Sample Preparation for Probe Electrospray Ionization Mass Spectrometry
Summary
This article introduces sample preparation methods for a unique real-time analytical method based on the ambient mass spectrometry. This method lets us perform real-time analysis of the biological molecules in vivo without any special pretreatments.
Abstract
Mass spectrometry (MS) is a powerful tool in analytical chemistry because it provides very accurate information about molecules, such as mass-to-charge ratios (m/z), which are useful to deduce molecular weights and structures. While it is essentially a destructive analytical method, recent advancements in the ambient ionization technique have enabled us to acquire data while leaving tissue in a relatively intact state in terms of integrity. Probe electrospray ionization (PESI) is a so-called direct method because it does not require complex and time-consuming pretreatment of samples. A fine needle serves as a sample picker, as well as an ionization emitter. Based on the very sharp and fine property of the probe tip, destruction of the samples is minimal, allowing us to acquire the real-time molecular information from living things in situ. Herein, we introduce three applications of PESI-MS technique that will be useful for biomedical research and development. One involves the application to solid tissue, which is the basic application of this technique for the medical diagnosis. As this technique requires only 10 mg of the sample, it may be very useful in the routine clinical settings. The second application is for in vitro medical diagnostics where human blood serum is measured. The ability to measure fluid samples is also valuable in various biological experiments where a sufficient volume of sample for conventional analytical techniques cannot be provided. The third application leans toward the direct application of probe needles in living animals, where we can obtain real-time dynamics of metabolites or drugs in specific organs. In each application, we can infer the molecules that have been detected by MS or use artificial intelligence to obtain a medical diagnosis.
Introduction
Mass spectrometry (MS) is a technological realization of reductionism; it reduces the object of analysis to a unit that can be interpreted on the basis of molecular species or cascades. Therefore, it is a representative method of analytical chemistry. It is made up of four processes: ionization, analysis, detection, and spectral acquisition. Because ionization of the molecule is the first process in mass spectrometry, it generally restricts the form of the analytes to be processed. Most ionization procedures require the destruction of the structure, morphology, and real-time biological processes of organic samples. For example, electrospray ionization (ESI) MS requires that the samples be in a liquid state for efficient ionization1. Samples, therefore, must go through a complex biochemical preparation, which alters the composition of molecules. Alternatively, while matrix-assisted laser desorption ionization (MALDI) MS can reconstruct molecular maps of thin sectioned tissue2,3, its ionization efficiency is too low to detect all molecules in the samples, and it is particularly poor at analyzing fatty acids. Considering these limitations, probe electrospray ionization (PESI)4 can be used to observe the real-time changes in biological systems in situ without destroying the structural integrity5, while the biological organism being observed is technically in a living state. A very fine needle is used in this case that serves simultaneously as a sample picker and an ion emitter. This means that the complex sample pretreatment sequences can be bypassed to obtain mass spectra that reflect the molecular components of the living system in situ.
There are several other ionization methods that rival PESI-MS. One is rapid evaporative ionization mass spectrometry (REIMS)6. This technique works well during surgery because it is assembled with an electrical knife and collects the ion plume generated during incision. While REIMS is very useful for the surgery, it is essentially a destructive method that requires the electrical ablation of the tissue. Therefore, it is not useful for the detailed analysis of cells and tissues in a preparative sample or in laboratory analyses. Moreover, because it collects a large amount of plume containing tissue debris, it requires lengthy maintenance of the devices after each use, thus limiting the use of this machine to special surgical procedures. A similar method, called laser desorption ionization mass spectrometry (LDI-MS)7, is another technique that is noninvasive and useful for the surface analysis. Because this technique is good at scanning the surface of a specimen, it achieves comprehensive two-dimensional analysis like MALDI imaging mass spectrometry8,9. However, because LDI-MS is only applicable to the surface analysis, PESI-MS is advantageous for analyzing the samples e.g., within the tissue. Another technique, the MasSpec Pen10, was reported to achieve high specificity and sensitivity in diagnosing thyroid cancer, but the diameter of the probe is in the order of mm and it is specific for the surface analysis, meaning that it cannot detect small nodules of cancer or deeply localized lesions. Moreover, as this method uses a microcapillary flow canal embedded in the probe pen, cross-contamination must be taken into consideration, similar to LDI-MS. Other techniques exist that have been applied to clinical settings, such as the flow probe and ionization form swab11, but they are not widespread.
