An Advanced Murine Model for Nonalcoholic Steatohepatitis in Association with Type 2 Diabetes
Summary
A simple and reliable diet-induced rodent animal model for nonalcoholic steatohepatitis (NASH) is described, achieved through non-SPF housing of the animals and administration of a specific high-fat diet. We describe identification of hepatic and adipose immune cell subsets to recapitulate human immunological conditions by exposing mice to environmental germs.
Abstract
Obesity is associated with chronic low-grade inflammation and insulin resistance, contributing to an increasing prevalence of chronic metabolic diseases, such as type 2 diabetes and nonalcoholic steatohepatitis (NASH). Recent research has established that pro-inflammatory immune cells infiltrate obese hypertrophic adipose tissue and liver. Given the emerging importance of immune cells in the context of metabolic homeostasis, there is a critical need to quantify and characterize their modification during the development of type 2 diabetes and NASH. However, animal models that induce pathophysiological features typical of human NASH are sparse.
In this article, we provide a detailed protocol to identify immune cell subsets isolated from liver and adipose tissue in a reliable mouse model of NASH, established by housing high-fat diet (HFD) mice under non-specific pathogen-free (SPF) conditions without a barrier for at least seven weeks. We demonstrate the handling of mice in non-SPF conditions, digestion of the tissues and identification of macrophages, natural killer (NK) cells, dendritic cells, B and T cell subsets by flow cytometry. Representative flow cytometry plots from SPF HFD mice and non-SPF mice are provided. To obtain reliable and interpretable data, the use of antibodies, accurate and precise methods for tissue digestion and proper gating in flow cytometry experiments are critical elements.
The intervention to restore physiological antigen exposure in mice by housing them in non-SPF conditions and unspecific exposure to microbial antigens could provide a relevant tool for investigating the link between immunological alterations, diet-induced obesity and related long term complications.
Introduction
Obesity is a multifactorial disorder and a major risk factor for developing heart disease, stroke, nonalcoholic steatohepatitis (NASH), type 2 diabetes (T2D) and some types of cancer. The prevalence of obesity is rapidly increasing globally. Today, 2.1 billion people — nearly 30% of the world’s population — are either obese or overweight1. Obesity-associated insulin resistance can lead to T2D, when exhausted pancreatic islet beta cells fail to compensate for the increased need for insulin to maintain glucose homeostasis2.
Adipose tissue is composed of various cell types including adipocytes, endothelial cells, fibroblasts and immune cells. During progression of obesity, changes in the number and activity of immune cells can lead to low-grade inflammation of hypertrophic adipose tissue3,4. Specifically, it has been found that excessive energy intake, accompanied by chronically elevated levels of blood glucose, triglycerides and free fatty acids, leads to adipocyte hypoxia, endoplasmic reticulum stress, impaired mitochondrial function and enhanced cytokine secretion, resulting in the activation of pro-inflammatory adipose immune cells5,6. Past research has mainly focused on innate immunity, but more recently adaptive immune cells (T and B cells) have emerged as important regulators of glucose homeostasis. They possess inflammatory (including CD8+ T cells, Th1, and B cells) or primarily regulatory functions (including regulatory T (Treg) cells, Th2 cells) and can both exacerbate or protect against insulin resistance7,8,9.
Furthermore, several mechanisms were proposed to explain how obesity increases steatohepatitis, including increased production of cytokines by adipose tissue10. NASH, the progressive form of nonalcoholic fatty liver disease and a major health burden in developed countries, is histologically characterized by ballooned hepatocytes, lipid accumulation, fibrosis and pericellular inflammation and may progress to cirrhosis, end stage liver disease or hepatocellular carcinoma. Several regimen (for instance the methionine and choline deficient diet11) are known to induce NASH-like liver pathology in non-human animal models, but most of these approaches do not recapitulate human conditions of NASH and its metabolic consequences as they either require specific gene knockout, non-physiological dietary manipulations or lack insulin resistance typical of human NASH. Moreover, our understanding of the underlying mechanisms of metabolic diseases is currently based on experiments carried out with laboratory mice housed under standard specific pathogen free (SPF) conditions. Those barrier facilities are abnormally hygienic and do not consider the microbial diversity humans have to encounter, which may account for difficulties in the translation process of animal studies to clinical approaches12,13,14.
To investigate the different immune cell subsets in adipose tissue and liver during the development of insulin resistance and NASH in an advanced mouse model reproducing human immunological conditions, mice were housed in individual cages in semi sterile conditions without a barrier. Mice housed under antigen exposed conditions developed NASH-like liver pathology already after 15 weeks of high-fat diet (HFD) feeding13. Compared to age-matched SPF mice they developed macrovesicular steatosis, hepatic infiltration and activation of immune cells.
