Fast and Efficient Expression of Multiple Proteins in Avian Embryos Using mRNA Electroporation
Summary
We report messenger RNA (mRNA) electroporation as a method that permits fast and efficient expression of multiple proteins in the quail embryo model system. This method can be used to fluorescently label cells and record their in vivo movements by time-lapse microscopy shortly after electroporation.
Abstract
We report that mRNA electroporation permits fluorescent proteins to label cells in living quail embryos more quickly and broadly than DNA electroporation. The high transfection efficiency permits at least 4 distinct mRNAs to be co-transfected with ~87% efficiency. Most of the electroporated mRNAs are degraded during the first 2 h post-electroporation, permitting time-sensitive experiments to be carried out in the developing embryo. Finally, we describe how to dynamically image live embryos electroporated with mRNAs that encode various subcellular targeted fluorescent proteins.
Introduction
Electroporation is a physical transfection method that uses an electrical pulse to create transient pores in the plasma membrane, allowing substances like nucleic acids or chemicals to pass into the cytosol. Electroporation is widely used to deliver DNA into bacteria, yeast, plants, and mammalian cells1,2,3. It is routinely used to introduce genetic payloads into target cells and tissues within the developing avian embryo to study the genetic control of development or label migrating populations of cells4,5,6,7. However, several experimental limitations also exist with DNA electroporation8. For instance, DNA electroporation often introduces highly variable numbers of expression vectors per cell and subsequently the mRNAs and proteins they encode. This variability can lead to considerable cell-cell heterogeneity that complicates both image analysis and data interpretation9,10. Additionally, proteins from DNA electroporation only begin to express ~3 h post-electroporation and do not reach the maximum efficiency in cell number and fluorescence intensity until 12 h, likely due to the time required to transfer into the nucleus and complete both transcription and translation in vivo11.
In contrast, mRNA transfection has been effectively used in a variety of model systems, including Xenopus laevis oocytes by microinjection12,13, reprogramming human stem cells by mRNA lipofectamine transfection14, and electroporating recalcitrant neural stem cells in adult mice15. We tested the ability of mRNA electroporation to efficiently label cells during early avian embryonic development using in vitro synthesized mRNAs that encode distinct fluorescent proteins (FPs). For our studies, we used the pCS2+ vector, a multipurpose expression vector that is commonly used for expressing proteins in Xenopus and zebrafish embryos. The SP6 and T7 RNA polymerase promoters in the pCS2+ permit the synthesis of mRNA and protein from any cloned gene when used in an in vitro transcription/translation system.
Here, we demonstrate that mRNA electroporation allows fast and efficient expression of fluorescent proteins (FPs) in gastrulating quail embryos. We designed and generated many of the expression vectors used in these studies. For example, we subcloned the LifeAct-eGFP gene16 into the pCS2+ vector17 to express from the CMV promoter and SP6 promoter. The inserted gene lies downstream of the SP6 promoter and upstream of the SV40 poly(A) tail18. In embryos co-electroporated with mRNA and DNA, FPs encoded from in vitro transcribed mRNAs were first detected within 20 min of electroporation, whereas FPs from DNA expression vectors were detected only after 3 h. Multiple mRNAs encoding for nuclear, Golgi, and membrane proteins can be electroporated into an embryo simultaneously, resulting in the quick and efficient expression of multiple proteins in individual cells. Finally, using an in vivo fluorescence recovery after photobleaching (FRAP) assay, we show that a majority of the electroporated mRNAs decay within 2 h. Thus, fast initial protein production combined with limited new protein translation makes mRNA electroporation a valuable technique when temporal control of expression is necessary.
Protocol
All animal procedures were carried out in accordance with approved guidelines from the Children’s Hospital Los Angeles and the University of Southern California Institutional Animal Care and Use Committees.
1. Generation pCS2-based Expression Vectors
- To clone pCS2.Lifeact-eGFP, prepare the vector backbone by digesting 2 µg of pCS2.CycB1-GFP (a construct containing a different insert) with BamHI (10 U) and BsrGI (10 U) in appropriate digestion buffer (see Table of Materials) diluted to 1x in a total reaction volume of 50 µL for 1 h at 37 °C (see Figure 1 for the schematic of cloning procedure).
- Dephosphorylate the free 3’OH ends of the vector backbone from the restriction enzyme reaction by adding Shrimp Alkaline Phosphatase (1 U). Incubate for 30 min at 37 °C.
- Run the whole mixture in a 1% agarose gel/1x Tris Acetate EDTA (TAE) buffer at 90 V for ~50 min and then stain the gel in a 0.5 µg/mL solution of ethidium bromide in 1x TAE buffer for 15 min with gentle rocking. Also, make sure to load molecular weight markers in a free lane to help determine DNA sizes.
- Using a DNA safe UV Transilluminator to avoid nicking DNAs, cut out the vector backbone (expected band size of 4kb) from the agarose gel quickly and isolate the DNA fragment from the gel piece using a gel purification kit using manufacturer’s instructions.
- Simultaneously with step 1.1, prepare the insert by digesting 2 µg of pEGFP-N1-Lifeact with BglII (10 U) and BsrGI (10 U) in appropriate digestion buffer diluted to 1x in a total reaction volume of 50 µL for 1 h at 37 °C. Run the whole mixture in a 1% agarose gel to isolate the 800 bp band as previously explained in step 1.1.2-1.1.3.
- After both the vector and insert are purified and quantified on the spectrophotometer, combine 50 ng of the vector and 38 ng of the insert (1:3 molar ratio of vector to insert) in T4 DNA ligase buffer prior to adding T4 DNA ligase to catalyze the ligation reaction. Additionally, set up a no DNA control, a vector only control, and an insert only control. Incubate all reactions at room temperature for 30 min.
- Transform 1 µL of the ligation mixture(s) into competent E. coli DH10. Also, transform water only negative control and 20 pg of pUC19 positive control. Spread bacteria onto Luria Broth (LB) agar plates containing 50 µg/mL ampicillin and incubate overnight at 37 °C.
- On the following morning, count the colonies on each plate.
NOTE: Ideally, there should be no colonies on the negative control plates, >100 colonies on the vector+insert ligation plates, and fewer colonies on the vector-only plate. - Pick at least 8 bacterial colonies from the agar plate transformed with the vector+insert ligation mixture and inoculate them using sterile toothpicks or pipet tips into 2 mL of liquid LB broth containing 50 µg/mL Ampicillin.
- Extract DNA from each of the clones using commercial plasmid miniprep kits.
- Run a diagnostic restriction digest using 500 ng of DNA from each clone using NotI (10 U) and BamHI (10 U) in the appropriate digestion buffer diluted to 1x for 1 h at 37 °C and run the digested DNA on a 1% agarose gel in 1x TAE buffer. Expected band sizes for pCS2.Lifeact-eGFP positive clones are 3.9 kb and 978 bp.
- Transform 1 pg-100 ng of DNA from the positive clone into competent E. coli DH10, and prepare 3-4 miniprep reactions as detailed in previous steps to obtain a sufficient amount of DNA for in vitro transcription reaction (10 µg minimum).
2. Preparation of mRNA by In Vitro Transcription
- Linearize 10 µg of pCS2.LifeAct-eGFP on the 3’ end of the insert with NotI (10 U) in an appropriate digestion buffer diluted to 1x in a total reaction volume of 50 µL at 37 °C overnight.
NOTE: To prevent RNase contamination and reduce mRNA degradation, wear gloves while handling mRNA samples. - Purify the DNA using a mixture of phenol: chloroform: isoamyl alcohol (25:24:1, v/v).
- Add 150 µL of RNase-free water to make the total volume of the digestion reaction equal to 200 µL. Then, add 200 µL of phenol: chloroform: isoamyl alcohol and vortex the mixture for 20 s.
- Centrifuge in a microfuge at max speed (18,400 x g) for 2 min. Carefully remove the top aqueous phase that contains the linearized DNA. Repeat this step to remove additional impurities and be careful not to disrupt the white precipitate that may form between the bottom and top liquid phases.
- Precipitate linearized DNA by adding 1/10 volume 3M sodium acetate (RNase-free) and 2.5 volumes of 100% ethanol. Leave the mixture at -20 °C for >30 min, then pellet the DNA by centrifuging at max speed (18,400 x g).
- Wash the DNA pellet with 70% ethanol, then air-dry the pellet for >5 min.
