thiocyanate and guanidinium chloride, which are harmful if inhaled or swallowed and irritate the skin and eyes. Contact with acids releases very toxic gas. If they come in contact with the skin or eyes, rinse with abundant water. If swallowed, rinse the mouth with abundant water. Do not induce vomiting. Always seek medical advice. - Add 120 µL of elution buffer (Table of Materials) to a 0.5 mL tube.
- Put 50 µL of magnetic particles (Table of Materials) and 1 mL of ethanol 96% into a 5 mL tube which constitute the Binding Buffer.
- HAdV and MS2 detection reagents
NOTE: Two different nucleic acid extraction reactions may be analyzed simultaneously in each PCR assay if an 8-well thermocycler is used. For all PCR assays, prepare molecular biology water aliquoted into 1.5 mL tubes.- HAdV detection
- Prepare a mix containing 10 µL of 22.5 µM AdF forward primer, 10 µL of 22.5 µM AdR reverse primer, and 5 µL of 11.25 µM AdP probe (Table 2).
- Add 2.5 µL of the mix to tubes 1 to 8 of a tube strip. Let it dry at room temperature, close the tubes, and keep them protected from the light.
- Cut the strip dividing it into two strips: tubes 1-5 and tubes 6-8.
- Prepare a serial dilution of DNA suspensions containing the nucleotide sequence of the amplified HAdV region. Air-dry 1 µL of suspensions containing 1 x 105, 1 x 107, and 1 x 109 GC/mL into tubes 6-8.
NOTE: Keep the tubes containing the primers, probe, and the air-dried suspensions protected from light.
- MS2 detection
- Prepare a mix containing 10 µL of 25 µM pecson-2F forward primer, 10 µL of 25 µM pecson-2R reverse primer, and 2.5 µL of 25 µM PecP-2 probe (Table 2).
- Add 2.25 µL of the mix to tubes 1 to 8 of a tube strip. Let it dry at room temperature, close the tubes, and keep them protected from the light.
- Cut the strip dividing it into two strips: tubes 1-5 and tubes 6-8.
- Proceed as in step 1.4.1.4 but use a standard suspension designed for MS2 instead.
- HAdV detection
2. Viral concentration
NOTE: Use gloves at all times during the sample collection, reagent preparation, flocculation, flocs collection, and waste disposal procedures. Wear protective glasses during the reagent preparation procedure.
- Sample collection
- Collect a 10 L water sample in a flat bottom bucket with a lid. Collect a minimum of two replicates for each sample.
NOTE: It is important to use clear buckets to visualize the pellet. If the water sample contains suspended material (e.g., sand, algae), let it sediment and, then, collect the water supernatant in a new bucket. - Register the collected volume, as well as the location and collection date, for each water sample.
- Put a magnetic stirrer connected to a battery under a bucket support and place the bucket containing the water sample on its support.
NOTE: Look for a flat surface in a fresh place and keep the buckets containing the water samples from direct sunlight.
- Collect a 10 L water sample in a flat bottom bucket with a lid. Collect a minimum of two replicates for each sample.
- Reagents preparation
- pH adjustment reagents
- Pour the citric acid powder and the sodium hydroxide pellets into the pots containing 25 and 50 mL of distilled water, respectively.
- Gently shake by hand until the citric acid and the sodium hydroxide are completely dissolved.
CAUTION: Do not pour the water over the sodium hydroxide pellets. Instead, pour the sodium hydroxide pellets over the water.
- Preflocculated skimmed milk
- Place a stand-up bag on a magnetic stirrer and pour 500 mL of distilled water in it. Drop a stirring magnet inside the bag and turn on the magnetic stirrer.
- Pour the contents of a sea salt sachet and of a skimmed milk tube into the stand-up bag and stir for 5 min at medium speed to dissolve them.
- Add 3 mL of citric acid to the PSM solution using a Pasteur pipette to make sure the pH lies between 3.4 and 3.6. Measure the pH of the PSM solution using pH indicator strips. Add smaller amounts of citric acid and sodium hydroxide as the pH gets closer to 3.5.
- Turn off the magnetic stirrer and make sure the flocs are visible. Use a flashlight to help to visualize the flocs.
- pH adjustment reagents
- Flocculation
- Drop a stirring magnet into the water, set the magnetic stirrer at maximum speed, and turn it on. Make sure the stirring magnet starts spinning.
- Add 10 mL of distilled water to a process control tube. Invert the tube a few times to rehydrate the viral stock and pour the solution into the water.
- Pour the contents of one sample conditioning sachet into the water and stir for 5 min.