This manuscript describes a robust flow cytometry analysis to define and count immune cell subsets from mouse adipose tissue and liver in a model of NASH. Flow cytometry analysis allows the detection of multiple parameters of individual cells simultaneously in contrast to RT-PCR or immunohistochemistry approaches.
In summary, our study offers a mouse model of short-term HFD for investigating the development of insulin resistance and NASH and the underlying mechanisms that also exhibits fidelity to the human condition.
Protocol
This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the Animal Welfare Act under the supervision of our institutional Animal Care and Use Committee. Animal protocols were conducted according to institutional ethical guidelines of the Charité Berlin, Germany, and were approved by the Landesamt für Gesundheit und Soziales and comply with the ARRIVE guidelines.
1. Diet-induced Animal Model of Steatohepatitis
- Transfer mice (C57Bl/6J, male) to non-SPF housing at the age of 4 weeks and expose them continuously to a broad range of environmental pathogens/antigens. Guarantee the exposure by daily handling of the laboratory animals as antigens are distributed by air passage in this way.
- Maintain mice (C57Bl/6J, male, 12 weeks old) in open caging systems with conventional filters on a 12 h:12 h light/dark cycle at a temperature of 22 °C. Do not irradiate or autoclave experimental diet and bedding. Access animal housing facility without mask or hairnet. Open doors between animal rooms. Do not use an airshower.
- Guarantee daily handling of the laboratory animals and switch rooms on a regular basis so that antigens are distributed by air passage. Complement dirty housing with daily non-specific microbial exposure to antigens from bedding of mammalian laboratory animals housed in rooms next to the laboratory mice.
- Measure proportions of CD44–CD62L– effector memory CD4+ and CD8+ T cells in blood and spleen using flow cytometry (as described in refefence13).
NOTE: In this regard we defined an increase of 20% of effector memory CD8+ T cells from CD8+ T cells as evidence of sufficient microbial exposure. - Start HFD (60 kJ% from fat, 19 kJ% from proteins and 21 kJ% from carbohydrates and ad libitum consumption of water with 6% sucrose content with five-week old C57Bl/6J male mice for 7-15 weeks. The HFD should contain 60% kJ from fat (as described above) in order to induce the development of insulin resistance throughout the course of the experiment.
NOTE: Hematoxylin staining should be performed and histological characteristics as hepatocyte ballooning, Mallory Denk bodies, immune cell infiltration and macrovesicular steatosis must be found to demonstrate NASH-like liver pathology (as shown in reference13).
2. Preparation of Reagents and Solutions
- Prepare 70% ethanol, phosphate buffered saline (PBS, without calcium and magnesium) supplemented with 0.5% BSA and ACK (Ammonium-Chloride-Potassium) lysis buffer.
- Staining buffer
- Dissolve 10 mL of fetal calf serum (FCS) in 500 mL of PBS to obtain 2% FCS PBS. Place staining buffer on ice before use.
- Store solution in a plastic bottle at 4 °C.
- Collagenase solution for adipose tissue digestion
- Dissolve 2.5 g of bovine serum albumin (BSA) in 500 mL of PBS to obtain a 0.5% BSA/PBS.
- Dissolve 74.5 g of CaCl2 in 10 mL of 0.5% BSA/PBS to obtain a 10 mM CaCl2 solution.
- Add 1 mg of collagenase type II (see Table of Materials) to each mL of 0.5% BSA with 10 mM CaCl2 PBS.
- Prepare 3 mL of collagenase digest solution per g of adipose tissue sample. Prepare fresh collagenase solution for each isolation.
- Collagenase solution for liver tissue digestion
- Dissolve 2.5 g of BSA in 500 mL of Hank´s Balanced Salt Solution (HBSS) to obtain 0.5% BSA HBSS.
- Add 10 mL of FCS in 500 mL of 0.5% BSA/HBSS to obtain 2% FCS 0.5% BSA/HBSS.
- Add 0.5 mg of collagenase type IV (see Table of Materials) to each mL of 2% FCS 0.5% BSA/HBSS.
- Add 0.02 mg of DNase per mL of 2% FCS 0.5% BSA/HBSS collagenase solution.
- Prepare 13 mL of collagenase digest solution per liver tissue sample.
- Prepare fresh collagenase solution for each isolation.
3. Generation of Single cell Suspensions
- Adipose tissue digestion
- Euthanize mice via isoflurane anesthesia followed by cervical dislocation. Spray the chest with 70% ethanol. Carefully make a 5-6 cm central incision through the integument and abdominal wall along the entire length of the rib cage to expose the pleural cavity and heart.
NOTE: Do not damage underlying organs and keep scissors tips up. - Inject at least 10 mL of 0.9% saline solution in the apex of the left ventricle using a 26 G needle.