- Dissolve the DNA pellet in 5 µL of RNase-free water. Quantify the DNA by spectrophotometer.
NOTE: The expected DNA concentration is ~0.5-1 µg/µL with an A260/A280 ratio between 1.7-2.0.
- Use 1 µg of the linearized pCS2.LifeAct-eGFP DNA for in vitro transcription (IVT). Follow the manufacturer’s instructions in the commercial kit (see Table of Materials) to include 10 µL of cap analog (final concentration 0.8 mM) and NTPs (final concentration 1 mM for ATP, CTP, and TTP; 0.2 mM for GTP), 2 µL of 10x reaction buffer (final concentration 1x), 2 µL of SP6 RNA polymerase, and RNase-free water up to 20 µL.
- Incubate the IVT mixture for about 2 h or longer, depending on transcript size.
NOTE: 2 h incubation works well for a 3 kb transcript, but overnight incubation works better for transcripts that are greater than 5 kb. - To remove free nucleotides from the transcription reaction, add 30 µL of LiCl RNA precipitation solution (7.5 M lithium chloride, 50 mM EDTA) to precipitate the mRNA. Vortex the mixture briefly and store at -20 °C for 30 min. Spin the mRNA down for 15 min in a microfuge at max speed (18,400 x g) and rinse with 70% ethanol (RNase-free).
- Incubate the IVT mixture for about 2 h or longer, depending on transcript size.
- Dissolve the synthesized mRNA in 15 µL of RNase-free water. Dispense the synthesized mRNA at 5 µL/tube (3 tubes total) and place at -80 °C for long-term storage. Quantitate the mRNA with a spectrophotometer.
NOTE: Expected yield is 15-20 µg of mRNA (~1 µg/µL). 1 µL (1 µg/µL) is sufficient for an experiment with ~10 embryos, assuming that the mRNA is diluted from 1 µg/µL to 500 ng/µL and that each embryo is injected with ~200 nL. Each tube should, therefore, contain enough mRNA for ~5 experiments. - Run ~300 ng of mRNA on a 1% agarose gel in 1x TAE buffer after cleaning all gel equipment with the RNase decontamination solution (e.g., RNase Away) and ensure that the mRNA appears as one band (multiple bands may be present if secondary structures form) and that no smear appears in the gel lane, which indicates RNA degradation.
3. Preparation of mRNA Electroporation Mix
- Thaw and dilute mRNA at the desired concentration (250-500 ng/µL in RNase-free water works well for all mRNAs tested in this work). Add 1/10 volume of colored dye (see Table of Materials, RNAse-free, 0.1% final concentration) to help visualize the mRNA injection site and to properly place the electrodes.
NOTE: If DNA and RNA are combined to perform a co-electroporation, ensure that DNA is free from contaminating RNases by using phenol-chloroform extraction and ethanol precipitation prior to mRNA and DNA mixture.- Be sure to prepare a negative control using a mock (no RNA added) electroporation solution. If possible, prepare a positive control using pre-validated mRNA (mRNA that has been shown to produce FP successfully in previous experiments).
NOTE: The negative control is essential for all imaging experiments to establish background fluorescence levels for normalization of data. The positive control is especially important when working with newly transcribed mRNA by helping to confirm that the electroporation settings worked.
- Be sure to prepare a negative control using a mock (no RNA added) electroporation solution. If possible, prepare a positive control using pre-validated mRNA (mRNA that has been shown to produce FP successfully in previous experiments).
- Store the mRNA electroporation solution on an ice slurry to prevent degradation until ready to proceed with electroporation.
4. Electroporate mRNAs into Living Quail Embryos
- Collect freshly laid fertilized quail eggs daily and store at 13 °C in a humidified refrigerator for no longer than 1 week. Incubate the quail eggs at 38 °C until the desired embryonic developmental stages19,20,21.
NOTE: HH3 to HH5 were used in this study for both static and dynamic imaging. For HH3 embryos, leaving the eggs at room temperature for 2 h prior to harvesting makes the isolation process much easier as the embryos are generally more resistant to physical manipulations when cooled down. - Isolate and prepare embryos according to the EC culture system22. Collect at least 5 embryos per condition, including at least one to serve as a negative control (not electroporated).
- Gently break and pour the egg into a 10 cm Petri dish. Remove the majority of the thick albumin with a transfer pipette and remove the remaining thick albumin around the embryo by gently wiping the surface of the yolk with a tissue wipe to ensure that the embryo sticks tightly to the paper ring.
- Lay precut filter paper (see Table of Materials) upon the embryo and use scissors to smoothly cut around the perimeter of the embryo.
- Use a Pasteur pipette to underlie the embryo with PBS, using gentle streams to vacate any yolk sticking to the embryo.
NOTE: This step is critical when working with younger (<HH3) embryos because these embryos tend to stick more to the yolk and will often detach from the vitelline membrane during subsequent washing steps. - Slowly pull the embryo/paper ring at an oblique angle up and off the yolk into a Petri dish filled with PBS for further cleaning. Once most of the yolk has been removed, place the embryo ventral side up on a 35 mm Petri dish covered with a semi-solid mixture of agar/albumen.
- Prepare six to eight 10 cm-long glass microcapillaries (O.D. = 1.2 mm) by using a glass micropipette puller instrument.
- Place the embryo ventral side up in the electroporation chamber filled with PBS. Using a glass microcapillary, inject a bolus of 200 nL of the mRNA or DNA/mRNA electroporation mix into the cavity between the epiblast and vitelline membrane covering the desired region.
NOTE: The entire anterior area for pellucida and some area for opaca was electroporated in most of the experiments shown in this manuscript. - Place the positive and negative electrodes (platinum flat square electrode; side length of 5 mm) on top and bottom of the embryo respectively and electroporate using the following pulse sequence: five square electric pulses of 5 V, 50 ms duration with 100 ms intervals using an in vivo electroporator. Ensure that the distance between the electrodes is ~5 mm.
NOTE: Optimizing the electroporation parameters is crucial to avoid conditions that can kill the fragile embryonic cells. Parameters of voltage, pulse length, pulse intervals, and the number of pulses for DNA and mRNA should be considered for various electroporation devices. - Incubate the electroporated embryos at 38 °C to the desired developmental stage.
NOTE: A fluorescent dissecting stereoscope (see Table of Materials) helps to screen transfected vs. non-transfected embryos. - If the embryos are to be statically imaged, fix them in 4% paraformaldehyde in PBS for 1 h at room temperature or overnight at 4 °C.
- Remove the embryos from the vitelline membrane by smoothly cutting around the perimeter of the filter paper with scissors and peeling off the shiny membrane on the dorsal surface with sharp forceps gently.
- Wash the fixed embryos in PBS/Triton (0.1%) 2x for 5 min and continue with in situ hybridization or immunostaining if desired.
- Finally, stain the embryo in 0.5 µg/µL DAPI in PBS/Triton (0.1%) for at least 30 min at room temperature. Wash embryos in PBS/Triton 2x for 5 min and clear the embryo in SCALE-U2 solution23 overnight.
- To analyze the efficiency of electroporation (see Figure 2), use the binary and particle analysis tool and DAPI channel on ImageJ to obtain nuclear outlines from all cells in the image.
- To use the binary tool on ImageJ, use a single z-slice in the DAPI channel containing a majority of cells and click Process > Binary > Make Binary. To separate nearby cells, click Process > Binary > Watershed. Obtain cell outlines by clicking Analyze > Analyze Particles, size set to 100-500 (µm2).
- Ensure that a majority of the cells are outlined in the DAPI channel and save the cell outlines by clicking More > Save on the ROI manager pop-up window.
- Use these outlines to obtain fluorescence intensity values for the mRNA and DNA channel by opening the previously saved file on a single z-slice in the mRNA or DNA channel and then click Measure on the ROI manager.
- Finally, filter these intensity values, counting cells with <6,000 fluorescence intensity as non-transfected and cells with >6,000 fluorescence intensity as transfected.
5. Image FPs Encoded by Electroporated mRNAs
- Choose the healthiest and best electroporated embryo for the dynamic imaging experiments after looking at all electroporated embryos under the fluorescent dissecting stereoscope.
- Continue to incubate the other electroporated embryos and the non-electroporated embryo (the negative control) in a separate incubator while imaging the chosen embryo in case this embryo dies during the experiment.