- Measure the pH of the water sample using pH indicator strips. Add a few drops of citric acid or sodium hydroxide to the water sample to make sure the pH lies between 3.4 and 3.6. Add smaller amounts of citric acid and sodium hydroxide as the pH gets closer to 3.5.
- Add 100 mL of the PSM solution using a 100 mL container.
- Seal the bucket with the lid, set the magnetic stirrer at minimum speed, and leave the water stirring for 8 h.
- Turn off the magnetic stirrer and let the water still for at least 5 h to allow the flocs to settle.
- Floc collection
- Build a siphoning system composed of a pipette controller, two pipettes, and a plastic tube, to aspirate the water supernatant for discharging the water on the flocs.
- Attach a 10 mL tape-end pipette to the pipette controller. Then, attach the tip of the pipette to the plastic tube.
- Remove the filter of a 10 mL open-tip pipette using tweezers and attach the pipette to the plastic tube.
- Place an empty bucket on the floor.
- Immerse the tip of the open-tip pipette in the water. Make sure to keep it close to the water surface to avoid disturbing the flocs. Aspirate the water supernatant until it reaches the tape-end pipette.
- Pinch the plastic tube by the end attached to the tape-end pipette and detach it from the pipette.
- Place the tube in an empty bucket. Release the pressure on the tube to let the water flow into the empty bucket.
- Pinch the plastic tube to stop the water flow when the water level is about to reach the stirring magnet and move the pipette away from the bucket.
- Shake the bucket to resuspend the flocs and pour them into a 500 mL stand-up plastic bag. Allow the flocs to settle for 1 h more.
- Carefully aspirate the water supernatant using a 50 mL pipette and a manual pipette controller, avoiding disturbing the flocs.
- Aspirate the flocs using a 100 mL pipette and transfer them to a 50 mL graduated centrifuge tube. Note down the final volume of sample concentrate and transfer 1 mL of it to a 5 mL tube containing the preservative solution, using a Pasteur pipette.
NOTE: It is important that the centrifuge tube is graduated so the final volume of the concentrated sample can be measured as accurately as possible and registered. - Perform the nucleic acid extraction in the field. Alternatively, store the preservative solution containing the viruses at 20-30 °C for up to 15 days or ship them to a reference laboratory for further analysis.
- Build a siphoning system composed of a pipette controller, two pipettes, and a plastic tube, to aspirate the water supernatant for discharging the water on the flocs.
- Waste disposal and material reuse
- Add the contents of a neutralizing agent sachet to the bucket containing the discarded water. Mix the water, and then, leave it still for 30 min.
- Dispose of the treated water as general waste.
- Place the pipettes and the plastic tube inside the bucket. Fill the bucket with water and pour in the contents of one neutralizing agent sachet.
- Wash them for at least 30 min to sterilize them and rinse them with abundant clean water to remove any remaining neutralizing agent.
3. Nucleic acid extraction
NOTE: Use gloves and always wear protective glasses during the nucleic acid extraction procedure.
- Magnetic pipette operation
- Get familiarized with the operation of the magnetic pipette before performing the extraction.
- Nucleic acid extraction
- Arrange the tubes containing the reagents required for the extraction in a rack, from left to right: binding buffer, washing buffers, and elution buffer. Set the appropriate number of tubes according to the number of samples or replicates being analyzed.
- Transfer 1 mL of sample concentrate to the tube containing the binding buffer, using a disposable Pasteur pipette. Invert the tube 8x to 10x to homogenize the solution.
- Incubate the solution at room temperature for 10 min while continuously mixing, either using a solar-powered orbital or manually.
- Collect the magnetic particles using the magnetic pipette and a clean tip, which can be reused for the three washing buffers.
- Release the magnetic particles into the tube containing 500 µL of washing buffer 1. Wash them by gently shaking the solution with the tip for 30 s and collect them.
- Transfer the magnetic particles to the tube containing 600 µL of washing buffer 2. Wash them by gently shaking the solution with the tip for 30 s and collect them.
- Transfer the magnetic particles to the tube containing 200 µL of washing buffer 3. Wash them by gently shaking the solution with the tip for 30 s and collect them.
- Release the magnetic particles into the tube containing the elution buffer and discard the tip.
- Incubate the solution at room temperature for 5 min while continuously mixing, using the solar-powered orbital.
NOTE: Mixing the magnetic particles in the elution buffer is a crucial step. In case of lacking an orbital, continuously mix by hand during the incubation time. - Replace the tip on the magnetic pipette by a new one and collect the magnetic particles from the elution buffer. Make sure to completely remove the magnetic particles from the elution buffer since they can interfere with molecular detection methods.
