Application of Biolayer Interferometry (BLI) for Studying Protein-Protein Interactions in Transcription
Summary
Interactions of transcription factors (TFs) with the RNA polymerase are usually studied using pulldown assays. We apply a Biolayer Interferometry (BLI) technology to characterize the interaction of GrgA with the chlamydial RNA polymerase. Compared to pulldown assays, BLI detects real-time association and dissociation, offers higher sensitivity, and is highly quantitative.
Abstract
A transcription factor (TF) is a protein that regulates gene expression by interacting with the RNA polymerase, another TF, and/or template DNA. GrgA is a novel transcription activator found specifically in the obligate intracellular bacterial pathogen Chlamydia. Protein pulldown assays using affinity beads have revealed that GrgA binds two σ factors, namely σ66 and σ28, which recognize different sets of promoters for genes whose products are differentially required at developmental stages. We have used BLI to confirm and further characterize the interactions. BLI demonstrates several advantages over pulldown: 1) It reveals real-time association and dissociation between binding partners, 2) It generates quantitative kinetic parameters, and 3) It can detect bindings that pulldown assays often fail to detect. These characteristics have enabled us to deduce the physiological roles of GrgA in gene expression regulation in Chlamydia, and possible detailed interaction mechanisms. We envision that this relatively affordable technology can be extremely useful for studying transcription and other biological processes.
Introduction
Transcription, which produces RNA molecules using DNA as template, is the very first step of gene expression. Bacterial RNA synthesis begins following the binding of the RNA polymerase (RNAP) holoenzyme to a target promoter1,2. The RNAP holoenzyme (RNAPholo) is comprised of a multi-subunit catalytic core (RNAPcore) and a σ factor, which is required for recognizing the promoter sequence. Transcription activators and repressors, collectively termed TFs, regulate the gene expression through the binding components of the RNAPcore, σ factors, and/or DNA. Depending on the organism, a significant portion of its genome may be devoted to TFs that regulate transcription in response to physiological needs and environmental cues3.
Chlamydia is an obligate intracellular bacterium responsible for a variety of diseases in humans and animals4,5,6,7,8. For example, Chlamydia trachomatis is arguably the number one sexually transmitted pathogen in humans worldwide, and a leading cause of blindness in some underdeveloped countries4,5. Chlamydia has a unique developmental cycle characterized by two alternating cellular forms termed the elementary body (EB) and reticulate body (RB)9. Whereas, EBs are capable of survival in an extracellular environment, they are incapable of proliferation. EBs enter host cells through endocytosis and differentiate into larger RBs in a vacuole in the host cytoplasm within hours post-inoculation. No longer infectious, RBs proliferate through binary fission. Around 20 h, they start to differentiate back to the EBs, which exit the host cells around 30-70 h.
Progression of the chlamydial developmental cycle is regulated by transcription. Whereas a supermajority of the nearly 1,000 chlamydial genes are expressed during the midcycle during which RBs are actively replicating, only a small number of genes are transcribed immediately after the entry of EBs into the host cells to initiate the conversion of EBs into RBs, and another small set of genes are transcribed or increasingly transcribed to enable the differentiation of RBs into EBs10,11.
The chlamydial genome encodes three σ factors, namely σ66, σ28 and σ54. σ66, which is equivalent to the housekeeping σ70 of E. coli and other bacteria, is responsible for recognizing promoters of early and mid-cycle genes as well as some late genes, whereas σ28 and σ54 are required for the transcription of certain late genes. Several genes are known to carry both a σ66-dependent promoter and a σ28-dependent promoter12.