NOTE: Successful perfusion is noted by blanching of the liver. - Open the peritoneal cavity with scissors and cut out the perigonadal fat pads on each side with fine curved scissors. Dissect gonadal tissues with fine curved scissors and weigh adipose tissue.
- Perform the mechanical dissociation using scissors and cut adipose tissue into fine pieces in a Petri dish at 4 °C. Transfer adipose tissue to 50 mL conical centrifuge tubes and rinse Petri dish with 1 mL of 0.5% BSA/PBS.
- Add 3 mL of adipose tissue digest solution (as prepared in step 2.3) per gram of adipose tissue. Incubate adipose tissue solution at 37 °C for 20 min under gentle shaking (200 rpm).
- Add 5 mL of 0.5% BSA/PBS per gram of adipose tissue and place on ice. Triturate the solution numerous times with a 10 mL serological pipette and pass it through a strainer (100 µm) with the aid of a plunger.
- Centrifuge at 500 x g for 10 min at 4 °C.
- Remove the floating adipocyte fraction by pipetting. Resuspend the cell pellet (stromal vascular fraction) in 1 mL of ACK lysis buffer (see Table of Materials). Add 10 mL of 2% FCS PBS.
- Centrifuge at 500 x g for 10 min at 4 °C.
- Decant supernatant and resuspend cell pellet in 250 µL of 2% FCS PBS.
- Count the number of viable cells on a hemocytometer using Trypan blue exclusion.
- Euthanize mice via isoflurane anesthesia followed by cervical dislocation. Spray the chest with 70% ethanol. Carefully make a 5-6 cm central incision through the integument and abdominal wall along the entire length of the rib cage to expose the pleural cavity and heart.
- Liver tissue digestion
- Store liver in a conical centrifuge tube filled with PBS and transport on ice.
- Perform the mechanical dissociation using syringe stamps in a Petri dish at 4 °C. Put dissected liver tissue in a 50 mL conical centrifuge tube containing 10 mL of warm liver digest solution. Rinse the Petri dish with 3 mL of liver digest solution.
- Incubate liver tissue solution at 37 °C for 20 min under gentle shaking (200 rpm).
- Add 20 mL of HBSS. Triturate the solution numerous times with a 10 mL serological pipette and pass it through a strainer (100 µm) with the aid of a plunger.
- Centrifuge at 500 x g for 10 min at 4 °C. Decant the supernatant and resuspend the cell pellet in 20 mL of HBSS.
- Centrifuge at 30 x g for 1 min at room temperature to remove the hepatocyte matrix. Discard the cell pellet.
- Centrifuge the supernatant at 500 x g for 10 min at 4 °C. Resuspend the pellet in 10 mL of 33% low viscosity density gradient medium solution (see Table of Materials) in HBSS followed by centrifugation (800 x g; 30 min; room temperature; no brake).
- Resuspend the pellet in 1 mL of ACK lysis buffer and incubate for 4 min at room temperature. Then add 10 mL of HBSS.
NOTE: To discard the supernatant aspirate the supernatant with interphase (hepatocytes) very carefully with a transfer pipette (as much as possible without taking the pellet with immune cells and erythrocytes). - Pass cells through a 30 μm strainer in a 15 mL conical centrifuge tube and centrifuge at 500 x g for 10 min at 4 °C. Decant supernatant and resuspend cell pellet in 250 µL of 2% FCS PBS.
- Count the number of viable cells on a hemocytometer using Trypan blue exclusion.
4. Surface Staining
- Prepare antibody mix for T-cell-subsets (Panel 1) and innate immune cells (Panel 2) as described in Table 1 and Table 2. Volumes are optimized for one sample (100 µL) regarding to antibody concentrations.
NOTE: Lymphocytes and innate immune cells present differences in autofluorescence and should be analyzed separately. - Use up to 3 x 106 cells in 100 µL in a polystyrene FACS tube for surface staining. To block Fc receptors add 10 µL of anti-CD16/CD32 antibody (diluted 1:100) and incubate for 10 min on ice. Use an unstained negative control sample to adjust side scatter (SSC) and forward scatter (FSC) to determine the location of the negative cell population.
- Vortex and add the appropriate volume of antibody mixture for Panel 1 and Panel 2. Add 1 µL of viability dye to each sample to allow for live and dead cell discrimination. Incubate for 20 min at 4 °C and protected from light.
- Wash two times with 2 mL of 2% FCS PBS and centrifuge at 300 x g for 5 min at 4 °C.
- Resuspend the cell pellet in 300 µL of 2% FCS PBS and store at 4 °C until FACS analysis.
NOTE: Before starting the measurement pass cells through 30 µm cell strainer into polystyrene FACS tube and vortex. Flow cytometry was performed with labeled cells stored in 2% FCS PBS at 4 °C for 1-3 h. Cells should be analyzed as soon as possible for optimized results.