- For the dynamic imaging, use the whole-mount ex ovo avian embryo culture as previously described24,25,26 with an inverted confocal microscope.
NOTE: The microscope is equipped with an onstage incubator (see Table of Materials) that maintains the temperature at 36 °C during imaging. It was observed during the microscope set-up that embryos incubated at 36 °C survive longer than at higher temperatures, possibly because the laser may cause local heating on the embryo. Readers should determine the optimum on-stage incubation temperature for their own microscope set-up.- To dynamically image and visualize embryogenesis, clean the electroporated embryo briefly with PBS to remove any bubbles that may form on the dorsal side of the embryo during the electroporation process by moving the embryo around using forceps in a PBS clean solution.
- Place the clean embryo directly onto an imaging dish containing a thin layer of albumen-agar (~150 µL) making sure not to generate any bubbles on the dorsal surface of the embryo22.
- To ensure survival for long-term imaging, add a small moist rolled-up piece of tissue paper at the inside edges of the imaging dish and seal the dish using paraffin film to minimize evaporation during the imaging and incubation.
- Move this dish quickly to the pre-warmed stage of a confocal microscope and locate the colored dye in the embryo using the brightfield channel (PMT laser 20%), which identifies the injected and electroporated region.
- Set imaging software to the desired objective (10 or 20x), dichroic mirror (488 nm for GFP nm, 561 for RFP), emission spectra (499-562 nm for GFP, 570-695 nm for RFP), and turn on an appropriate laser (488 nm for GFP, 561 nm for RFP).
NOTE: Electroporated mRNA was translated into proteins that were seen within 20 min (see Figure 3). The imaging metadata used for most of the images in this paper were: an inverted confocal microscope with a 20x objective (see Table of Materials); pixel dwell time, ~1.5 μs; mean of 4-line scans.- Click Live on the imaging software and adjust the laser power to a setting that is appropriate for the fluorescence intensity depending on each microscope laser power. Start by imaging the embryo using 1% laser power, 800 gain, and increase the laser power slowly by 1% increments until saturated pixels are seen.
- Follow this by decreasing the laser power slightly until no saturated pixels are seen anymore.
NOTE: The laser power chosen for the beginning of an imaging session of an embryo may be good for earlier time points but not ideal at later time points if the fluorescence of the cells becomes much brighter or dimmer over time. To address this, image the embryo at slightly lower power settings initially since the electroporated cells generally get brighter over the first 6 hours after electroporation (see Figure 3A-E for quantification of signal increase). If the later images are saturated, continue to image with the original imaging settings, but take an additional image immediately after with weaker imaging settings (smaller pinhole or weaker laser power). - Image the embryo every 3-5 min in order to track individual cell migration across different time points. For this work, the images were z-stacks (~50 µm thick) of the entire electroporated area, plus some extra room towards the bottom of the z-stack in case the embryo sinks into the agarose bed throughout the imaging session.
- Observe how fast the cells are moving by checking the first few time points of the first movie. If the cells are moving at a fast rate (meaning that they will exit the area of the imaged region within a couple more time points), consider expanding the zoom of the imaged area (1x à 0.8x) or imaging a different region.
NOTE: Embryonic regions in the area pellucida move much more rapidly than those in the area opaca. Additionally, younger embryos (HH3, HH5) often contain cells that are undergoing more rapid movement compared to older embryos (>HH7). - After imaging the electroporated region of the embryo, image an un-electroporated region of the same embryo to determine autofluorescence levels (should be minimal if low laser power <10% is used to image the embryo).
6. Fluorescence Recovery After Photobleaching (FRAP) to Assay mRNA Integrity
NOTE: An in vivo fluorescence recovery after photobleaching (FRAP) assay can be used to determine how long transfected mRNA could be translated into FPs. The following protocol outlines a FRAP experiment to detect the half-life of H2B.Citrine mRNA in an electroporated embryo.
- Perform FRAP experiments on the inverted confocal microscope using a 20x 0.8 NA objective and a completely open pinhole.
- After confirming electroporation of H2B-Citrine on the stereomicroscope and setting the embryo on the pre-heated stage on the inverted confocal microscope (see step 5.2.4), photobleach most of the cellular fluorescence from H2B-Citrine at a variety of time points (45 min, 2 h, and 5 h post-electroporation) by using the 405 nm laser with 70% laser power, 100 iterations, scan speed 4, which leaves only 5% of fluorescence remaining.
NOTE: This process should take a couple of minutes. - Continue to incubate the embryos on the stage at 36 °C after photobleaching.
- After confirming electroporation of H2B-Citrine on the stereomicroscope and setting the embryo on the pre-heated stage on the inverted confocal microscope (see step 5.2.4), photobleach most of the cellular fluorescence from H2B-Citrine at a variety of time points (45 min, 2 h, and 5 h post-electroporation) by using the 405 nm laser with 70% laser power, 100 iterations, scan speed 4, which leaves only 5% of fluorescence remaining.
- Be sure to pay attention to actively dividing cells within the photobleached region, which indicate that the photobleached cells have not completely died post-treatment.
- Acquire post-bleach images (z-stacks of the electroporated region) for up to 30 min at regular time intervals (3 or 5 min) using the confocal microscope.
NOTE: If available, use a transgenic H2B-XFP line as a positive control for ensuring cell survival in the photobleached region. The rate of fluorescence recovery for the FP encoded by the electroporated mRNA should decrease, but that for the FP encoded via transgene should remain consistent throughout the entire movie. - To ensure that the imaging conditions do not deleteriously affect embryo survival, concurrently incubate electroporated embryos that are not photobleached, which may serve as a control for imaging.
- To quantitate the photobleaching results for mRNA decay post-electroporation, track the cell fluorescence over time (3 or 5 min) by measuring the fluorescence intensity of the center (a 7.5 µm circle) of each cell using ImageJ. Measure this for all cells within the photobleached region that are not undergoing mitosis and have been completely photobleached.
NOTE: Consider omitting the mitotic cells from the quantification since mitotic nuclei have stronger fluorescence than interphase nuclei due to chromatin condensation. - Plot the fluorescence intensity over time post-bleach at a variety of time points (45 min, 2 h, and 5 h).
Representative Results
mRNA electroporation is more efficient than DNA electroporation
We used pCS2+.H2B-Citrine to prepare in vitro transcribed mRNA. Since DNA electroporation is usually performed at 1-2 µg/µL, we used an equimolar concentration of mRNA (calculated to be around 0.25-0.5 µg/µL for H2B-Citrine) for mRNA electroporation. We first tested the electroporation efficiency of pCS2+.H2B-Citrine DNA compared to H2B-Citrine mRNA (in vitro transcribed from the SP6 promoter of pCS2+.H2B-Citrine) by electroporating DNA or mRNA separately into HH5 quail embryos then examining electroporation efficiency at 12 h post-electroporation. Although DNA electroporation leads to brighter fluorescence in some electroporated cells, the efficiency of DNA electroporation was visibly lower compared to the widely-expressed mRNA encoded FPs (Figure 2A and B).
To account for inherent embryo-to-embryo variability in the electroporation protocol, an equimolar combination of mRNA encoding H2B-Citrine and DNA expressing H2B-mKate2 was electroporated into the ectoderm of HH5 embryos and observed 12 h post-electroporation. When comparing DNA and mRNA co-electroporation in the same embryo, DNA expressing FP was also significantly and consistently less efficient than that of mRNA (Figure 2C). By quantifying all embryos electroporated with only mRNA, DNA, or a combination of the two, we found that mRNA transfects ~75% of cells in a given region, while DNA only transfects ~25% of cells in a given region (Figure 2D). The spread in the expression of fluorescent proteins encoded by mRNA or DNA was not significantly different in all co-electroporated embryos (Figure 2E).