PESI is extreme miniaturization of ESI, wherein the capillary of the nano-electrospray converges on a solid needle with a tip curvature radius of several hundred nm. Ionization takes place in the extremely restricted area of the needle tip by forming a Taylor cone, on which samples remain until ionization of all the fluid on the tip is completed12. If the analyte stays on the tip of the metal needle, excess charge is continuously generated at the interface between the metal needle and the analytes. Therefore, sequential ionization of molecules occurs depending on their surface activity. This property makes the needle tip a kind of chromatogram, separating the analytes depending on their surface activity. More technically, molecules with the stronger surface activity come to the surface of the Taylor cone and are ionized earlier than those with weaker surface activity, which adhere to the surface of the needle until the end of the ionization process. Thus, complete ionization of all molecules picked up by the needle is achieved13. Moreover, because this technique does not involve the addition of superfluous solvent to the sample, several hundreds of femtoliters are sufficient to get mass spectra strong enough for further analysis14. These properties are advantageous for the analysis of intact biological samples. However, a major disadvantage of PESI-MS lies in the discontinuity in ionization because of the reciprocating movement of the needle along the vertical axis, similar to a sawing machine. Ionization only takes place when the tip of the probe reaches the highest point when the height of the ion orifice is aligned on the horizontal axis. Ionization ceases while the needle picks up samples, and so the stability of ionization is not equal to that in conventional ESI. Therefore, PESI-MS is not an ideal method for proteomics.
To date, PESI-MS has been applied chiefly to the analysis of biological systems, covering a broad range of fields from basic research to clinical settings. For example, the direct analysis of human tissue prepared during the surgery was able to reveal the accumulation of triacylglycerol in both renal cell carcinoma15 and pharyngeal squamous carcinoma16. This method can also measure liquid samples, such as blood, to focus on the lipid profile. For example, some molecules have been delineated during dietary changes in rabbits; it was reported that some of these molecules decreased at very early stages of the experiments, indicating the high sensitivity and usefulness of this system for clinical diagnosis17. Furthermore, direct application to a living animal allowed the detection of biochemical changes of the liver after just one night of fasting5. Zaitsu et al.18 revisited this experiment5 and analyzed the metabolic profiles of the liver in almost the same way, with results that reinforced the stability and reproducibility of our original method. Furthermore, we were able to discriminate the cancer tissue from surrounding non-cancerous liver in mice using this technique19. Therefore, this is a versatile mass spectrometry technique that is useful in various settings, both in vivo and in vitro. From another standpoint, the PESI module can be made to fit various mass spectrometers by adjusting the mounting attachment. In this short article, we introduce the basics and examples of applications (Figure 1), including applications with living animals5.
According to the regulations and laws in each country, parts of this protocol will need to be revised to meet the criteria of each institution. Application to the living organism is the most interesting and challenging because it can provide biochemical or metabolic changes in tissues or organs in living animals in situ. While this application was approved by the institutional committee for animal care at the University of Yamanashi, in 20135, another round of approval will now be necessary because of recent changes in regulations for the animal experiments. Several modifications in the experimental scheme are, therefore, advisable. Regarding the mass spectra obtained in experiments, taking the fluctuations of mass spectra between each measurement into account, there is no spectral information sharing system that is common to the nucleotide sequencing community. Care must be taken when the operator handles the needle to avoid needle-stick accidents, especially when removing the needle from the needle holder. A special device for detaching the needle is very useful for this purpose. Since the compartment of the PESI module is an airtight, closed chamber, leakage of the ion plume does not occur if the mass spectrometer is operated according to the instructions.
Protocol
The institutional committee for animal care at the University of Yamanashi approved all the protocols and the use of experimental animals stated herein. Human sample usage was approved by the institutional ethics board at the University of Yamanashi.
1. Solid tissue preparation
NOTE: Samples must be kept on ice after their removal from the animal or human body to preserve the tissue freshness. If measurements do not immediately follow dissection, it is recommended to store tissue at -80 °C. It is not advisable to place the tissue in any kind of buffer or saline, because they may extract certain contents from the tissue. Tissue that has been fixed with aldehydes or embedded in paraffin/wax or cryogel is not suitable for MS measurements.