- Discard the tip with the particles attached to it. Proceed with the PCR analysis, ship the elution buffer to a reference laboratory, or store the elution buffer in at 4 ˚C for further analysis.
4. Viral detection with a battery-operated eight-tube real-time thermocycler
NOTE: Use gloves and always wear protective glasses during the nucleic acid detection procedure. Replace the gloves for new ones when performing a new PCR experiment to avoid cross-contamination.
- HAdV quantitative PCR
- Add 14 µL of nuclease-free water to tubes 2, 4, and 5.
- Add 4 µL of PCR mix to tubes 1-5.
- Add 14 µL of nucleic acid extraction from sample A to tube 1 and 14 µL from sample B to tube 3.
- Add 2 µL of extracted nucleic acids from sample A to tube 2 and 2 µL from sample B to tube 4. Close the five-tube strip.
- Add 14 µL of nuclease-free water to tubes 6, 7, and 8.
- Add 4 µL of this mix to tubes 6, 7, and 8, and close them.
- Put both strips in the thermocycler and select the appropriate run (Table 3).
- Discard the water tube and keep the extracted nucleic acid in the freezer, if possible, or, if not, in a fresh place protected from light in case a second test needs to be performed.
- Clean the micropipettes, the rack, the working material, and all material properly with a nucleic acid cleaner, and discard the plastic bag containing the used tip, gloves, and tubes.
CAUTION: Nucleic acid remover is a very flammable liquid and steam. Do not breathe in the spray mist and avoid any contact of the liquid with the eyes or the skin. Otherwise, immediately rinse with abundant water and seek medical advice. - Collect the results and analyze the obtained data.
- MS2 quantitative PCR
- Add 14 µL of nuclease-free water to tubes 2, 4, and 5.
- Add 4 µL of PCR mix and 0.5 µL of reverse transcriptase enzyme to tubes 1-5.
- Add 14 µL of nucleic acid extraction from sample A to tube 1 and 14 µL from sample B to tube 3.
- Add 1 µL of extracted nucleic acids from sample A to tube 2 and 1 µL from sample B to tube 4. Close the five-tube strip.
- Add 15.5 µL of nuclease-free water to tubes 5, 6, 7, and 8, and close them.
- Add 4 µL of PCR mix and 0.5 µL of reverse transcriptase enzyme to tubes 6, 7, and 8.
- Put both strips in the thermocycler and select the appropriate run (Table 3).
- Proceed as described in steps 4.1.8 to 4.1.10.
Representative Results
Method development
This procedure has been developed in the Laboratory of Viruses Contaminants of Water and Food with the collaboration of GenIUL and Oxfam Intermón. It comprises of three different steps. The first one, the viral particle concentration, is an adaptation of a skimmed milk flocculation method previously described12,17,18. The original method was modified as to be independent of a power supply, simpler, and without centrifugation steps.
The recovery of the VirWaTest concentration method was tested in HAdV and MS2 bacteriophage-spiked groundwater samples. The viral recovery of the VirWaTest concentration and extraction method was estimated to be 3.01% to 18.02% for MS2 and 17.52% to 44.22% for HAdV. These recoveries were calculated from the values obtained by quantitative PCR during the development of the method compared to the initial concentrations of HAdV and MS2 in the water samples after spiking them with known concentrations of these viral stocks.
The VirWaTest magnetic nucleic acid extraction was compared to a commercial RNA mini kit (e.g., QIAamp Viral RNA Mini Kit), a column-based extraction method used in the laboratory, by testing 33 river and groundwater samples spiked with HAdV and MS2 and concentrated by skimmed milk flocculation. The comparison results showed that the VirWaTest method recovery was higher in 23/33 cases for HAdV, as well as for MS2 (Table 4). A Wilcoxon test showed p-values of 0.0005569 for HAdV and 0.02791 for MS2. VirWaTest nucleic acid extraction provides significantly higher viral recoveries than the commercial one.
Detection of HAdV in environmental water samples concentrated by the VirWaTest method
To test the developed method for viral concentration in the field, the Oxfam Water, Sanitation and Hygiene (WASH) team, located in Banghi (RCA) in March 2017, collected and concentrated viruses from five well water samples.
Also, in the area of Pedernales (Ecuador), which was affected by earthquakes in 2016 and 2017, six well water samples were collected by an Oxfam WASH team in February 2017, and its viruses were concentrated by the VirWaTest concentration method. Viral concentrates from both settings were sent to the laboratory in Barcelona for VirWaTest nucleic acid extraction and viral quantification.