Protein expression is quicker in mRNA electroporation than DNA electroporation
The transfected mRNA should lead to faster protein production since it can be immediately recognized by cytosolic translation machinery as seen in Figure 3. DNA must be translocated to the nucleus, transcribed, and the mRNA transported back to the cytosol where it is recognized by cytosolic translation machinery, expected to take more time. To directly compare mRNA versus DNA expression rates of protein production, we carried out a series of time course experiments in which we co-electroporated an equimolar combination of DNA (pCMV.H2B-mKate2) and mRNA (H2B-Citrine) into the ectoderm of HH5 embryos. We detected mRNA-encoded FP expression around 22 min post-electroporation, which continues increasing in relative fluorescence intensity for ~6 h (Figure 3A-E, Supplemental. Movie S1b). In contrast, DNA-encoded FP expression is first seen at ~1.5 h post-electroporation (Figure 3A’-E’, Supplemental. Movie S1c) and continues increasing in brightness until 6 h when the movie was stopped. Because the DNA electroporated cells were saturated by the 6 h mark, we reimaged this condition with weaker imaging conditions (Figure 3F-F’’). Across all time points of this time lapse (22 min to 6 h), mRNA transfects several times more cells than DNA (Supplemental. Movie S1a, the total efficiency of electroporation through brightfield and DAPI are not shown because these images are taken from a time-lapse of a wild type quail embryo). Based on this, we conclude that mRNA electroporation leads to earlier expressed protein.
Co-electroporation of multiple mRNAs is highly efficient
Next, we sought to further test the efficiency of mRNA electroporation by electroporating multiple mRNAs into a region of interest. Co-electroporation of multiple DNAs had previously been shown to be relatively inefficient, showing with the number of multi-labeled being 25%, 14%, and 10% for 2, 3, and 4 DNA constructs electroporated, calculated as a proportion of the total number of cells11. We first electroporated two mRNAs that encode spectrally distinct FPs (Turquoise2-Golgi/H2B-Citrine) into HH5 embryos and obtained high transfection efficiency in all electroporated regions. We used this double-electroporation to capture movies of radial expansion between area pellucida and opaca in a HH4 gastrulating embryo, imaged 2 h post-electroporation (Supplemental Movie S2a, b, and c). The H2B-Citrine is localized within the cell nucleus and permits cell proliferation to be traced27. The Turquoise2-Golgi FP appears to show subcellular polarity within the embryonic cells but does not appear to correlate with speed or direction of moving ectoderm cells in vivo28. This movie (Supplemental Movie S2) shows that cells electroporated with multiple mRNAs can continue normal cellular activity without any apparent defects.
Furthermore, co-electroporation of 4 mRNAs – Turquoise2-Golgi (to visualize Golgi apparatus), LifeAct-eGFP (to visualize F-actin), H2B-Citrine (to visualize nucleus), and membrane-Cherry FP (to visualize membranes) — resulted in 87% transfection efficiency of all 4 mRNAs in all electroporated regions (Figure 4). LifeAct-EGFP and H2B-Citrine were separated by the linear unmixing processing tool in the commercial software.
FRAP assay shows the rapid degradation of electroporated mRNA during the first two hours post-electroporation.
It is well-known that naked mRNAs are quickly degraded by cellular RNAases29. We postulated that mRNA electroporation might be useful for loss or gain-of-function experiments by allowing fast and efficient expression of proteins in a developing embryo. Therefore, it would be helpful to quantitate the rate of mRNA decay after mRNA electroporation, since mRNA degrades faster than DNA in cells. In previous reports of mRNA electroporation in adult mouse neural stem cells, around 80% mRNA was identified 2 h post-electroporation by qRT-PCR, yet little to no mRNA was detected by 24 h post-electroporation15. We sought to further quantify mRNA expression post-electroporation by designing an in vivo FRAP assay to detect mRNA levels in electroporated cells.
Photobleaching is the irreversible destruction of the fluorophore that can occur when the fluorophore (fluorescent proteins in our case) is in an excited state, which eliminates fluorescence during observation30,31. Any emitted light from fluorescent proteins (FPs) that can be detected in photobleached cells above baseline levels should, therefore, represent newly translated FPs encoded by intact, transfected mRNAs. Therefore, we sought to photobleach the electroporated cells to eliminate all existing FP-derived fluorescence at a variety of time points and track subsequent fluorescence recovery.
Using optimized photobleaching settings as described in the protocol section, we found that photobleaching initially reduces the fluorescence intensity in all photobleached cells by ~95% when assayed immediately after the photobleaching event. This was done at 45 min, 2 h, and 5 h post-electroporation (Figure 5A-C). The fluorescence recovery in each cell within the photobleached region was tracked over a 30 min time period after the photobleaching at each time point by imaging the same region at regular time intervals (3 or 5 min). Our data suggests that there is a significant decrease in transfected mRNA levels between 45 min and 5 h post-electroporation. This is shown by the large decrease in FRAP measured at 5 h compared to 45 min post-electroporation. To highlight the fluorescence recovery in the bleached cells especially in the 5 h time point, we enhanced the brightness in Figure 5A-C using the brightness/contrast tool in ImageJ and show these modified images in Figure 5D-F. Interestingly, there is a large heterogeneity in fluorescence recovery from cell-to-cell within the photobleached area at 2 h because some cells recover fluorescence faster than other cells (Figure 5E’’, see arrowheads). However, all cells within the 5 h photobleached area recover fluorescence slower (Figure 5F’’) than the photobleached cells at 2 h. We also detect actively dividing cells within our photobleached region, suggesting that cells have not died as a result of the photobleaching process (Figure 5E’’).
We quantify these results in Figure 6A by showing the normalized signal intensity of each cell within the photobleached areas over a 30 min period after the photobleaching event. For each cell, the fluorescence values were normalized against the signal intensity of that cell immediately after photobleaching. This normalization was done for all cells across all time points imaged (45 min, 2 h, 5 h). The normalized values of fluorescence recovery of the cells within the same photo bleached region were then pooled together and graphed over time after photobleaching event; therefore, each point in the graph represents ~35 cells. The unnormalized data is also shown in Figure 6B, in which each line represents a cell with recovering fluorescence signal intensity immediately after photobleaching. This data overall suggests that by 5 h, all cells have depleted most of their mRNA, with a large portion of this drop occurring between 45 min and 2 h post-electroporation.
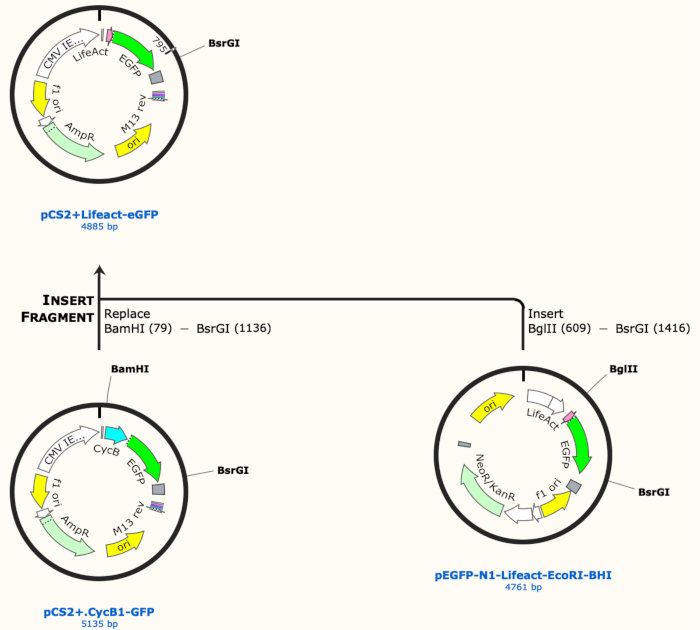
Figure 1: Schematic of cloning procedure to make pCS2+LifeAct-eGFP construct. Please click here to view a larger version of this figure.