- Cut the tissue specimen to approximately 2 x 2 x 2 mm using a scalpel or knife. Alternatively, punch out the sample with a trepan (a disposable biopsy punch, bore size 3 mm) used for the skin examination. In this case, trim the length of the column to 2 mm.
NOTE: Any tissue can be analyzed using this method. In this experiment, liver15 and kidney19, have been successfully analyzed to obtain mass spectra. Generally, parenchymal organs give good mass spectra, while those that have fibrous components do not. - If the tissue is contaminated with blood, wash briefly with ice-cold phosphate-buffered saline.
NOTE: To minimize the postmortem tissue damage, complete this step, as soon as possible, at room temperature. If measurements are not made immediately, snap freeze the tissue in liquid nitrogen and store at -80 °C. - Place the cut or punched out sample (approximately 10 mg) into a plastic microtube and add 100 µL of 50% ethanol.
NOTE: The exact weight of samples is not determined because the mass spectrometry used in this system does not provide quantitative data. Approximately 10 mg is optimal for the parenchymal tissue. - Homogenize the sample using a micropestle.
- Place 10 µL of the homogenate into the well of the cartridge (Figure 2).
- Place the cartridge in the ionization chamber of the mass spectrometer and install the stainless-steel needle probe that will be used for the sample ionization (Figure 2).
NOTE: Probe needles are supplied by the manufacturer. They are made of stainless steel and have a curvature radius of approximately 400 nm. - Close the chamber lid firmly to automatically activate the safety device.
- Start the on-board computer by clicking the Start icon to analyze. Using the onscreen panels, apply a voltage of 2.3 kV to the needle to generate the electrospray and ensure that the frequency of needle is 3 Hz.
- Wait for 30 s for the measurement to be completed.
NOTE: The session automatically ceases when data acquisition is completed. The quality of the analysis is monitored by a total ion chromatogram (TIC). The measurement time is defined to obtain the representative mass spectra and can be shortened or extended. - Dispose of the cartridge and needle in a biohazard disposer after measuring each sample.
NOTE: Only one sample can be measured at a time. It is not necessary to calibrate the machine before every measurement; calibration depends on the regular check-up by the supplier once every six months. - Analyze the mass spectra and export the mass spectral text data using the software associated with the mass spectrometer (see Table of Materials) as stated in the steps below (Figure 3).
- Click on the lcd file in the data file browser window of the software.
- Choose and click on a single peak on the mass spectrum window and to automatically depict an extracted ion chromatogram (EIC).
- Check the TIC and EIC and click on the average spectrum icon to select the range-of-time window for generating the mass spectrum.
NOTE: In this analysis, the target molecules appear in the mass-to-charge ratio (m/z) window (Figure 3). Furthermore, because the duration of the ionization per single stroke of needle movement is very short, all the mass spectra obtained are essentially averaged spectra over 10 s (300 scans). - Generate a text file containing m/z and ion intensity by clicking on the Export tab for further analysis. This text file can be stored in any folder.
NOTE: Any RNA preservatives will affect the native spectral pattern of samples. In addition, it is advisable to handle and store tissue without any liquid (such as phosphate-buffered saline) to prevent the elution of molecular components from the tissue into the liquid during storage. Ideally, fresh or fresh frozen samples without any treatment are used.
2. Body fluids (serum) preparation
NOTE: This whole procedure is almost identical to that used for solid tissue. The cartridge for the fluid sample is available from the manufacturer. Because the contamination by red blood cells (RBCs) can greatly diminish the efficiency of spectral acquisition of the intended component (plasma or serum), be sure to eliminate all RBCs by centrifuging before measurements.
- Take 10 µL of the serum sample and put it into a 1.5 mL microtube.
NOTE: Both fresh and stored serum can be used. - Add 190 µL of 50% ethanol to the 1.5 mL tube, then vortex for 2 min at room temperature.
- Centrifuge the liquid at 15,000 x g for 1-5 min at 4 °C.
- Transfer 10 µL of supernatant into the well of the cartridge.
- Place the cartridge in the ionization chamber of the machine and install the stainless steel needle probe that will be used for sample ionization (Figure 2).
- Close the chamber lid firmly to automatically activate the safety device.
- Start the on-board computer by clicking the Start icon to ionize.