In Ecuador, naturally occurring HAdV were detected in six out of the six samples analyzed, with concentration values ranging from 3.27 x 101 to 1.80 x 102 GC/L, whereas one out of the five samples collected and concentrated in Banghi (RCA) tested positive for HAdV, at a concentration of 3.46 x 102 GC/L (Table 5).
MS2, added to all tested samples as internal process control, was detected in all samples tested, showing that the method was correctly performed from concentration to detection.
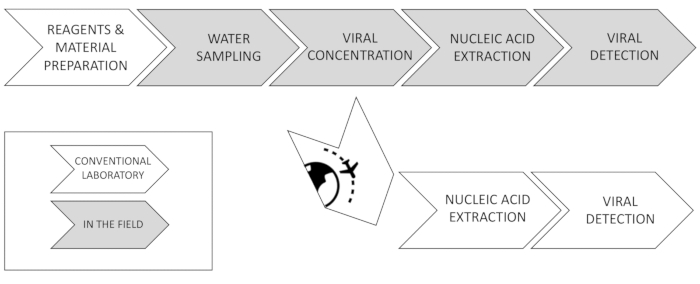
Figure 1: VirWaTest method. Overview of the steps the VirWaTest method is composed of. Please click here to view a larger version of this figure.
| Materials | Concentration | Nucleic Acid Extraction | Detection |
| Equipment | Magnetic Stirrer (2 Units) Battery (2 Units) Battery/Stirrer Connectors (2 Units) Power Adapters (2 Units) Bucket Support (2 Units) Rope (1 Unit) Stirring Magnet (3 Units) Tweezers (1 Unit) Silicon Tubing (1 Unit) Pipette Controller (1 Unit) Marker (1 Unit) |
Magnetic Pipette (1 Unit) Solar Rotary Platform (1 Unit) Multi-Size Tube Rack (1 Unit) Timer (1 Unit) |
Thermocycler (1 Unit) Battery (1 Unit) Computer Tube Rack (1 Unit) 0.5 μL – 10 μL Micropipette (1 Unit) 2 μL – 20 μL Micropipette (1 Unit) |
| Consumables and Reagents (for every 2 water samples/replicates) | Bucket (3 Units) pH Indicator Strips Tape-End 10 mL Pipette (2 Units) Open-Tip 10 mL Pipette (2 Units) Tape-End 50 mL Pipette (2 Units) Tape-End 100 mL Pipette (2 Units) Pasteur Pipette (4 Units) Stand-Up Plastic Bag (4 Units) Plastic Container with 25 mL of distilled water (1 Unit) Plastic Container with 50 mL of distilled water (1 Unit) 100 mL Empty Container (1 Unit) Gloves (6 Pairs) Process Control (2 Tubes) Tube with 10 mL of distilled water (2 Units) Citric Acid (1 Tube) Sodium Hydroxide (1 Tube) Skimmed Milk (1 Tube) Citric Acid Sachet (1 Unit) Distilled Water (1 500 mL Bottle) Sample Conditioning (2 Sachets) Preservative Solution (2 Tubes) Neutralizing Agent (4 Sachets) |
Magnetic Pipette Tips (2 Units) Pasteur Pipette (2 Units) Binding Buffer (2 Tubes) Washing Buffer 1 (2 Tubes) Washing Buffer 2 (2 Tubes) Washing Buffer 3 (2 Tubes) Elution Buffer (2 Tubes) Gloves (6 Pairs) |
10 μL Micropipette Tips (1 box) 20 μL Micropipette Tips (1 box) DNA qPCR Mix (1 Tube) RNA qPCR Mix (1 Tube) Reverse Transcriptase Enzyme (1 Tube) Molecular Biology Water (1 Tube) HAdV Tube Strip with primers, probe and standard (1 Unit) MS2 Tube Strip with primers, probe and standard (1 Unit) Nucleic Acid Remover Gloves (6 Pairs) |
Table 1: VirWaTest contents. Equipment, consumables, and reagents that have to be prepared for the concentration, extraction, and detection of viruses in two water samples/replicates.
| Virus | Primers | Genbank Accession No. | Position | Sequence (5’‒3’) | Length | Reference | ||
| Human Adenovirus (HAdV) | AdF | J01917.1 | 18869‒18887 | CWTACATGCACATCKCSGG | 19 | Hernroth et al., 200215 | ||
| AdR | 18919‒18937 | CRCGGGCRAAYTGCACCAG | 19 | |||||
| AdP1 | 18890‒18916 | 6-FAM-CCGGGCTCAGGTACTCCGAGGCGTCCT-BMN-Q535 | 27 | |||||
| MS2 Bacteriophage (MS2) | pecson-2F | NC_001417.2 | 344‒363 | AAGGTGCCTACAAGCGAAGT | 20 | Pecson et al., 200916 | ||
| pecson-2R | 659‒678 | TTCGTTTAGGGCAAGGTAGC | 20 | |||||
| PecP-2 | 369‒388 | 6-FAM-ATCGTGGGGTCGCCCGTACG-BHQ-1 | 20 | |||||
Table 2: Oligonucleotides for quantitative PCR experiments. Primers and probe designed to bind to nucleic acid sequences of HAdV and MS2.