5. Flow Cytometry Compensation, Acquisition, and Gating
- Run unstained negative control sample to set the FSC and SSC and adjust voltages of the flow cytometer to detect leucocyte populations and to distinguish between debris and viable cells. Exclude debris and dead cells.
NOTE: Viability of the cell suspensions from the liver might be lower than the viability from the cell suspensions from perigonadal adipose tissue due to an additional density separation step. - Run single stained control samples for multi-color compensation.
NOTE: Antibody capture beads could also be used to adjust for spectral overlap if the number of cells of a cell population of interest is too low to compensate using cells. To detect possible autofluorescence signals of the cells fluorescence minus one (FMO) controls for every antibody are recommended but were not applied in this protocol. - Start the measurement, collect appropriate number of events (at least 50,000 events) and record experimental data.
- Export FCS data files for analysis and set the gating strategy. Gate on CD45+ leucocytes to identify subsequent cell populations.
Representative Results
The protocol described allows the characterization of surface markers of innate and adaptive immune cells isolated from murine perigonadal adipose tissue and liver in a model of diet-induced NASH. In this model, NASH was induced by administration of HFD plus sucrose (6%) in drinking water for 7 to 15 weeks in C57Bl/6J mice, as previously reported13. Importantly, mice were housed in semi sterile conditions and, thus, exposed to environmental antigens throughout the experiment. HFD fed mice housed in SPF conditions and chow diet mice housed under SPF and non-SPF conditions served as controls. HFD feeding resulted in significant body weight gain already after 7 weeks in both groups. A significant difference in body weight was first evident at week 4 (P < 0.001) and remained significant throughout the following experimental weeks. However, no significant differences in weight gain were shown between SPF and non-SPF mice after 7 weeks13. The stromal vascular fraction and immune cells from perigonadal adipose tissue and livers of male mice fed a HFD for 7 weeks were isolated by collagenase digestion. Immune cells were labeled with fluorophore-conjugated primary antibodies and proportions of T cells, B cells, macrophages, NK cells, dendritic cells and granulocytes were quantified via flow cytometry analysis. Gating for T cell subpopulations, including doublet discrimination, and viability staining, in murine perigonadal fat is illustrated in Figure 1. CD45+ leucocytes are first gated for CD4 and CD8 and subsequently gated for CD44 and CD62L to discriminate between naïve, central memory, effector memory and effector T cells. CD44+ cells were then further characterized with CD127, KLRG1 and PD-1. Figure 2 provides the gating strategy for analyzing B cells, granulocytes, NK cells, macrophages and dendritic cells.
Representative results of extracellular staining of T cells within murine liver of HFD exposed mice compared to HFD SPF mice are demonstrated in Figure 3A. Indeed, a higher percentage of effector memory CD4+ and CD8+ T cells can be detected in HFD exposed mice, whereas intrahepatic naïve CD4+ and CD8+ T cells were found to be considerably lower in exposed compared to SPF mice at week 7 (Figure 3A).
To validate these results, the hepatic inflammatory response associated with antigen exposure was investigated by hematoxylin/eosin staining of liver sections of 15 weeks fed mice13. Severe steatosis, including increase of large lipid droplets resulting in macrovesicular steatosis, lobular inflammation, hepatocellular ballooning and destroyed lobular structure, was found in HFD exposed mice while only some SPF mice displayed a mild fat accumulation in the liver (Figure 3B). As reported in Figure 3C, the percentage of NK cells was higher in perigonadal adipose tissue of HFD exposed mice, whereas HFD SPF mice showed higher percentages of dendritic cells. No significant differences were found for macrophages and monocytes. Altogether, these results confirm more severe hepatic steatosis in HFD exposed mice and minor differences in adipose tissue between HFD exposed and SPF mice.
Table 1 shows the antibodies used in panel 1. Antibodies used in flow cytometry analysis for extracecullar staining of B cells, macrophages, NK cells, dendritic cells and granulocytes are depicted in Table 2.