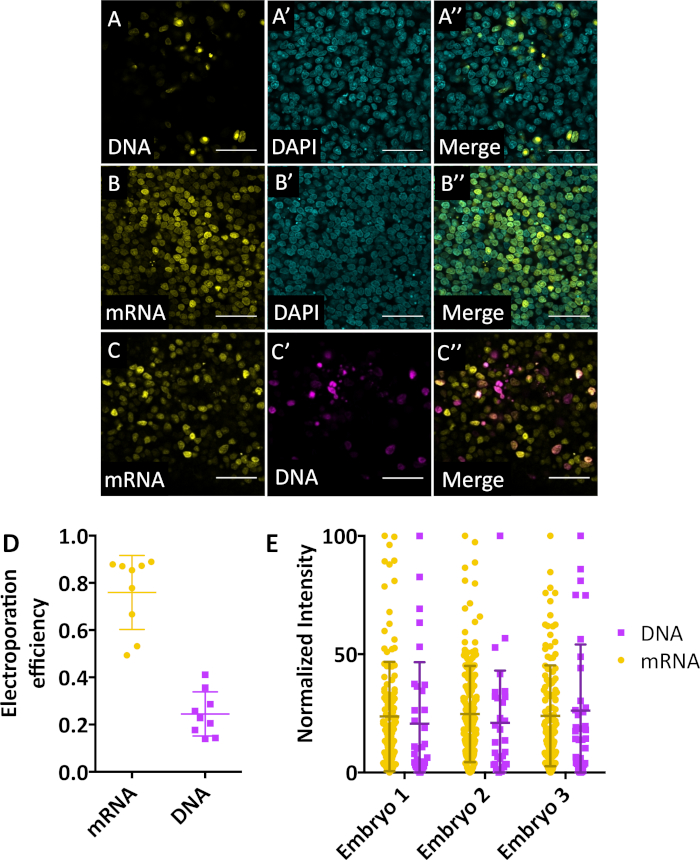
Figure 2: mRNA electroporation is more efficient than DNA electroporation. (A-A’’) DNA expressing pCS2.H2B-Citrine was electroporated into the ectoderm of HH5 embryos. Fluorescence was observed 12 h post-electroporation. Scale bar = 50 µm. (B-B’’) mRNA expressing H2B-Citrine was electroporated into the ectoderm of HH5 embryos. Fluorescence was observed 12 hours post-electroporation. Scale bar = 50 µm. (C-C’’) Equimolar combination of DNA (pCMV.H2B-mKate2) and mRNA (H2B-Citrine) was electroporated into HH5 embryos and observed 12 h post-electroporation. Three embryos were tested per condition (n = 2 experiments). Scale bar = 50 µm. (D) Three embryos from A, B, and C (9 total embryos) were quantified for electroporation efficiency using particle analysis tool on ImageJ. A 200 µm region from each embryo was quantified (at least 150 cells were counted per region). Middle dash represents mean, while top and bottom bars represent standard deviation from the mean. (E) Three co-electroporated (DNA and mRNA) embryos are analyzed for spread of signal across all electroporated cells in a given 200 µm region. Each embryo had around 100 cells positive for mRNA electroporation and 30 cells positive for DNA electroporation. Middle dash represents mean, while top and bottom bars represent standard deviation from the mean. There is no significant difference between the spread of mRNA vs. DNA electroporation, but mRNA electroporation is much more efficient than that of DNA. (A-C) Dimensions = 425.10 x 425.10 x 55 µm. Pixel dwell 2.55 µs. Average line 4. Bits per pixel = 16. Microscope metadata = inverted confocal microscope, 20x objective (see Table of Materials) (A) Ex: 488 nm (0.05%), Emission Track S1 = 508-579 nm; (B) Ex = 488 nm (5.0%), Emission Track S1 = 508-579 nm; (C) Ex = 488 nm (5.0%), 561 nm (5.0%), Emission Track S1 = 508-579 nm; S2 = 606-695 nm Please click here to view a larger version of this figure.
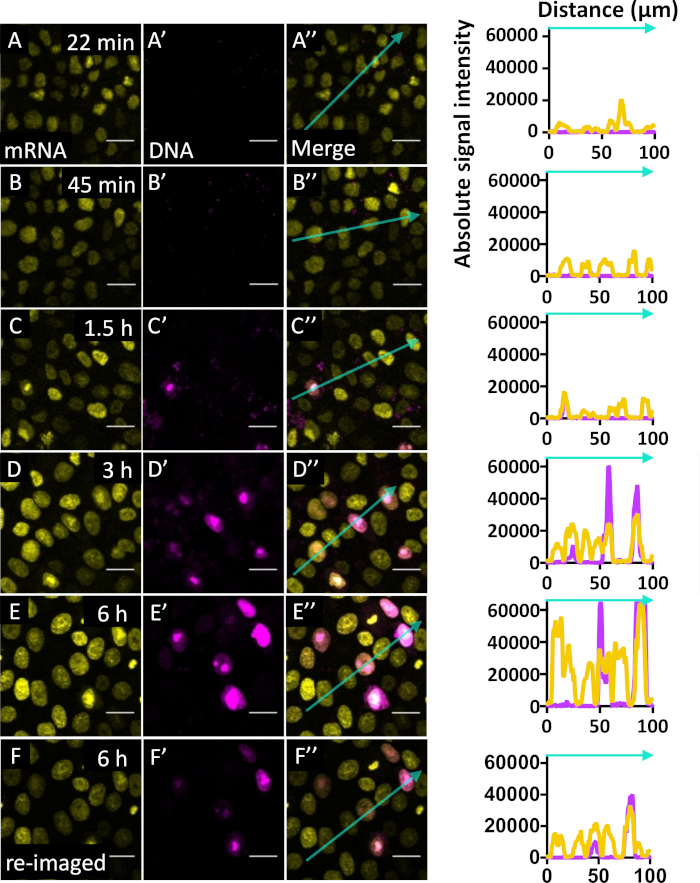
Figure 3: mRNA electroporation shows expression by 22 min post-electroporation and is much quicker than DNA electroporation. Representative images of time course after mRNA (H2B-Citrine) and DNA (pCMV.H2B-mKate2) electroporation with plot profile analysis at 22 min (A-A’’), 45 min (B-B’’), 1.5 h (C-C’’), 3 h (D-D’’), and 6 h (E-E’’) at the start of imaging (n = 1). Plot profile analysis is provided for each time point shown across the drawn arrow in the merged image. Each peak in the plot profile represents the nucleus of an electroporated cell. The cells generally become brighter over the 6 h interval of the movie, indicating that mRNA is still being translated into new fluorescent protein. For the 6 hour time point, images of the same region captured using identical imaging conditions (E-E’’) and weaker imaging conditions (F-F’’) are shown because the images of the DNA electroporated cells were highly saturated using the initial imaging conditions. Scale bar = 20 µm. Dimensions: 425.10 x 425.10 x 55 µm. Image is shown as a maximum intensity projection across all time points imaged. Pixel dwell 2.55 µs. Average line 4. Bits per pixel = 16. Microscope metadata = inverted confocal microscope, 20x objective (see Table of Materials) Ex = 488 nm (15%), 594 nm (5.0%) for initial; Ex = 488 nm (7.5%), 594 nm (0.5%) for last time point (Figure 2F), Emission Track S1 = 499-562 nm; S2 = 605-661 nm. Please click here to view a larger version of this figure.
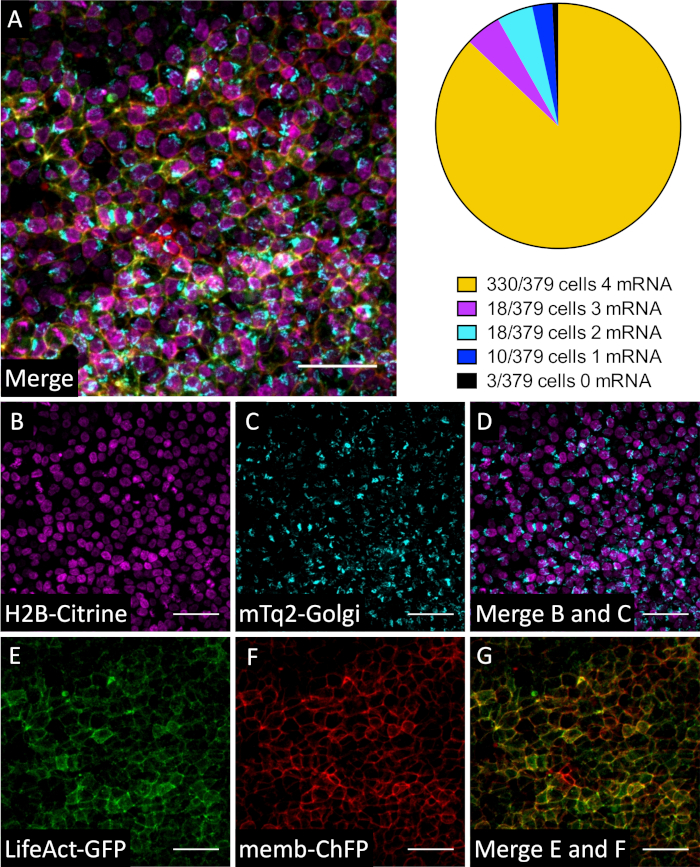
Figure 4: Co-electroporation of four mRNAs targeting different sub-cellular structures is highly efficient. The ectoderm of a HH3 embryo was electroporated with mRNAs that encode mTurquoise2-Golgi, LifeAct-eGFP, H2B-Citrine, and membrane-Cherry. The embryo was fixed and imaged at 4 hours post-electroporation. Most cells are electroporated within the electrode targeted region with all four mRNAs (87%). Representative figures shown (3 embryos each, n = 2 experiments). Scale bar = 50 µm. (A) Merge all four mRNAs (mTurquoise2-Golgi, LifeAct-eGFP, H2B-Citrine, and membrane-Cherry). (B) Only H2B-Citrine channel (Ex = 488 nm (0.7%), Em = 455-499 nm). (C) Only mTurquoise2-Golgi channel (Ex = 458 nm (22.0%), Em = 455-499 nm). (D) Merge H2B-Citrine and mTurquoise2-Golgi shows that Golgi envelopes one side of the nucleus in most cells. (E) Only LifeAct-eGFP channel (Ex = 488 nm (0.7%), Em: 455-499 nm). (F) Only Membrane-Cherry channel (Ex = 594 nm (37.1%), Em = S2: 570-695 nm). (G) Merge LifeAct-eGFP and membrane-Cherry shows overlap in most cells. Dimensions = 425.10 x 425.10. Pixel dwell 1.58 µs. Average line 4. Bits per pixel = 16. Microscope metadata = inverted confocal microscope, 20x objective (see Table of Materials) Ex = 405 nm (2.4%), 458 nm (22.0%), 488 nm (0.7%), 594 nm (37.1%), Emission Track S1 = 455-499 nm; S2 = 570-695 nm. Please click here to view a larger version of this figure.