| Temperature | Time | Cycles | Temperature | Time | Cycles |
| 95 °C | 12 min | 1 | 55 °C | 15 min | 1 |
| 95 °C | 10 min | 1 | |||
| 95 °C | 15 s | 40 | 95 °C | 15 s | 40 |
| 60 °C | 1 min | 60 °C | 1 min |
Table 3: Thermal conditions for quantitative PCR experiments. Temperature, time, and cycles used for HAdV and MS2 amplification.
| QIAgen | VirWaTest | QIAgen | VirWaTest | ||
| Median | 2.53 x 103 | 2.43 x 103 | 4.74 x 104 | 5.13 x 104 | |
| Mean | 1.74 x 104 | 3.12 x 104 | 4.24 x 104 | 5.97 x 104 | |
| SD | 5.66 x 105 | 2.23 x 106 | 4.49 x 104 | 1.63 x 105 | |
| p-value (Wilcoxon) for HAdV: 0.0005569 | |||||
| p-value (Wilcoxon) for MS2: 0.02791 | |||||
| Virus tested | Samples Tested | QIAgen | VirWa Test | ||
| HAdv | 33 | 10 | 23 | ||
| MS2 | 33 | 10 | 23 | ||
Table 4: VirWaTest nucleic acid extraction development. Median, mean, and SD values obtained when comparing the Qiagen and VirWaTest extraction methods in 33 groundwater samples for HAdV and MS2.
| Country | Site | Sample | HAdV GC/L |
| RCA | Eau de la SODECA | 1 | ND |
| Ile Bongossoua, Village 1 | 2 | ND | |
| Ile Bongossoua, Village 3 | 3 | ND | |
| Bangui, Puits Quartier Fondo | 4 | ND | |
| Bangui, Puits Quartier Yambassa | 5 | 3.64 x 102 | |
| Ecuador | Borehole A | 6 | 7.96 x 101 |
| Borehole B | 7 | 1.80 x 102 | |
| Borehole C | 8 | 8.88 x 101 | |
| Well water A | 9 | 6.06 x 101 | |
| Well water B | 10 | 1.12 x 102 | |
| Well water C | 11 | 3.27 x 101 |
Table 5: Quantification of HAdV using the VirWaTest Method. Quantitative PCR results, expressed in GC/liter, for HAdV quantified in water samples collected and concentrated from RCA and Ecuador.
Discussion
The VirWaTest method enables the concentration of viruses and nucleic acid extraction from water samples at the point of use by non-experienced users. It is an affordable, rapid, and simple protocol. The concentration is based on the principle of organic flocculation using skimmed milk, by which the low pH and high conductivity conditions make skimmed milk proteins aggregate into flocs the viruses adsorb to. When the flocs sediment, it is easy to collect them, making it possible to concentrate 10 L of water, whereas traditional ultracentrifugation cannot deal with big water volumes.
The method has been modified for being practicable at the point of sampling without using any electric equipment except for a battery-operated magnetic stirrer. Some other approaches based on water filtration have been adapted to the concentration of viruses and can also be performed at the point of use. However, suspended material present in water samples often clogs the filters; thus, the small volume that may be concentrated with these systems is a serious limitation.
This is the first description of a method for concentrating viruses from water samples at the point of use, regardless of the turbidity of the sample. The VirWaTest concentration method allows several samples to be processed simultaneously if the appropriate material is available. Moreover, skimmed milk flocculation has been shown to be useful for bacteria and parasite concentration19.
The preparation of the appropriate material is crucial for performing the procedure properly. Consider the number of water samples to be analysed and prepare the reagents and material before moving to the place where the test is needed for the preparation of the reagents and material. Several samples can be processed at the same time if the appropriate amount of material is available.
Before starting the concentration of a water sample, a known concentration of a viral stock will be used to spike the sample as process control. This method uses a dried viral stock which is rehydrated with distilled water before being added to the water sample at the point of use. This is useful to rule out false-negative results at the end of the procedure and gives an indication of the performance of the method.
Viral concentrates obtained with the VirWaTest may be further tested in the field applying the VirWaTest nucleic acid extraction and detection methods or, alternatively, concentrates may be sent to a reference laboratory at room temperature. The preservative solution added to the concentrate enables viruses to remain stable for up to 2 weeks (unpublished results).