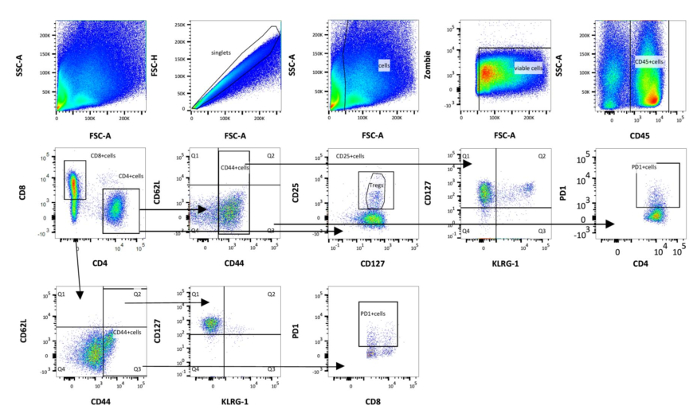
Figure 1: Schematic representation of gating strategy used in flow cytometry analysis of adipose immune cells (panel 1). First, the cells are gated on singlets. Then, debris is excluded by using forward scatter area (FSC-A) and side scatter area (SSC-A) and choosing the correct size. Cells are further characterized by the expression of CD45. Viable cells are selected using alive/dead cell marker that is an amine reactive fluorescent dye that is non-permeant to live cells, but permeant to the cells with compromised membranes. T cells were divided into cytotoxic T cells (CD8+) and T-helper cells (CD4+) and subdivided into naïve (CD44+CD62L–), central memory (CD44+CD62L+), effector memory (CD44+CD62L–) and effector (CD44–CD62L+) T cells. Finally, CD44+ cells are gated on KLRG1, CD127 and PD-1. Please click here to view a larger version of this figure.
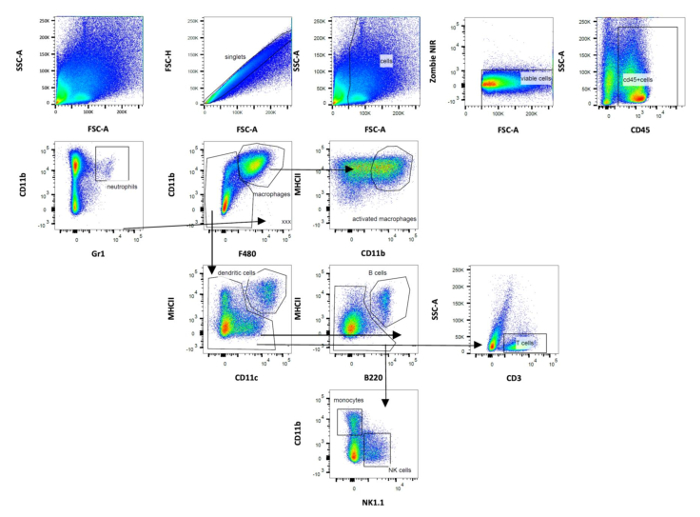
Figure 2: Schematic representation of gating strategy used in flow cytometry analysis of adipose immune cells (panel 2). Gating of dendritic cells was based on CD11b and CD11c expression. The following other populations were defined: B cells, NK cells, macrophages, monocytes and granulocytes. Data were analyzed after acquisition with the appropriate software. Please click here to view a larger version of this figure.
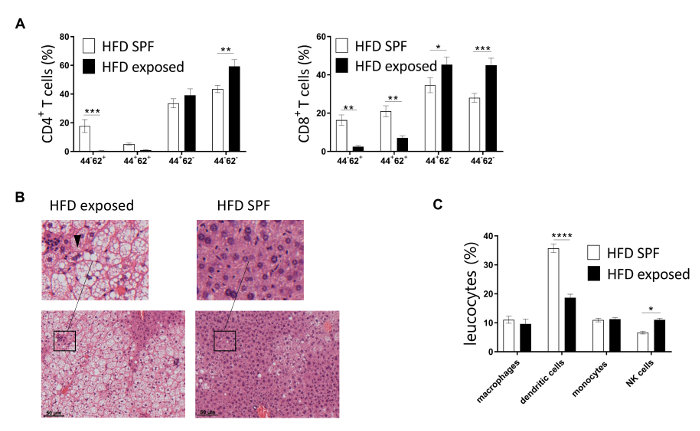
Figure 3: Representative flow cytometry analysis of T cells isolated from murine perigonadal adipose tissue and liver. (A) CD4+ and CD8+ T cells of murine livers from 7 week HFD mice maintained under SPF and exposed conditions were analyzed via flow cytometry. (B) Representative staining of 15 week HFD SPF and exposed mice. Infiltration of immune cells and ballooned hepatocytes (arrowhead) illustrate NASH. (C) Percentages of adipose innate immune cells as percentage of leukocytes in HFD mice maintained under SPF or exposed conditions for 7 weeks. n = 6-10 mice per group. Significance was determined using two-way ANOVA multiple measurement. Data are represented as mean ± SEM. ** P <0.01, *** P < 0.001. This figure has been modified from refefence13. Please click here to view a larger version of this figure.
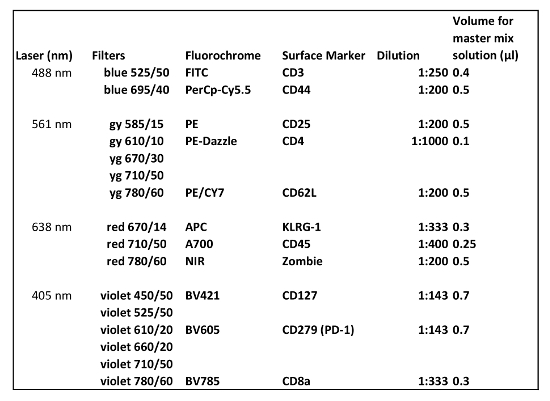
Table 1: Antibodies used in flow cytometry for extracellular staining (Panel 1). The amount of antibody described is for the analysis of one sample.