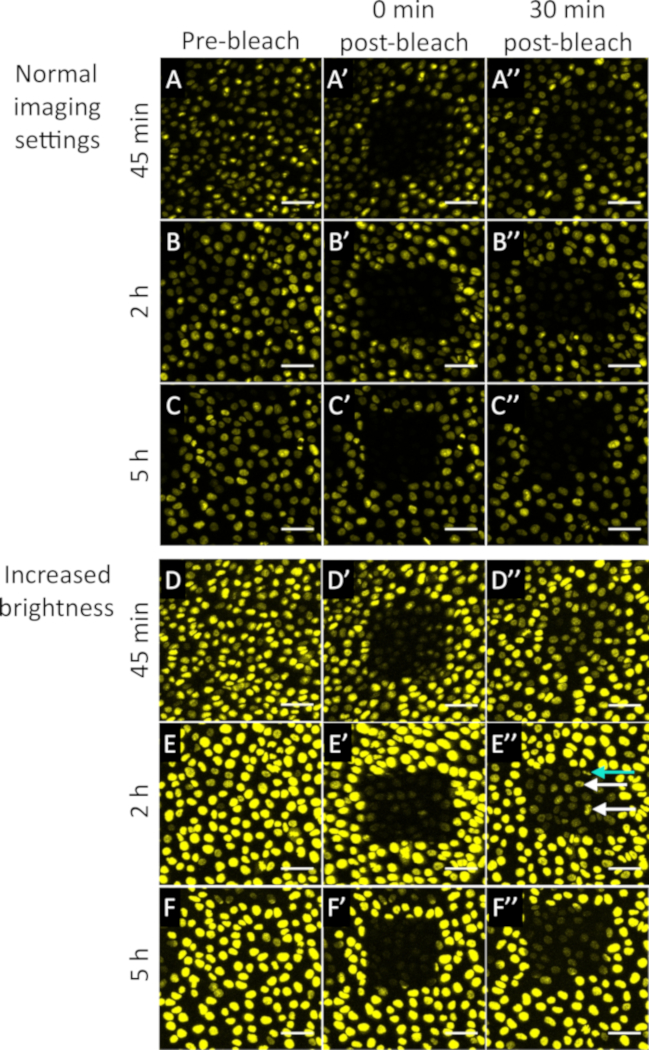
Figure 5: Photobleaching assay shows rapid degradation of electroporated mRNA during the first five hours post-electroporation. Photobleaching of a 100 µm² region containing cells electroporated with mRNA expressing H2B-Citrine is performed at 45 min, 2 h, and 5 h post-electroporation. The embryo was imaged at regular time intervals after the photobleaching (3 or 5 min). Photobleaching conditions are as follows: 70% 405 nm laser, 100 iterations, about 5 min duration for entire region. Scale bar 50 = µm. (A-A’’) 45 min pre- and post-bleach. B shows embryo immediately after photobleaching and C shows embryo 30 min post-bleach. Scale bar = 50 µm. (B-B’') 2 h pre- and post-bleach. The perimeter of the square is shifted because the cells within the photobleached region moved slightly during the bleach period. (C-C’’) 5 h pre- and post-bleach. (D-D’’) Same images as A-A’’ with enhanced brightness (maximum value adjusted to 11550 to enhance brightness post-collection). (E-E’’) Same images as B-B’’ with enhanced brightness (maximum value adjusted to 11550 to enhance brightness post-collection). The white arrowheads point to cells that have higher fluorescence recovery than surrounding cells. The cyan arrowhead points to a photobleached cell that has recently undergone mitosis. (F-F’’) Same images as C-C’’ with enhanced brightness (maximum value adjusted to 11550 to enhance brightness post-collection). Dimensions = 425.10 x 425.10 x 42 µm. Image is shown as a maximum intensity projection across all time points imaged. Pixel dwell 1.58 µs. Average line 4. Bits per pixel = 16. Microscope metadata = inverted confocal microscope, 20x objective (see Table of Materials) Ex = 488nm (1.5%), Emission Track S1 = 508-553 nm Please click here to view a larger version of this figure.
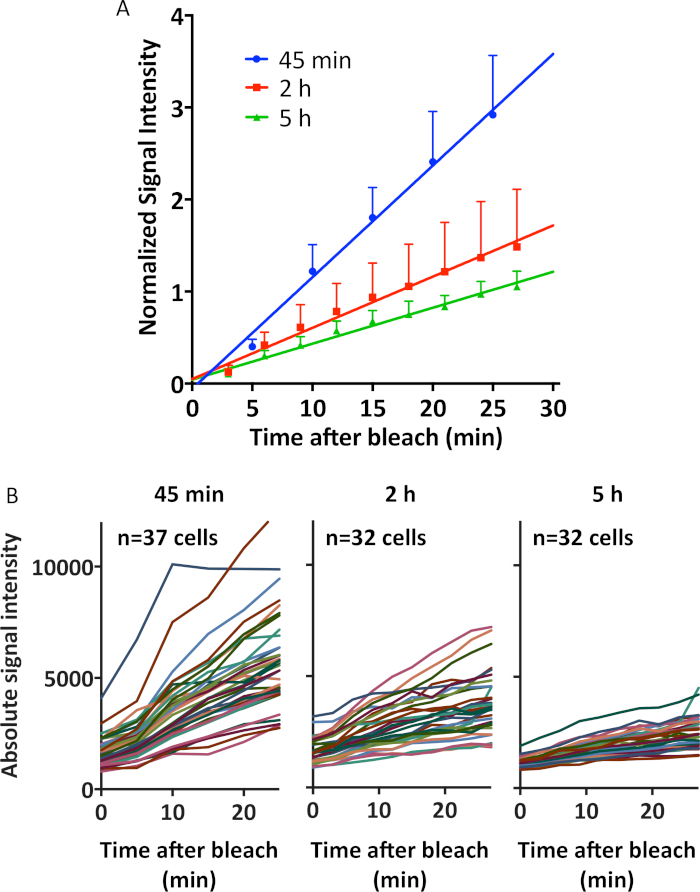
Figure 6: Quantification of the fluorescence recovery in photobleached cells. (A) The fluorescence intensity of all cells within the bleached regions (~35 cells) were tracked for each single cell during each time period post-bleach. There is a large drop in rate of recovered signal intensity from 45 min to 2 h, followed by a smaller drop from 2 h to 5 h. The top error bar is shown for each point, which represents standard deviation from the mean. The line shown is a linear regression line. R² for 45 min, 2 h, and 5 h is 0.83, 0.58, and 0.84 respectively. The top half of the error bar is shown for each point. (B) The fluorescence intensity of all cells within the bleached regions (~35 cells) were tracked for each single cell during each time period post-bleach and graphed without normalization. There is a large drop in rate of recovered signal intensity from 45 min to 2 h, followed by a smaller drop from 2 h to 5 h. Please click here to view a larger version of this figure.