Despite a complicated developmental cycle, only a small number of TFs have been found in chlamydiae13. GrgA (previously annotated as a hypothetical protein CT504 in C. trachomatis serovar D and CTL0766 in C. trachomatis L2) is a Chlamydia-specific TF initially recognized as an activator of σ66-dependent genes14. Affinity pulldown assays have demonstrated that GrgA activates their transcription by binding both σ66 and DNA. Interestingly, it was later found with that GrgA also co-precipitates with σ28, and activates transcription from σ28-dependent promoters in vitro15. To investigate whether GrgA has similar or different affinities for σ66 and σ28, we resorted to using BLI. BLI assays have shown that GrgA interacts with σ66 at a 30-fold higher affinity than with σ28, suggesting that GrgA may play differential roles in σ66-dependent transcription and σ28-dependent transcription15.
BLI detects the interference pattern of white light that reflects from a layer of immobilized protein on the tip of a biosensor and compares it to that of an internal reference layer16. Through the analysis of these two interference patterns, BLI can provide valuable and real-time information about the amount of protein bound to the tip of the biosensor. The protein that is immobilized to the tip of the biosensor is referred to as the ligand, and is generally immobilized with the help of a common antibody or epitope tag (e.g., a poly-His- or biotin-tag) that has an affinity for an associated particle (such as NTA or Streptavidin) on the tip of the biosensor. The binding of a secondary protein, referred to as the analyte, with the ligand at the tip of the biosensor creates changes in the opacity of the biosensor and therefore results in changes in interference patterns. When repeated over different concentrations of the analyte, BLI can provide not only qualitative but also quantitative information about the affinity between the ligand and analyte16.
To the best of our knowledge, we were the first to employ BLI to characterize protein-protein interactions in transcription15. In this publication, we demonstrate that a GrgA fragment, which was previously shown to be required for σ28-binding, indeed mediates the binding. This manuscript focuses on steps of the BLI assays, and generation of BLI graphs and parameters of binding kinetics. Methods for the production (and purification) of ligands and analytes are not covered here.
Protocol
1. Preparation of proteins
- Use a dialysis bag (with an appropriate cut-off size) to dialyze each protein to be used for BLI assays (including both the His-tagged ligand and the analyte) against 1,000 volumes of the BLI buffer (25 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2, 0.1 mM DTT, pH 8.0, pre-chilled to 4 ˚C) at 4 ˚C for 4 h.
NOTE: BLI assays require the ligand to be present at concentrations that saturate the binding sites on the biosensor and the analyte to be highly purified so that the molar concentrations of the analyte that react with the ligand is known. Methodologies for the expression and purification of His- and Strep-tagged proteins are not covered here, but can be found in previous publications14,15. Although this system does not require the ligand to be in highly purified form, it is essential to dialyze even unpurified ligands to the BLI buffer in order to minimize shifts in white-light interference patterns caused by any buffer changes during the assay. - Switch to fresh BLI buffer and continue the dialysis for another 4 h.
2. Biosensor hydration and assay set-up
- Approximately 10 min prior to the start of an assay, pipette 200 µL of the BLI buffer into a PCR tube.
- Remove a Ni-NTA-biosensor from the original packaging by holding the wide portion of the biosensor using a gloved hand.
- Place the biosensor over the PCR-tube such that only the glass tip of the biosensor is submerged in the BLI buffer.
- Keep the biosensor tip submerged for at least 10 min to ensure full hydration.
- Verify that the glass tip of the Ni-NTA-biosensor does not touch anything other than the BLI buffer during the above step.
NOTE: This protocol uses a Ni-NTA-biosensor in conjunction with a His-tagged ligand. If needed, a SA-Streptavidin-biosensor can be used in conjunction with a biotinylated ligand instead if: (i) both the ligand and analyte carry a His tag or (ii) neither of them does.
- Verify that the glass tip of the Ni-NTA-biosensor does not touch anything other than the BLI buffer during the above step.
- Turn the BLItz machine on.
- Ensure that the machine is connected to the computer through a USB data output port at the back of the machine.
- On the computer, open the associated software (e.g., BLItz Pro), and click on Advanced Kinetics on the left-hand side of the screen.
- On the software, type out all appropriate information about the experiment (including the Experiment Name, Description, Sample ID, and Protein Concentration) under each respective heading.