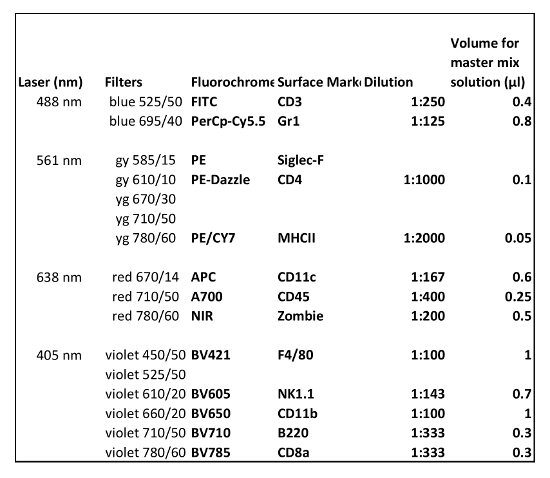
Table 2: Antibodies used in flow cytometry for extracellular staining (Panel 2). The amount of antibody described is for the analysis of one sample.
Discussion
Steatohepatitis has a strong association with metabolic abnormalities such as obesity, insulin resistance and dyslipidemia15. Multiple studies indicate that adipose tissue inflammation can drive the pathogenesis of type 2 diabetes, including altered levels of cells of both the innate and adaptive immune system4,5,16,17. In addition, it has been found that obesity modulates the activation of immune pathways, which can lead to liver complications18. There is an increasing interest in the characterization of immune cell phenotypes in adipose and liver inflammatory response because each immune cell subpopulation contributes in a different way to the development of insulin resistance and steatohepatitis.
This method gives detailed information about how to isolate and quantify relative amounts of immune cells from perigonadal adipose tissue and liver. Furthermore, the methods show how to maintain HFD fed mice in non-SPF conditions in order to induce steatohepatitis. The protocol can be used to study immune cell functions and to investigate associations of specific immune cells between different tissues in a microbiologically normalized environment.
In our present study, HFD feeding of non-SPF mice resulted in the development of insulin resistance and steatohepatitis, whereas HFD SPF mice developed insulin resistance, mild hepatic steatosis, but not steatohepatitis as determined by immunohistochemistry. Moreover, adipose and liver immune cell subsets differed significantly between HFD exposed and SPF mice, which confirms our understanding of the significance of different housing conditions. In summary, we could show that microbial exposure to commensal flora could influence the immunological properties of C57Bl/6 mice dramatically. Previous work has shown that the diversity and function of the gut microbiome is affected by diet-induced obesity19,20 and that gut barrier function and intestinal permeability depend on housing conditions21. We aim to address the link between gut colonization and adipose and liver inflammation in an upcoming publication.
Several points have to be considered during the planning stage of the proposed methods. First, single-cell suspensions are required for all flow cytometry assays and cell viability may be affected by the level of mechanical tissue dissociation and the duration of enzymatic tissue digestion. In order to avoid the destruction of the antibody epitope and to maximize the yield of functionally viable, dissociated cells, the type of tissue, age of the animal, genetic modifications, concentrations of enzymes, temperature and incubation times should be taken into consideration.
Second, our protocol has been optimized to quantitatively determine different immune cell phenotypes from obese HFD fed mice with adipose tissue inflammation and steatohepatitis. As tissue integrity, the number of immune cells and, thus, the quality of tissue digestion, might be affected by inflammatory processes, the protocol and collagenase used should be optimized for other tissues than liver or adipose tissue in specific experiments relating to dissection methods, incubations times or the collagenase used to digest the tissue. Third, it is important for the protocol that different experimental groups are included each day to eliminate effects due to day-to-day variation of the flow cytometry assay (temperature, incubation times, etc.).