Supplementary movie 1: mRNA electroporation leads to earlier protein expression than DNA electroporation. H2B-Citrine mRNA and H2B-mKate2 DNA electroporated into ectoderm of HH5 embryo. Region imaged is at the border of extra-embryonic and embryonic tissue. Scale bar = 50 µm. (a) Both H2B-Citrine and H2B-mKate2 channel shown (Ex = 488 nm (15%), 594 nm (5.0%), Em: 499-562 nm; S2: 605-661 nm). (b) Only mRNA H2B-Citrine channel shown (Ex = 488 nm (15%), Em = 499-562 nm). (c) Only H2B-mKate2 DNA channel shown (Ex = 594 nm (5.0%), Em = 605-661 nm). Dimensions = 850.19 x 850.19 x 110 µm. Image is shown as a maximum intensity projection across all time points imaged. Pixel dwell 2.55 µs. Average line 4. Bits per pixel = 16. Microscope metadata: inverted confocal microscope, 20x objective (see Table of Materials) Ex = 488 nm (15%), 594 nm (5.0%), Emission Track S1 = 499-562 nm; S2 = 605-661 nm. Please click here to download this file.
Supplementary movie 2: Co-electroporation of mRNAs permits distinct cell behaviors to be dynamically studied. Radial expansion of cells between the embryonic and extra-embryonic border are electroporated with mRNA (H2B-Citrine and mTurquoise2-Golgi) and imaged during outward migration. This movie was imaged by confocal microscopy from 2 to 5 h post-electroporation by imaging every 6 min. Scale bar = 50 µm. (a) Both H2B-Citrine and mTurquoise2-Golgi channels shown (Ex = 458 nm (28%), 488 nm (5.0%), Em = 446-526 nm; S2: 508-553 nm). (b) Only H2B-Citrine channel shown (Ex = 488 nm (5.0%), Em = S2: 508-553 nm). (c) Only mTurquoise2-Golgi channel shown (Ex = 458 nm (28%), Em = 446-526 nm). Dimensions = 425.10 x 425.10 x 32.5 µm. Image is shown as a maximum intensity projection across all time points imaged. Pixel dwell 0.79 µs. Average line 4. Bits per pixel = 16. Microscope metadata = inverted confocal microscope, 20x objective (see Table of Materials) Ex = 458 nm (28%), 488 nm (5.0%), Emission Track S1 = 446-526 nm; S2 = 508-553 nm. Please click here to download this file.
Discussion
In this protocol, we provided step by step instructions on how to precisely microinject and electroporate mRNA into the cells of gastrulating quail embryos. We demonstrated that in vitro synthesized mRNA electroporation allows fast and efficient expression of fluorescent proteins (FPs) in gastrulating quail embryos (Figure 2 and 3). Fluorescence from H2B-citrine protein translated from electroporated mRNAs could be detected by confocal microscopy within ~20 min and increased in FP fluorescence for hours (Figure 3, Supplemental Movie 1). This is surprisingly fast and close to the presumed maturation time for citrine32,33. FPs expressed from DNA vectors were detected after 2-3 h (Figure 3, Supplemental. Movie 1), which is similar to previous reports8,34,35. Various FPs have unique maturation times, so advanced planning will assure that the ideal FP is chosen in terms of the spectral distinction and maturation kinetics desired for the experiment. Additionally, the synthesized mRNAs more efficiently co-transfect embryonic cells than DNA vectors11. The higher transfection efficiency results in more cells being transfected in the region of interest with single (Figure 2) or multiple populations of mRNAs (Figure 4).
Stability of mRNA is highly regulated and can be affected by polyadenylation, splicing, translation, secondary structure, and untranslated regions to name a few36,37. The half-lives of both endogenous and synthesized mRNA within cells can vary from minutes to days36,38. We used in vivo fluorescence recovery after photobleaching (FRAP) to bleach existing mRNA-encoded H2B-citrine proteins and observed the most appreciable H2B-citrine fluorescence recovery during the first 2 hours post-electroporation (Figure 5 and 6). Different mRNAs will undoubtedly have different half-lives based on their length, internal sequences, 5’ and 3’ modifications36,37, so each must be assayed if desired.
Vital imaging typically requires a trade-off between optimum conditions for high-quality imaging and fast, less-toxic imaging39,40. When imaging, it is advisable to use as low laser power as possible to minimize photobleaching and photodamage to the embryonic cells (see notes for step 5.3 of the protocol)40. Altering the microscope setting for fast collections (e.g., faster scan speeds, less averaging) often can help without losing necessary image resolution41. Finally, care should be taken in handling and manipulating the embryos during harvesting, electroporation, and imaging (see step 4). The early-stage embryos are delicate and can be easily damaged.
Overall, the precise timing and accurate localization of mRNA electroporation permit perturbation experiments that capitalize on the rapid expression, co-transfection efficiency, and quick decay of mRNA transcripts. Over-expression or misexpression of a gene of interest can be accomplished through electroporation of synthesized mRNAs. Tittering the concentration for each mRNA is crucial, as electroporating too much mRNA may cause non-specific cellular toxicity, while too little mRNA may not label desired cells or induce phenotypic changes adequately. A plethora of vectors generated for in vitro synthesis of mRNAs that encode FP fusions markers for visualization or altered gene products for perturbations of diverse biological processes already exist from various animal model systems. Many of these constructs designed for in vitro mRNA synthesis can be quickly and inexpensively obtained from non-profit plasmid repositories.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank David Huss for helpful insights into this work. This work was supported in part by the Rose Hills Foundation Summer Research Fellowship (2016-2018) and USC Provost’s Undergrad Research Fellowship to M.T., the Saban Research Institute Intramural Training Pre-Doctoral Award to M.D., and the University of Southern California Undergraduate Research Associates Program award to R.L.
Materials
| BamHI-HF | New England Biolabs | R3136L | |
| BglII | New England Biolabs | R0144S | |
| BsrG1-HF | New England Biolabs | R3575S | |
| NotI-HF | New England Biolabs | R3189L | |
| SalI-HF | New England Biolabs | R3138L | |
| Phenol:Chloroform:Isoamyl Alcohol | Thermo Fisher | 15593031 | |
| SP6 mMessage Machine in vitro transcription kit | Thermo Fisher | AM1340 | |
| Fast Green FCF | Sigma Aldrich | F7252 | |
| Triton X-100 | Sigma Aldrich | 93443 | 4-(1,1,3,3-Tetramethylbutyl)phenyl-polyethylene glycol, t-Octylphenoxypolyethoxyethanol, Polyethylene glycol tert-octylphenyl ether |
| DAPI | Sigma Aldrich | D9542 | 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride, 4′,6-Diamidino-2-phenylindole dihydrochloride, DAPI dihydrochloride |
| Whatman No.1 filter paper | Sigma Aldrich | WHA1001125 | |
| glycerol | Sigma Aldrich | G9012 | |
| Urea | Sigma Aldrich | 51457 | |
| pmTurquoise2-Golgi | Addgene | 36205 | pmTurquoise2-Golgi was a gift from Dorus Gadella (Addgene plasmid # 36205 ; http://n2t.net/addgene:36205 ; RRID:Addgene_36205) |
| pmEGFP-N1-LifeAct | Nat. Methods 2008;5:605-7. PubMed ID: 18536722 | ||
| pCS2.Lifeact-mGFP | Addgene | This paper | |
| pCS.H2B-citrine | Addgene | 53752 | pCS-H2B-citrine was a gift from Sean Megason (Addgene plasmid # 53752 ; http://n2t.net/addgene:53752 ; RRID:Addgene_53752) |
| pCS.memb-mCherry | Addgene | #53750 | pCS-memb-mCherry was a gift from Sean Megason (Addgene plasmid # 53750 ; http://n2t.net/addgene:53750 ; RRID:Addgene_53750) |
| Zeiss LSM-780 inverted microscope | Carl Zeiss Microscopy GmbH | The LSM-780 is a confocal and multi-photon microscope that offers the sensitivity required for vital imaging work. Equipped with a motorized stage, an autofocus device, and a full stage-top blackout incubator, the 780 is an excellent microscope for high-end live cell/embryo imaging. The high-sensitivity 32-channel Quasar detector allows for spectral imaging, linear unmixing, and high color count (>4) image acquisition. Excitation can be performed with 6 lines single photon lasers (405, 458, 488, 514, 564 and 633 nm), Chameleon (Coherent) 2-photon laser (range from 690nm to 1000nm), and run with ZEN 2011 SP7 (Black) system software. | |
| CUY-21 EDIT in vivo electroporator | Bex Co., Ltd. | ||
| Platinum flat square electrode, side length 5 mm | Bex Co., Ltd. | LF701P5E | |
| Olympus MVX10 FL Stereo Microscope | Olympus LifeScience | ||
| XM10 Monochrome camera | Olympus LifeScience | ||
| Phosphate-Buffered Saline (PBS) for HCR (10×, pH 7.4) | To prepare 1 L of a 10× stock solution, combine 80 g of NaCl (Sigma-Aldrich S3014), 2 g of KCl (Sigma-Aldrich P9541), 11.4 g of Na2HPO4 (anhydrous; Sigma-Aldrich S3264), and 2.7 g of KH2PO4 (anhydrous; Sigma-Aldrich P9791). Adjust the pH to 7.4 with HCl, and bring the final volume to 1 L with ultrapure H2O. Avoid using CaCl2 and MgCl2 in PBS for HCR. It is important that the PBS for HCR is prepared as an RNase-free solution (e.g., via diethylpyrocarbonate [DEPC] treatment). | ||
| 1.37 M NaCl | |||
| 27 mM KCl | |||
| 80 mM Na2HPO4 20 mM KH2PO4 | |||
| PBS/Triton | Add 1 mL of Triton X-100 (Sigma Aldrich 93443) and 100 mL of 10× PBS to 890 mL of ultrapure distilled H2O. Filter the solution through a 0.2-μm filter and store it at 4 ̊C until use. | ||
| 1× phosphate-buffered saline (PBS) (DEPC-treated; pH 7.4) | |||
| 0.1% Triton X-100 |
References
- Neumann, E., Schaefer-Ridder, M., Wang, Y., Hofschneider, P. H. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO Journal. 1 (7), 841-845 (1982).