- Discard the sample cartridge and needle as described in 1.10 once measurements have been taken.
- Analyze the obtained sets of EIC as stated below (Figure 3).
- Open the lcd file on the data browser.
- Click on a single peak to display the TIC and EIC.
- Select the range-of-time window for generating mass spectra using the average spectrum icon.
- Export the generated file containing both the m/z and ion intensity of a corresponding peak by clicking the export tab on the monitor.
NOTE: This procedure can be applied to saliva, urine, and other body fluids as well.
3. Preparation for in vivo PESI-MS in living organism
NOTE: In this section, an application to monitor the metabolic profile of 5-Fluoro-2'-deoxyuridine (5-FdU) in a living mouse model is introduced. Use aseptic conditions throughout.
- Infuse 100 µL of 100 mM 5-FdU solution into the tail vein of a 2-month-old mouse (approximately 20 g weight).
NOTE: There is no preference for sex, age, or mouse strain. While this protocol was approved at the time that we carried out the experiment5, the feasibility of this experiment depends on the ethical restrictions of the country in which it will be performed. - Anesthetize the mouse following your approved animal care protocol. Assess the depth of anesthesia by the lack of response to tail pinching.
NOTE: If the usage of sodium pentobarbital is not permitted by the ethical committee for animal experiments, alternative methods may be used, such as ketamine hydrochloride. - Shave the abdominal cavity using an electric razor and apply vet ointment to the eyes.
- Put the anesthetized mouse in a supine position and fix the paws onto the plastic plate using mending tape. Scrub the surgical site with scrubs of 70% alcohol three times.
- Cut open the abdominal cavity using scissors. First, cut the skin laterally (10 mm) from just above the diaphragm. Laterally cut the muscle of the abdominal body wall and peritoneum (ca. 7 mm) and hold the wound open using stainless wire to expose the liver surface.
NOTE: There is no need to perfuse the surface of the liver. - Apply the tip of the probe needle to the surface of the liver. Adjust the depth of the needle to approximately 0.5 mm deep to optimize ionization efficiency, checking the TIC and spectral pattern simultaneously. Set the high voltage to 2.3 kV and frequency to 3 Hz on the screen of the control panel.
- Remove the animal from the plastic plate.
- Suture the wound using a surgical gut suture and return the mouse to the cage. Do not leave the mouse unattended until it has regained consciousness to maintain sternal recumbency. Do not return the mouse to the company of other animals until it has fully recovered.
- Perform the time course of ion intensity in EIC and the procedure of EIC depiction as in 1.11.1 to 1.11.4.
Representative Results
As depicted in Figure 3, the data obtained by PESI-MS technique are the mass spectra, whose m/z range from 10 to 1,200 in this system. While one can detect molecules up to m/z 2,000, there were few peaks obtained using this technique over the mass range of m/z 1,200. Therefore, we analyzed peaks from m/z 10 to 1,200. There were conspicuous groups of peaks around m/z 800 and 900; the former represents cell membrane components, such as phosphatidyl choline (PC) species, and the latter represents triacyl glycerol (TAG). Figure 4 shows the mass spectra from a normal human kidney and a renal cell carcinoma (RCC). The mass spectra were obtained using the direct tissue method explained in protocol 1. Because the clear cell type RCC accumulates TAG in the cytoplasm, a group of spectra around m/z 900 is very pronounced. The result in Figure 4 was obtained by the extraction method introduced in Figure 1 for solid samples.
Because the lipid metabolism of hepatocellular carcinoma changes during the disease progression20, it is possible to monitor the stages of the disease by routine biopsy combined with PESI-MS. The result in Figure 5 depicts the differences in spectral pattern between normal liver tissue and HCC. Especially, there are several remarkable spectra of TAG in HCC. If the resolution of the mass spectrometer is superior, it is possible that the molecular annotation of each spectrum will allow the elucidation of the molecular mechanism of cancer, as well as normal cellular physiology.