- Click on Biosensor Type and choose Ni-NTA from the drop-down menu.
- Under the Run Settings heading, verify that the Shaker is set to Enable.
- Under the Step Type List heading, verify that there are 5 items listed: Initial Baseline, Loading, Baseline, Association, and Dissociation.
NOTE: The duration of each step can be changed from default as needed. For optimal results, use a minimum of 30 s for Initial Baseline and Baseline; and 120 s for Association and Dissociation. The duration of the Loading step (ranging from 120 to 240 s) will depend upon the concentration of the ligand and affinity of the His-epitope tag on the ligand to the Ni-NTA-biosensor.
- Remove the hydrated Ni-NTA-biosensor from the PCR tube and affix it to the biosensor mount on the machine by sliding the wide portion of the biosensor onto the mount.
NOTE: Do not let the biosensor dry out during the experiment. - Place a 0.5 mL black microcentrifuge tube into the tube holder of the machine and pipette 400 µL of the BLI buffer into it.
- Verify that the slider of the machine is positioned such that the tube holder is situated in front of the black arrow on the machine.
- Close the cover of the machine such that the biosensor tip becomes submerged in the buffer in the microcentrifuge tube.
- Click Next on the software to begin recording the Initial Baseline.
3. Loading of ligand onto biosensor
- After the Initial Baseline step has finished recording, open the cover of the machine.
- Move the slider to the right such that the drop holder (instead of the tube holder) is situated in front of the black arrow.
- Pipette 4 µL of a dialyzed His-tagged ligand (from Step 1.1) onto the drop holder and close the cover of the machine.
NOTE: The optimal concentration of the ligand to be used may vary for each protein. A concentration between 1.0 to 2.0 mg/mL is usually adequate to saturate the NTA at the tip of the biosensor in 240 s. - On the software, click Next to begin Loading.
4. Washing away additional ligand
- After the Loading step has finished recording, open the cover of the machine.
- Move the slider to the left such that the tube holder is once again situated in front of the black arrow.
- Close the lid of the machine and ensure that the biosensor tip is submerged into the BLI buffer of the tube in the tube holder.
- Click Next once again on the software to begin recording the Baseline.
5. Association of analyte to ligand
- After the Baseline step has finished recording, open the cover of the machine.
- Remove the drop holder and clean it by pipetting out any protein and rinsing it with double-deionized water (ddH2O) for a total of 5 times.
- Use a tissue wipe to clean the surface of the drop holder after the wash.
- Replace the drop holder back onto the machine.
- Move the slider on the machine to the right such that the drop holder is once again situated in front of the black arrow.
- Pipette 4 µL of a dialyzed analyte (from Step 1.1) onto the drop holder and close the cover of the machine.
- On the software, click Next to begin Association.
6. Dissociation of analyte from ligand
- After the Association step has finished recording, open the cover of the machine.
- Move the slider on the machine to the right such that the tube holder is once again situated in front of the black arrow.
- On the software, click Next to begin Dissociation.
- After the Dissociation step has finished recording, open the cover of the machine.
- Remove the drop holder and tube holder.
- Rinse both with ddH2O thoroughly to wash away any protein.
- Remove the biosensor and discard it safely.
7. Repeating interactions with different concentrations
- Repeat Steps 2-7 for the same ligand-analyte pair using different analyte concentrations.
NOTE: The concentration of the analyte may need to be adjusted across several runs before obtaining optimal results. In our experience, a ratio of 1:5:10 of analyte concentrations, starting with 75 nM, is usually adequate.
8. Analyzing the data using the software
- Once all runs have finished, save the data on the software by clicking File and then Save Experiment As on the left side of the screen.
- Under the Run Data heading, select Step Correction and Fitting (1:1) and click Analyze to generate kinetic data.
- To extract the quantitative data into a worksheet and generate graphs, click on Export to CSV and save the recorded data as a .csv file. Open the .csv file using spreadsheet software.