However, there are some important limitations of the described method. In order to discriminate positive versus negative signals to properly adjust the gates to identify the cells positive for specific surface markers in the experimental samples, Fluorescence Minus One (FMO) controls (an FMO control contains all the fluorochromes in a panel, except for the one that is being measured) should have been used. In this experiment the data could be properly compensated and the gating was clear, but we recommend to add FMO controls to the experimental setup, whenever possible. Moreover, FoxP3, which needs to be stained intracellularly requiring permeabilization of the cell membrane, should have been used to quantify regulatory T cells because it allows a more accurate definition than the use of CD25 and CD127 (Table 2). Even though flow cytometry allows the quantification of several surface markers simultaneously, their number is limited to 12 per sample due to overlap in emission spectra. To correct for spectral overlap, time-intensive fluorescence compensation is required. To account for this, strategic panel development or the use of mass cytometry techniques is required. Furthermore, difficulties could arise in housing the mice in non-SPF conditions with regard to specific animal housing guidelines, but the current small number of studies on non-SPF rodents shows that such studies are possible12,21. For example, separated animal facilities for SPF and non-SPF mice could be evaluated. The development of a human adult-like immune system is compromised in SPF mice12,21, resulting in limitations to translate treatment strategies from animal experiments to clinical studies.
In conclusion, we provide a detailed protocol for phenotypic characterization of murine adipose and hepatic immune cells using flow cytometry in a diet-induced animal model of NASH and insulin resistance. Considering the complex function and regulation of both innate and adaptive cells in the context of obesity and metabolic dysregulation, our proposed methods should help to deepen our understanding of immunological mechanisms to balance metabolic homeostasis and, thus, help to develop human therapeutics that can restore anti-inflammatory immune responses. Overall, the provided animal model serves as a rapid and robust model for assessing metabolic complications associated with obesity and can be applied to other disease models.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Anke Jurisch, Diana Woellner, Dr. Kathrin Witte and Cornelia Heckmann for assistance with experimental procedures and Benjamin Tiburzy from Biolegend for helpful comments on the gating strategy. J.S. was supported by the Helmholtz Grant (ICEMED). This study was supported by grants from the Clinical Research Unit of the Berlin Institute of Health (BIH), the “BCRT-grant” by the German Federal Ministry of Education and Research and the Einstein Foundation. K.S.-B. and H.-D.V. are funded by FOR2165.
Materials
| 100µm cell strainers | Falcon | 352340 | |
| 1ml syringe | BD | 309659 | |
| 26G x 5/8 needles | BD | 305115 | |
| 35mm Petri Dishes | Falcon | 353001 | |
| 40µm cell strainers | Falcon | 352340 | |
| ACK lysis buffer | GIBCO | A1049201 | |
| Alexa Fluor 700 anti-mouse CD45 | Biolegend | 103127 | AB_493714 (BioLegend Cat. No. 103127) |
| Analysis software | FlowJo 10.0.8 software | ||
| APC anti-mouse CD11c Antibody | Biolegend | 117309 | AB_313778 (BioLegend Cat. No. 117309) |
| APC anti-mouse KLRG1 (MAFA) Antibody | Biolegend | 138411 | AB_10645509 (BioLegend Cat. No. 138411) |
| BV421 anti-mouse CD127 Antibody | Biolegend | 135023 | AB_10897948 (BioLegend Cat. No. 135023) |
| BV421 anti-mouse F4/80 Antibody | Biolegend | 123131 | AB_10901171 (BioLegend Cat. No. 123131) |
| BV605 anti-mouse CD279 (PD-1) Antibody | Biolegend | 135219 | AB_11125371 (BioLegend Cat. No. 135219) |
| BV605 anti-mouse NK-1.1 Antibody | Biolegend | 108739 | AB_2562273 (BioLegend Cat. No. 108739) |
| BV650 anti-mouse/human CD11b Antibody | Biolegend | 101239 | AB_11125575 (BioLegend Cat. No. 101239) |
| BV711 anti-mouse/human B220 Antibody | Biolegend | 103255 | AB_2563491 (BioLegend Cat. No. 103255) |
| BV785 anti-mouse CD8a Antibody | Biolegend | 100749 | AB_11218801 (BioLegend Cat. No. 100749) |
| C57Bl/6J mice, male, 5 weeks old | Forschungseinrichtungen für experimentelle Medizin (FEM) | ||
| CaCl2 | Charité – Universitätsmedizin Berlin | A119.1 | |
| Collagenase NB 4G Proved Grade | SERVA | 11427513 | |
| Collagenase Typ I | Worthington | LS004197 | |