Real-time monitoring of drug metabolism can be performed using PESI. In our experiments, the pharmacodynamics of the anti-oncotic agent 5-FdU was monitored over 10 s after being injected through the tail vein of a mouse. Very rapid detection of spectra was observed only a few seconds after injecting the drug (Figure 6), implying the rapid transportation of the drug via the bloodstream to the liver. The intensity of the sodium adducts of 5-FdU waned with a brief defect of the ion signal in the recording at around 6-7 s after starting the measurement. This was because of the varying depth of the PESI needle in the liver parenchyma, which was the result of body movements of the anesthetized mouse. Therefore, care has to be taken to adjust the depth of the PESI needle for the real-time measurement of living animals. There is no recommended way to find the optimal depth of the needle, but it becomes apparent with practice.
Blood samples from three individuals were analyzed by PESI-MS. In this case, 10 µL of serum was mixed with 190 µL of 50% ethanol and then used for PESI-MS analysis. While the overall spectral patterns appear to share several common peaks (Figure 7), there are many minute differences in the spectra from these three people. They are a fingerprint of each person in terms of metabolic activities. For detailed information on molecular identities, it is necessary to use a high-end machine. The mass spectrometer used in the experiments did not provide a high enough resolution to identify the molecules.
If there is contamination by polymer compounds, the original spectra in Figure 4 give rise to overlaid periodic and conspicuous peaks that blur the spectra from the original sample (Figure 8). To demonstrate this experimentally, we added polypropylene glycerol triol (PPGT) to 2.5% to reproduce a case of contamination. Almost all the specific spectra seen in Figure 8 were masked by the addition of PPGT.
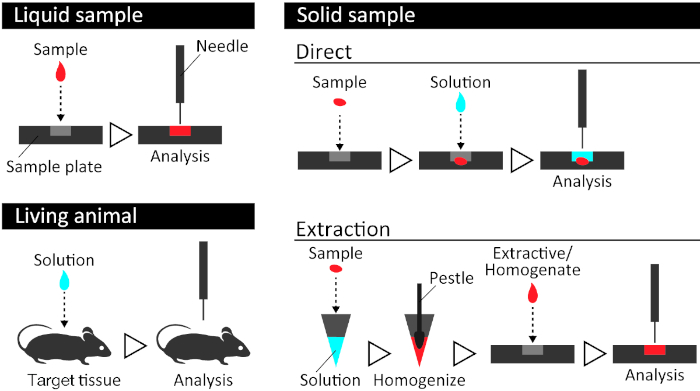
Figure 1: Schematic overview of a sample preparation flow. For samples, a specialized sample cartridge is ideal for stable data acquisition. Liquid sample analysis is the easiest way to perform PESI-MS, by simply placing the solution into the cartridge. There are two methods for applying this method to solid tissue. The direct application of PESI to the tissue is very easy and fast, while the extraction method enables to achieve a much more stable acquisition of spectra depending on the sample species. In this procedure, a small piece of the tissue sample is put in a microtube with 100 µL of 50% ethanol and homogenized with a micropestle. 10 µL of the resulting homogenate is then used for mass spectrometry. In the case of analyzing a living animal, 30 µL of 50% ethanol is placed on the serosal surface of the target organ, followed by measurement. Please click here to view a larger version of this figure.
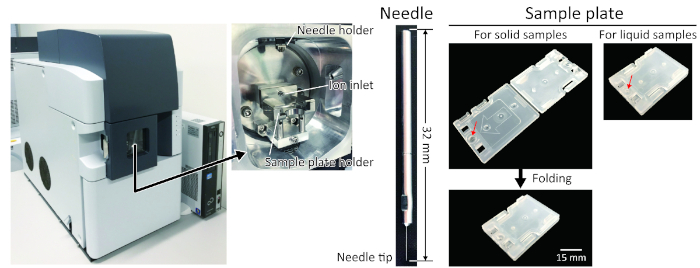
Figure 2: Overview and basic components of PESI-MS. A mass spectrometer equipped with a PESI module. The ionization part is installed in a closed chamber where the ion inlet, needle holder, and sample holder are placed. The needle is ready-made, and the disposable stainless-steel needle is fixed to the needle holder. It is set in the mass spectrometer just after placing the sample cartridge in the chamber. The average curvature angle of the needle is 400 nm. The cartridge for placing the sample is disposable and made of plastic polymers. It has a small well for placing the liquid sample (red arrow). After putting the solid sample into the well, the cassette is folded to pinch the sample and seal the well. For a liquid sample cartridge, a different one is used that does not require folding, and this is a recommended version that is superior to solid samples. Please click here to view a larger version of this figure.