- To most effectively show the Association and Dissociation kinetics, remove all plot points prior to the Baseline step, and normalize all subsequent plot points from the final Baseline value.
Representative Results
Through BLI assays, we previously established that binding of GrgA to σ28 is dependent on a 28 amino acid middle region (residues 138-165) of GrgA15. Accordingly, compared with N-terminally His-tagged full length GrgA (NH-GrgA), a GrgA deletion construct lacking this region (NH-GrgAΔ138-165) had a decreased association rate and an increased dissociation rate, leading to a 3 million-fold loss of overall affinity (Table 1). Here, we demonstrate that this middle region directly binds σ28 in the absence of the rest of the GrgA protein. In these experiments, the middle region tagged with an N-terminally His-tag (NH-GrgA138-165) was used as the ligand, which was first immobilized to the tip of a Ni-NTA biosensor (Figure 1A). After washing unbound NH-GrgA138-165 off the biosensor, real-time association with the analyte σ28 was recorded following the addition of σ28. Finally, the real-time dissociation was recorded following the wash. Recordings of experiments with three different analyte concentrations starting 30 s prior to ligand binding and ending 2 minutes after the beginning of wash are shown in Figure 1A. To better visualize the ligand-analyte interaction, we remove data prior to the addition of the ligand and reset the baseline to 0 to derive Figure 1B.
Values of kinetic parameters for interaction of the NH-GrgA138-165 fragment with σ28 are presented in Table 1. Compared to the NH-GrgA X σ28 interaction, the NH-GrgA138-165 X σ28 interaction displayed a trending statistically significant 60% reduction in ka, a highly statistically significant 64% increase in kd, and a highly statistically significant 3.5-fold increase in KD. These changes demonstrate that compared to NH-GrgA, NH-GrgA138-165 binds σ28 more slowly, dissociates from σ28 faster, and has a decreased overall affinity with σ28. Therefore, residues 138-165 in GrgA binds σ28 but with reduced affinity compared to full length GrgA.
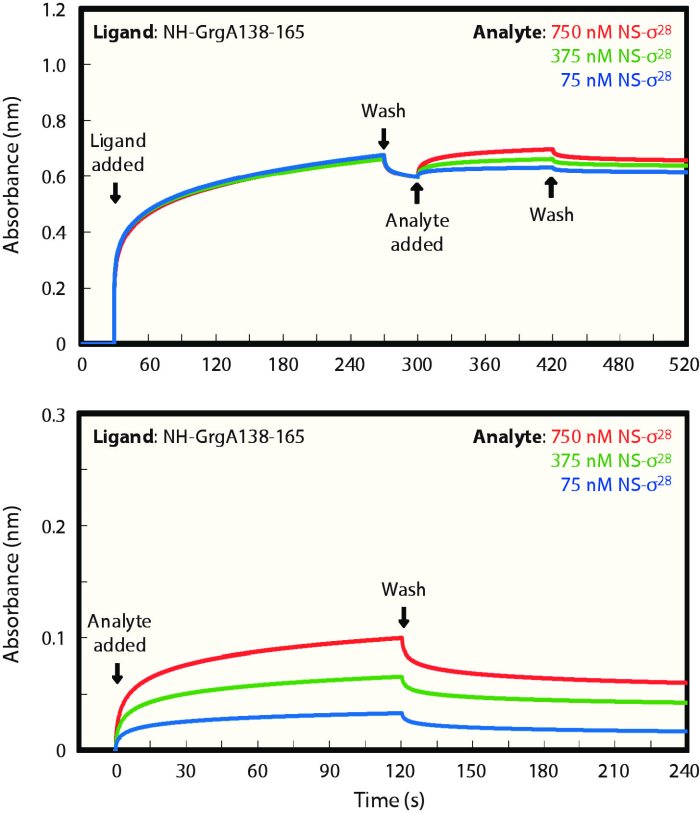
Figure 1: A 28 amino acid middle region of GrgA binds σ28 in vitro.