| Conical centrifuge tube 15ml | Falcon | 352096 | |
| Conical centrifuge tube 50ml | Falcon | 352070 | |
| DNAse | Sigma-Aldrich | 4716728001 | |
| Fetal bovine serum | Biochrom | S0115 | |
| Filter 30µm | Celltrics | 400422316 | |
| FITC anti-mouse CD3 Antibody | Biolegend | 100203 | AB_312660 (BioLegend Cat. No. 100203) |
| Flow cytometry | BD-LSR Fortessa | ||
| Forceps | Sigma-Aldrich | F4142-1EA | |
| HBSS | Bioanalytic GmBH | 085021-0500 | |
| High-fat diet | SSNIF | E15741–34 | 60 kJ% from fat, 19 kJ% from proteins, and 21 kJ% from carbohydrates |
| micro dissecting scissors | Sigma-Aldrich | S3146 | used for dissection purposes |
| PE anti-mouse CD25 Antibody | Biolegend | 101903 | AB_312846 (BioLegend Cat. No. 101903) |
| PE/Cy7 anti-mouse CD62L Antibody | Biolegend | 104417 | AB_313102 (BioLegend Cat. No. 104417) |
| PE/Cy7 anti-mouse I-A/I-E (MHCII) Antibody | Biolegend | 107629 | AB_2290801 (BioLegend Cat. No. 107629) |
| PE/Dazzle 594 anti-mouse CD4 Antibody | Biolegend | 100565 | AB_2563684 (BioLegend Cat. No. 100565) |
| Percoll solution | Biochrom | L6115 | |
| PerCP/Cy5.5 anti-mouse CD44 Antibody | Biolegend | 103031 | AB_2076206 (BioLegend Cat. No. 103031) |
| PerCP/Cy5.5 anti-mouse Gr-1 Antibody | Biolegend | 108427 | AB_893561 (BioLegend Cat. No. 108427) |
| Phosphate buffered saline | Gibco | 12559069 | |
| Round-Bottom Tubes with cell strainer cap | STEMCELL Technologies | 38030 | |
| TruStain fcX anti-mouse CD16/32 | Biolegend | 101301 | AB_312800 (BioLegend Cat. No. 101301) |
| Trypan Blue | Sigma-Aldrich | T6146 | |
| Zombie NIR Fixable Viability Kit | Biolegend | 423105 | viablity stain |
References
- Guh, D. P., et al. The incidence of co-morbidities related to obesity and overweight: A systematic review and meta-analysis. BMC Public Health. 9, 88 (2009).
- Prentki, M. Islet β cell failure in type. J Clin Invest. 116 (7), 1802-1812 (2006).
- Shoelson, S. E., Lee, J., Goldfine, A. B. Inflammation and insulin resistance. Journal of Clinical Investigation. 116 (7), 1793-1801 (2006).
- Kahn, S. E., Hull, R. L., Utzschneider, K. M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 444, 840 (2006).
- Exley, M. A., Hand, L., O’Shea, D., Lynch, L. Interplay between the immune system and adipose tissue in obesity. Journal of Endocrinology. 223 (2), R41-R48 (2014).
- Ferrante, A. W. Macrophages, fat, and the emergence of immunometabolism. Journal of Clinical Investigation. 123 (12), 4992-4993 (2013).
- Winer, D. A., et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nature Medicine. 17, 610 (2011).
- Onodera, T., et al. Adipose tissue macrophages induce PPARγ-high FOXP3(+) regulatory T cells. Scientific Reports. 5, (2015).
- Lackey, D. E., Olefsky, J. M. Regulation of metabolism by the innate immune system. Nature Reviews Endocrinology. 12, 15 (2015).
- Calle, E. E., Kaaks, R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature Reviews Cancer. 4, 579 (2004).
- Ibrahim, S. H., Hirsova, P., Malhi, H., Gores, G. J. Animal Models of Nonalcoholic Steatohepatitis: Eat, Delete, and Inflame. Digestive Diseases and Sciences. 61 (5), 1325-1336 (2016).
- Beura, L. K., et al. Recapitulating adult human immune traits in laboratory mice by normalizing environment. Nature. 532 (7600), 512-516 (2016).
- Sbierski-Kind, J., et al. Distinct Housing Conditions Reveal a Major Impact of Adaptive Immunity on the Course of Obesity-Induced Type 2 Diabetes. Frontiers in Immunology. 9 (1069), (2018).
- Japp, A. S., et al. Wild immunology assessed by multidimensional mass cytometry. Cytometry Part A. 91 (1), 85-95 (2017).
- Benedict, M., Zhang, X. Non-alcoholic fatty liver disease: An expanded review. World Journal of Hepatology. 9 (16), 715-732 (2017).
- McNelis, J. C., Olefsky, J. M. Macrophages, Immunity, and Metabolic Disease. Immunity. 41 (1), 36-48 (2014).
- Ferrante, A. W. The Immune Cells in Adipose Tissue. Diabetes, Obesity & Metabolism. 15, 34-38 (2013).
- Bertola, A., et al. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PloS One. 5 (10), e13577 (2010).
- Turnbaugh, P. J., Bäckhed, F., Fulton, L., Gordon, J. I. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host & Microbe. 3 (4), 213-223 (2008).
- Singh, R. K., et al. Influence of diet on the gut microbiome and implications for human health. Journal of Translational Medicine. 15 (1), 73 (2017).
- Müller, V. M., et al. Gut barrier impairment by high-fat diet in mice depends on housing conditions. Molecular Nutrition & Food Research. 60 (4), 897-908 (2016).