VirWaTest nucleic acid extraction is a magnetic particle-based method. It is easy and fast and allows several samples to be processed at the same time and shows equivalent and even better recovery efficiency than methods currently used for viral nucleic acid extractions. The nucleic acids may be sent to reference labs at room temperature or, if users are confident in performing molecular detection, a quantitative PCR assay can be performed at the point of use of the original water sample.
Also, if a small laboratory facility is available, the VirWaTest concentration protocol may be coupled to standard nucleic acid extraction kits that depend on a centrifuge and standard PCR-based methods that require a freezer to maintain the reagents but that use standard thermocyclers, which are less expensive than battery-operated ones.
However, since a power supply is sometimes not available, we have optimized detection assays for MS2 bacteriophages as process control and HAdV as a viral fecal indicator, and the methodology can be customized for any other virus of interest, such as hepatitis viruses, RoV, NoV, or others, to be run in the field without needing a freezer for maintenance of the reagents, nor conventional thermocyclers but only a battery-operated one. Several battery-operated thermocyclers are commercially available. Alternatively, if a power supply is available, other conventional qPCR equipment may be used. If an eight-tube thermocycler is used, up to two nucleic acid extractions (two different samples or two replicates from the same sample) can be tested in the same PCR assay. For each nucleic acid extraction, two different quantities will be tested to rule out enzymatic inhibition originated by the sample. The adaptation is based on the previous preparation of PCR tubes by air-drying primers, probes, and standard suspensions. Several lyophilized commercial qPCR solutions exist that could be used by applying the same procedure as described here. We have described one possibility that we considered to be easy to perform.
Comparison assays performed during the development of the method showed that the VirWaTest methods of concentration, extraction, and detection are efficient for the quantification of viruses in water samples.
The limit of detection (LOD) of the described procedure is variable since the volume collected after concentration is variable. Also, the LOD can be slightly different for different viruses. If a volume of approximately 10 L is collected, the LOD for HAdV would be around 1 x 102 viral GC/L. So, relatively small concentrations of HAdV can be detected by the VirWaTest method. In Pedernales (Ecuador), six out of the six samples tested presented HAdV, in concentrations close to the LOD, ranging from 3.27 x 101 to 1.80 x 102 GC/L. In Banghi (RCA), HAdV were detected in one out of the five samples analyzed, at a concentration of 3.46 x 102 GC/L. The presence of HAdV, as well as the presence of MS2 as the control process in the tested samples, shows the method is useful for the recovery of viral particles from water samples, even when performed by nonexperienced users.
Feedback obtained until now indicates that viral concentration is an easy procedure to perform, with no major problems when applied at the point of use of the samples, although 8 h and 5 h steps are required. It is important to note that these steps occur without any human assistance. Moreover, a faster method based on filtration is being developed as an alternative for nonturbid waters. However, flocculation seems to be, until now, the unique method that allows the concentration of samples presenting high turbidity. Viral concentrates may be then sent to any laboratory around the world at room temperature, which also makes it easier to test viruses since sampling areas do not always have good transportation services providing cooling conditions. Alternatively, the extraction and detection protocols presented here allow testing for viral detection at the point of use of the samples.
As far as we know, this is the first method reported to be useful for the concentration and testing for the presence of viruses in water samples in the field. Further efforts should be conducted to apply the procedure to the evaluation of the presence of human viral pathogens of interest in several other humanitarian crisis scenarios. Also, the user's feedback will be necessary to provide insights into the potential implementation to make the procedure friendlier.
Disclosures
The authors have nothing to disclose.
Acknowledgements
VirWaTest was a research project funded by the HIF (Humanitarian Innovation Funds) program of ELHRA (Enhancing Learning & Research for Humanitarian Assistance). The authors acknowledge the WASH teams who kindly collaborated in this study. The analysis of samples in Ecuador was funded by Oxfam Ecuador and Dirección de Investigaciones de la Universidad de las Americas (AMB.BRT.17.01). S. Bofill-Mas is a Serra-Hunter fellow at the University of Barcelona.