(A) Real-time changes in light interference patterns recorded by in four stages: (i) binding of NH-GrgA138-165 (Ligand) to a Ni-NTA biosensor, (ii) wash, (iii) binding of NS-σ28 (Analyte) at different concentrations to the immobilized NH-GrgA-138-165 (Ligand), and (iv) subsequent wash. (B) Enhanced visualization of ligand-analyte association and dissociation following removal of values in the first two stages from (A) and reset of the baseline. Panel B is modified from Desai et al., 201815. Please click here to view a larger version of this figure.
| Ligand | n | ka | kd | KD | References | |||||||||||
| 1/Ms | % control | 1/s | % control | M | % control | |||||||||||
| NH-GrgA | 8 | (1.5 ± 1.7) x 104 | 100 | (2.8 ± 0.8) x 10-3 | 100 | (2.2 ± 0.3) x 10-7 | 100 | Desai et al, 2018 | ||||||||
| NH-GrgAΔ138-165 | 2 | (5.6 ± 0.1) x 103 | 37 | (4.1 ± 0.3) x 102 | 1.5 x 107 | (6.9 ± 4.5) x 10-2 | 3.1 x 108 | Desai et al, 2018 | ||||||||
| p=0.125 | p<0.002 | p<0.001 | ||||||||||||||
| NH-GrgA138-165 | 3 | (6.0 ± 1.0) x 103 | 40 | (4.6 ± 0.4) x 10-3 | 164 | (7.7 ± 0.3) x 10-7 | 350 | This study | ||||||||
| p=0.074 | p=0.006 | p<0.001 | ||||||||||||||
Table 1: A mutant of GrgA, containing only amino acid residues 138-165, binds σ28 despite lower affinity compared to the full-length GrgA.
BLI assays were performed with Ni-NTA biosensors using His-tagged full-length GrgA or deletion mutants as ligands and purified Strep-tagged σ28 as an analyte. Graphs of recordings are shown in Figure 1. Values of kinetic parameters (averages ± standard deviations) were generated with the associated software17. ka (association rate constant) is defined as the number of complexes formed per s in a 1 molar solution of A and B. kd (dissociation rate constant) is defined as the number of complexes that decay per second. KD (dissociation equilibrium constant), defined as the concentration at which 50% of ligand binding sites are occupied by the analytes, is kd divided by ka. n, number of experimental repeats. p values were calculated using 2-tailed Student’s t tests. Kinetic parameters for NH-GrgA and NH-GrgAΔ138-165 were from Desai et al., 201815.
Discussion
Protein-protein interactions are crucial for the regulation of transcription and other biological processes. They are most commonly studied through pulldown assays. Although pulldown assays are relatively easy to perform, they are poorly quantitative and may fail to detect weak but biologically meaningful interactions. In comparison, by detecting real-time association and dissociation between a ligand and an analyte, BLI provides association and dissociation rate constants, as well as, overall affinity.
Compared to pulldown assays, BLI assays offer higher sensitivity. For example, GrgA-σ28 interactions are detected with lower nM concentrations of analytes by BLI but not by pulldown assays (unpublished data). Unlike pulldown, BLI does not rely on a detection antibody, which may significantly affect sensitivity.
More importantly, BLI analyses can provide mechanistic insights into the interaction between proteins, whereas pulldown assays cannot. This is exemplified by the interactions of σ28 with different GrgA constructs. Compared with NH-GrgA, NH-GrgAΔ138-165 and NH-GrgA138-165 suffer only a 60% loss in ka in binding σ28. These findings are consistent with our previous BLI data showing that GrgA lacking its N-terminal 64 residues has a decreased affinity with σ28, suggesting that the N-terminal sequence of GrgA contributes to σ28 binding. Although NH-GrgAΔ138-165 and NH-GrgA138-165 have similar ka values in binding σ28, the former has a 91,000-fold higher kd than the latter. These results indicate that binding of 138-165 triggers structural changes in GrgA, greatly stabilizing the complex.