- Potter, H. Electroporation in biology: methods, applications, and instrumentation. Analytical Biochemistry. 174 (2), 361-373 (1988).
- Shillito, R. D., Saul, M. W., Paszkowski, J., Muller, M., Potrykus, I. High-Efficiency Direct Gene-Transfer to Plants. Bio-Technology. 3 (12), 1099-1103 (1985).
- Cui, C., Rongish, B., Little, C., Lansford, R. Ex Ovo Electroporation of DNA Vectors into Pre-gastrulation Avian Embryos. CSH Protocol. 2007 (24), 4894 (2007).
- Voiculescu, O., Papanayotou, C., Stern, C. D. Spatially and temporally controlled electroporation of early chick embryos. Nature Protocol. 3 (3), 419-426 (2008).
- Voiculescu, O., Stern, C. D. Manipulating Gene Expression in the Chick Embryo. Methods Molecular Biology. , 105-114 (2017).
- Chuai, M., et al. Cell movement during chick primitive streak formation. Developmental Bioliogy. 296 (1), 137-149 (2006).
- Krull, C. E. A primer on using in ovo electroporation to analyze gene function. Developmental Dynamics. 229 (3), 433-439 (2004).
- Momose, T., et al. Efficient targeting of gene expression in chick embryos by microelectroporation. Developmental, Growth and Differentiation. 41 (3), 335-344 (1999).
- Nakamura, H., Watanabe, Y., Funahashi, J. Misexpression of genes in brain vesicles by in ovo electroporation. Developmental, Growth and Differentiation. 42 (3), 199-201 (2000).
- Teddy, J. M., Lansford, R., Kulesa, P. M. Four-color, 4-D time-lapse confocal imaging of chick embryos. Biotechniques. 39 (5), 703-710 (2005).
- Green, M. R., Maniatis, T., Melton, D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 32 (3), 681-694 (1983).
- Mimoto, M. S., Christian, J. L. Manipulation of gene function in Xenopus laevis. Methods in Molecular Biology. 770, 55-75 (2011).
- Luni, C., et al. High-efficiency cellular reprogramming with microfluidics. Nature Methods. 13 (5), 446-452 (2016).
- Bugeon, S., et al. Direct and efficient transfection of mouse neural stem cells and mature neurons by in vivo mRNA electroporation. Development. 144 (21), 3968-3977 (2017).
- Riedl, J., et al. Lifeact: a versatile marker to visualize F-actin. Nature Methods. 5 (7), 605-607 (2008).
- Turner, D. L., Weintraub, H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes and Development. 8 (12), 1434-1447 (1994).
- Krieg, P. A., Melton, D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Research. 12 (18), 7057-7070 (1984).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chick embryo. Journal of Morphology. 88 (1), 49-92 (1951).
- Eyal-Giladi, H., Kochav, S. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stage of the development of the chick. I. General morphology. 발생학. 49, 321-337 (1976).
- Ainsworth, S. J., Stanley, R. L., Evans, D. J. Developmental stages of the Japanese quail. Journal of Anatomy. 216 (1), 3-15 (2010).
- Chapman, S. C., Collignon, J., Schoenwolf, G. C., Lumsden, A. Improved method for chick whole-embryo culture using a filter paper carrier. Developmental Dynamics. 220 (3), 284-289 (2001).
- Hama, H., et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature Neurosciences. 14 (11), 1481-1488 (2011).
- New, D. A new technique for the cultivation of the chick embryo in vitro. Journal of Embryology and Experimental Morphology. 3, 320-331 (1955).
- Drake, C. J., Davis, L. A., Hungerford, J. E., Little, C. D. Perturbation of beta 1 integrin-mediated adhesions results in altered somite cell shape and behavior. 발생학. 149 (2), 327-338 (1992).
- Sato, Y., et al. Dynamic analysis of vascular morphogenesis using transgenic quail embryos. PLoS ONE. 5 (9), e12674 (2010).
- Kanda, T., Sullivan, K. F., Wahl, G. M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Current Biology. 8 (7), 377-385 (1998).
- Uetrecht, A. C., Bear, J. E. Golgi polarity does not correlate with speed or persistence of freely migrating fibroblasts. European Journal of Cell Biology. 88 (12), 711-717 (2009).
- Alberts, B., et al. . Molecular Biology of the Cell. , (2007).
- Arndt-Jovin, D. J., Jovin, T. M. Fluorescence labeling and microscopy of DNA. Methods Cell Biol. 30, 417-448 (1989).
- Jovin, T. M., Arndt-Jovin, D. J. Luminescence digital imaging microscopy. Annu Rev Biophysics and Biophysical Chemistry. 18, 271-308 (1989).
- Heikal, A. A., Hess, S. T., Baird, G. S., Tsien, R. Y., Webb, W. W. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine). Proceedings of the National Academy of Science U S A. 97 (22), 11996-12001 (2000).
- Iizuka, R., Yamagishi-Shirasaki, M., Funatsu, T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Analytical Biochemistry. 414 (2), 173-178 (2011).
- Nakamura, H., Katahira, T., Sato, T., Watanabe, Y., Funahashi, J. Gain- and loss-of-function in chick embryos by electroporation. Mechanism of Development. 121 (9), 1137-1143 (2004).
- Swartz, M., Eberhart, J., Mastick, G. S., Krull, C. E. Sparking new frontiers: using in vivo electroporation for genetic manipulations. 발생학. 233 (1), 13-21 (2001).
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Critical Reviews in Biochemistry and Molecular Biology. 39 (4), 197-216 (2004).
- Bevilacqua, P. C., Ritchey, L. E., Su, Z., Assmann, S. M. Genome-Wide Analysis of RNA Secondary Structure. Annual Review in Genetics. 50, 235-266 (2016).
- Harland, R., Misher, L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 102 (4), 837-852 (1988).
- Lippincott-Schwartz, J. The long road: peering into live cells. Nature Cell Biology. 12 (10), 918 (2010).
- Sato, Y., Lansford, R. Transgenesis and imaging in birds, and available transgenic reporter lines. Development Growth & Differentiation. 55 (4), 406-421 (2013).
- Goldman, R. D., Spector, D. L. . Live Cell Imaging: A Laboratory Manual. , (2005).