Materials
| 5x HOT FIREPol Probe qPCR Mix Plus (ROX) | Solis BioDyne | 08-14-00001 | Includes Solis Biodyne's 5x HOT FIREPol Probe qPCR Mix Plus (qPCR Mix), 50 Reactions |
| 8-Microtube Strips with Caps | dD Biolab | 840637 | Low Profile, Thin Walls, Adapted for Quantitative and Qualitative PCR |
| Aquagenx CBT E. coli Kit | Aquagenx, LLC | ECCBT10 | 10 Tests per Kit |
| Batteries and Power Adapters for Magnetic Stirrer | GenIUL | 900011674 | Includes 12V car power adapter |
| Bucket Support | GenIUL | 900011648 | Aluminium support |
| Bucket, 10 L | Cater4You | 10LTR | Polypropilene, Tamperproof, Clear color |
| Centrifuge Tube, 50 mL | LabBox | CTSP-E50-050 | Polypropylene, Sterile, Graduated, With Skirt |
| Citric Acid 1-Hydrate, 500 g | PanReac AppliChem | 1410181211 | Pure, Pharma Grade, 1 Kilogram |
| Clear PET Bottle | LabBox | FPET-500-088 | Clear Color, PET, Cap Not Included |
| Difco Skim Milk, 500 g | Becton Dickinson | 232100 | Dehydrated |
| DNA/RNA Shield, 250 mL | Zymo Research | R1100-250 | DNA/RNA Preservation Medium, 250 mL |
| Easy9 Pipette Controller | LabBox | EAS9-001-001 | 0.3 μm filter, Pipettes from 0.1 to 100 mL, Autoclavable silicone pipette holder |
| Eppendorf Tube, 0.5 mL | Eppendorf | 0030121023 | Polypropilene, Safe-Lock |
| Eppendorf Tube, 2 mL | Eppendorf | 0030120094 | Polypropilene, Safe-Lock |
| Eppendorf Tube, 5 mL | dD Biolab | 999542 | Polypropylene, Sterile, Graduated |
| Ethanol 96% V/V, 1 L | Panreac AppliChem | 361085-1611 | For UV, IR and HPLC |
| Laboratory Tweezers | LabBox | FORS-007-002 | Thin, Curved End, L= 120 mm |
| Magnetic Stirrer | GenIUL | 900017000 | Battery-powered |
| Marker | dD Biolab | 929203 | Black, Extra fine Tip, Water Resistant, Fast Drying, For Plastic and Glassware |
| Micro Rota-Rack for Microtubes | dD Biolab | 37782 | 4 Modules, L x W x H= 208 x 100 x 100 mm |
| Mini8 Real-Time PCR Cycler | Coyote Biosciences, China | Mini-8 | Portable, Works with 12V Power Supplies or External Batteries, Two channels, Capacity for 8 Tubes |
| NucliSens Lysis Buffer | Biomerieux | 200292 | Reagents for up to 48 Isolations, Store at Ambient Temperature |
| Open Tip Serological Pipette, 10 mL | Deltalab | 900136N | Sterile, Individually Wrapped (Paper/Plastic) |
| PE Screw Cap PP28 | LabBox | TP28-004-020 | For PET Bottles |
| pH Indicator Strip | LabBox | WSPH-001-001 | Range pH 2.8 to pH 4.4, 50 Strips per Pack |
| Plastic Test Tube | Quimikals | 300913 | Includes Cap |
| Polyethylene Pasteur Pipette | LabBox | PIPP-003-500 | Graduated, 7 mL Overall Volume, Non-Sterile |
| Polypropylene Screw Flask With Screw Cap, 150 mL | Deltalab | 409726 | Screw cap, Sterile, graduated up to 100 mL |
| Polypropylene Screw Flask With Screw Cap, 60 mL | Deltalab | 409526G | Screw cap, Sterile, Graduated up to 50 mL |
| Powder Powder Detergent | – | – | Regular Powder Soap for washing clothes |
| Power Cables for Magnetic Stirrer | GenIUL | 900011692 | Connection between batteries and magnetic stirrers |
| QuickPick Magnetic Tool | BioNobile | 24001 | Hand-held tool for magnetic particles |
| QuickPick Tips in Box | BioNobile | 24296 | RNase-Free, Autoclaved, 96 Units |
| QuickPick XL gDNA Magnetic Particles | BioNobile | SN51100 | 3.2 mL |
| Sea Salts | Sigma-Aldrich | S9883-500G | An artificial salt mixture closely resembling the composition of the dissolved salts of ocean water |
| Silicone Tubing | LabBox | SILT-006-005 | Roll of 5 Meters, Inner ø x Outer ø= 6 x 10 mm |
| Sodium Hydroxide Pellets, 98.5 – 100.5% | VWR Chemicals | 28244295 | Pellets, 1 Kg |
| Solar Rotary Platform | SOL-EXPERT Group | 70020 | Acrylic Plate, 10 RPM, Supports up to 300 Grams |
| SOLIScript 1-step Probe Kit | Solis BioDyne | 08-57-00250 | Includes Solis Biodyne's 5x One-Step Probe Mix (qPCR Mix) and 40x One-Step SOLIScript Mix (Reverse Transcriptase Enzyme), 250 Reactions |
| SPEEDTOOLS RNA Virus Extraction Kit | BioTools | 21.141-4197 | Includes BioTools's BAW Buffer (Washing Buffer 1), BAV3 Buffer (Washing Buffer 2 and 3) and BRE Buffer (Elution Buffer). |