With a longer history than BLI, surface plasmon resonance (SPR) can also quantify the real-time protein-protein interactions18,19. While the sensitivity of BLI is thought to be lower than that of SPR20, the former currently outperforms the latter in cost-effectiveness. For example, costs of SPR biosensors are much higher than those of BLI biosensors.
Due to the nature of the underlying principle of SPR, it is heavily influenced by the microfluidics of the media surrounding the protein. Therefore, experiments involving some SPR instruments require considerable perception on the part of the researcher to ensure optimal buffer conditions21,22,23,24. On the other hand, current BLI instruments feature a very limited temperature control range25 and, as such, are ill-fitted for determining thermodynamic parameters (such as enthalpy and Gibbs free energy) for a given interaction.
Glycerol, a commonly used cryoprotectant, is incompatible with BLI, despite its broad chemical compatibility. Therefore, it is critical to remove glycerol from the ligand and analyte by dialysis. The resulting glycerol-free proteins must be stored at 4 °C, which may lead to increased instability and inaccurate kinetic parameters. We recommend that BLI assays be performed soon after dialysis, particularly if inconsistent kinetic parameters are obtained from at different times. The exact time frame within which BLI assays should be completed will vary among proteins and be affected by their concentrations.
As with SPR, BLI has been used for small molecule screening26. Considering that newer BLI instruments offer high throughput options for screening, we envision that BLI can become very useful for the identification and characterization of small molecules that facilitate or interfere with protein-protein interactions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Institutes of Health (Grants # AI122034 and AI140167) and New Jersey Health Foundation (Grant # PC 20-18).
Materials
| BLItz machine | ForteBio | 45-5000 | |
| Dialysis tubing cellulose membrane | MilliporeSigma | D9652 | |
| Dip and Read Ni-NTA biosensor tray | ForteBio | 18-5101 | Ready-to-use Ni-NTA biosensors for poly-His-tagged Proteins |
| Drop holder | ForteBio | 45-5004 | |
| PCR tubes (0.2 mL) | Thomas Scientific | CLS6571 | |
| Microcentrifuge tubes (black) | Thermo Fisher Scientific | 03-391-166 | |
| Kimwipes | Thermo Fisher Scientific | 06-666A | |
| DTT | Thermo Fisher Scientific | R0861 | |
| EDTA | MilliporeSigma | E6758 | |
| MgCl2 | MilliporeSigma | M8266 | |
| NaCl | MilliporeSigma | S9888 | |
| Tris-HCl | GoldBio | T095100 |
References
- Vvedenskaya, I. O., et al. Interactions between RNA polymerase and the core recognition element are a determinant of transcription start site selection. Proceedings of the National Academy of Sciences of the U.S.A. 113 (21), E2899-E2905 (2016).
- Feklistov, A., Sharon, B. D., Darst, S. A., Gross, C. A. Bacterial sigma factors: a historical, structural, and genomic perspective. Annual Review of Microbiology. 68, 357-376 (2014).
- Visweswariah, S. S., Busby, S. J. Evolution of bacterial transcription factors: how proteins take on new tasks, but do not always stop doing the old ones. Trends in Microbiology. 23 (8), 463-467 (2015).
- Taylor, H. R., Burton, M. J., Haddad, D., West, S., Wright, H. Trachoma. Lancet. 384 (9960), 2142-2152 (2014).
- WHO. Global incidence and prevalence of selected curable sexually transmitted infections: 2008. Sexual and Reproductive Health Matters. 20, (2012).
- Schmidt, S. M., Muller, C. E., Mahner, B., Wiersbitzky, S. K. Prevalence, rate of persistence and respiratory tract symptoms of Chlamydia pneumoniae infection in 1211 kindergarten and school age children. The Pediatric Infectious Disease Journal. 21 (8), 758-762 (2002).
- De Puysseleyr, K., et al. Evaluation of the presence and zoonotic transmission of Chlamydia suis in a pig slaughterhouse. BMC Infectious Diseases. 14 (560), (2014).
- Hulin, V., et al. Host preference and zoonotic potential of Chlamydia psittaci and C. gallinacea in poultry. Pathogens and Disease. 73 (1), 1-11 (2015).
- Belland, R., Ojcius, D. M., Byrne, G. I. Chlamydia. Nature Review of Microbiol. 2 (7), 530-531 (2004).
- Belland, R. J., et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proceedings of the National Academy of Sciences of the USA. 100 (14), 8478-8483 (2003).
- Nicholson, T. L., Olinger, L., Chong, K., Schoolnik, G., Stephens, R. S. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. Journal of Bacteriology. 185 (10), 3179-3189 (2003).
- Tan, M., Tan, M., Bavoil , P. M. . Intracellular pathogens I: Chlamydiales. , 149-169 (2012).
- Domman, D., Horn, M. Following the Footsteps of Chlamydial Gene Regulation. Molecular Biology and Evolution. 32 (12), 3035-3046 (2015).
- Bao, X., Nickels, B. E., Fan, H. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of σ66. Proceedings of the National Academy of Sciences of the USA. 109 (42), 16870-16875 (2012).
- Desai, M., et al. Role for GrgA in regulation of σ28-dependent transcription in the obligate intracellular bacterial pathogen Chlamydia trachomatis. Journal of Bacteriology. 200 (20), (2018).
- Joy, C., et al. Label-Free Detection of Biomolecular Interactions Using BioLayer Interferometry for Kinetic Characterization. Combinatorial Chemistry & High Throughput Screening. 12 (8), 791-800 (2009).
- Abdiche, Y., Malashock, D., Pinkerton, A., Pons, J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Analytical Biochemistry. 377 (2), 209-217 (2008).
- Szabo, A., Stolz, L., Granzow, R. Surface plasmon resonance and its use in biomolecular interaction analysis (BIA). Current Opinion in Structural Biology. 5 (5), 699-705 (1995).
- Nelson, R. W., Krone, J. R. Advances in surface plasmon resonance biomolecular interaction analysis mass spectrometry (BIA/MS). Journal of Molecular Recognition. 12 (2), 77-93 (1999).
- Yang, D., Singh, A., Wu, H., Kroe-Barrett, R. Comparison of biosensor platforms in the evaluation of high affinity antibody-antigen binding kinetics. Analytical Biochemistry. 508, 78-96 (2016).
- Visentin, J., et al. Overcoming non-specific binding to measure the active concentration and kinetics of serum anti-HLA antibodies by surface plasmon resonance. Biosensors and Bioelectronics. 117, 191-200 (2018).
- Baba, A., Taranekar, P., Ponnapati, R. R., Knoll, W., Advincula, R. C. Electrochemical surface plasmon resonance and waveguide-enhanced glucose biosensing with N-alkylaminated polypyrrole/glucose oxidase multilayers. ACS Applied Materials & Interfaces. 2 (8), 2347-2354 (2010).
- Del Vecchio, K., Stahelin, R. V. Using Surface Plasmon Resonance to Quantitatively Assess Lipid-Protein Interactions. Methods in Molecular Biology (Clifton, N.J.). 1376, 141-153 (2016).
- Wang, D. S., Fan, S. K. Microfluidic Surface Plasmon Resonance Sensors: From Principles to Point-of-Care Applications. Sensors (Basel, Switzerland). 16 (8), 1175 (2016).
- Shah, N. B., Duncan, T. M. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. Journal of visualized experiments : JoVE. (84), e51383 (2014).
- Wartchow, C. A., et al. Biosensor-based small molecule fragment screening with biolayer interferometry. Journal of Computer-Aided Molecular Design. 25 (7), 669-676 (2011).