| SpinBar Octhaedral Stirring Magnet | dD Biolab | 045926 | Pivot Ring, L x ø = 38 x 8 mm, Blue |
| Tape-End Serological Pipette, 10 mL | Deltalab | PN10E1 | Sterile, Individually Wrapped (Paper/Plastic) |
| Tape-End Serological Pipette, 50 mL | Deltalab | 900043 | Sterile, Individually Wrapped (Paper/Plastic) |
| Termi-DNA-Tor – Nucleic Acid Remover | BioTools | 22001-4291 | Remover of nucleic acids, bacteria, fungi and mycoplasma from material and surfaces, 450 mL |
| Water Molecular Biology Reagent, 1L | Sigma-Aldrich | W4502-1L | Nuclease and Protease Free, 0.1 μm Filtered |
| Whirl-Pak Bag, 540 mL | Deltalab | 200361 | Stable bottom |
| Zip Lock Plain Bag | LabBox | BZIP-080-100 | Polyethylene, L x W= 120 x 80 mm |
References
- The Sphere Project. . The Sphere Project: Humanitarian Charter and Minimum Standards in Humanitarian Response. , (2011).
- Bartram, J., et al. . Water Safety Plan Manual:Step-by-step risk management for drinking-water suppliers. , (2009).
- World Health Organization. . Guidelines for Drinking-water Quality. , (2011).
- World Health Organization. . 25 Years Progress on Sanitation and Drinking Water. , (2015).
- Girones, R., et al. Molecular detection of pathogens in water – The pros and cons of molecular techniques. Water Research. 44 (15), 4325-4339 (2010).
- Rodriguez-Manzano, J., et al. Standard and new fecal indicators and pathogens in sewage treatment plants, microbiological parameters for improving the control of reclaimed water. Water Science and Technology. 66 (12), 2517-2523 (2012).
- Puig, M., et al. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Applied and Environmental Microbiology. 60 (8), 2963-2970 (1994).
- Carter, M. J. Enterically infecting viruses: Pathogenicity, transmission and significance for food and waterborne infection. Journal of Applied Microbiology. 98 (6), 1354-1380 (2005).
- Bofill-Mas, S., Pina, S., Girones, R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Applied and Environmental Microbiology. 66 (1), 238-245 (2000).
- Bofill-Mas, S., et al. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Applied and Environmental Microbiology. 72 (12), 7894-7896 (2006).
- Rames, E., Roiko, A., Stratton, H., Macdonald, J. Technical aspects of using human adenovirus as a viral water quality indicator. Water Research. 96, 308-326 (2016).
- Calgua, B., et al. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. Journal of Virological Methods. 153 (2), 79-83 (2008).
- Bofill-Mas, S., et al. Cost-effective method for microbial source tracking using specific human and animal viruses. Journal of Visualized Experiments. 58, 5-9 (2011).
- International Organization for Standardization. . ISO 10705-1:1995: Water quality – Detection and enumeration of bacteriophages – Part 1: Enumeration of F-specific RNA bacteriophages. , (1995).
- Hernroth, B. E., Conden-Hansson, A. C., Rehnstam-Holm, A. S., Girones, R., Allard, A. K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Applied and Environmental Microbiology. 68 (9), 4523-4533 (2002).
- Pecson, B. M., Martin, L. V., Kohn, T. Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: Advantages and limitations of an enzymatic treatment to reduce false-positive results. Applied and Environmental Microbiology. 75 (17), 5544-5554 (2009).
- Calgua, B., Barardi, C. R. M., Bofill-Mas, S., Rodriguez-Manzano, J., Girones, R. Detection and quantitation of infectious human adenoviruses and JC polyomaviruses in water by immunofluorescence assay. Journal of Virological Methods. 171 (1), 1-7 (2011).
- Bofill-Mas, S., et al. Cost-effective method for microbial source tracking using specific human and animal viruses. Journal of Visualized Experiments. (58), e2820 (2011).
- Gonzales-Gustavson, E., et al. Characterization of the efficiency and uncertainty of skimmed milk flocculation for the simultaneous concentration and quantification of water-borne viruses, bacteria and protozoa. Journal of Microbiological Methods. 134, 46-53 (2017).