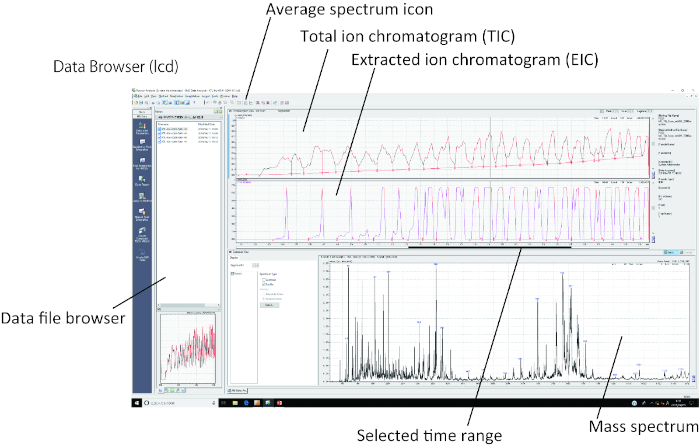
Figure 3: Graphic user interface (GUI) for PESI-MS. All the procedures for generating mass spectra from the total ion chromatogram (TIC) can be performed with the associated software. After opening the lcd file in the data browser, the TIC can be displayed, and the time range can be selected for generating mass spectra. The generated mass spectra can be exported as text data containing both mass-to-charge ratio (m/z) and ion intensity. Please click here to view a larger version of this figure.
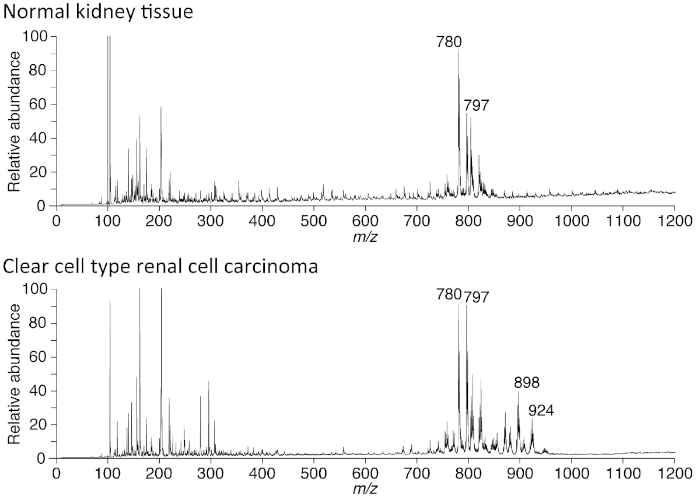
Figure 4: Human renal cell carcinoma shows characteristic peaks of neutral lipids. There were relatively strong peaks in the spectra from human renal cell carcinoma tissue (RCC) that were not identified in the surrounding non-cancerous tissue. These peaks chiefly represent triacylglycerol (TAG) at around mass-to-charge ratio (m/z) 900. Spectral acquisition was performed using the positive ion mode by direct analysis. High voltage was set to 2.3 kV. Please click here to view a larger version of this figure.
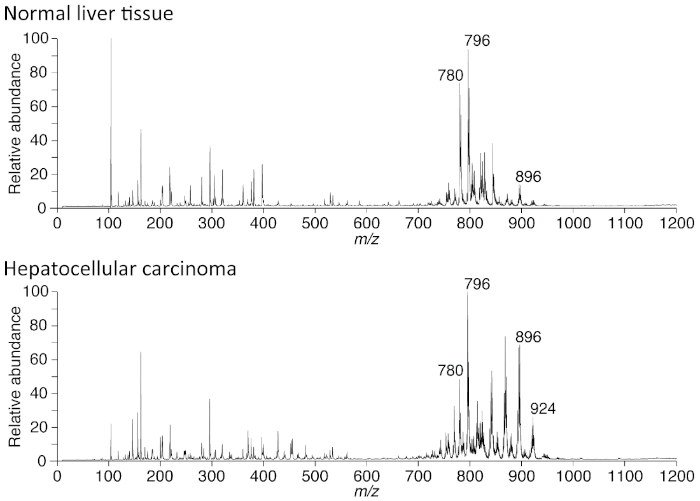
Figure 5: Example of data acquired in normal liver tissues and human hepatocellular carcinoma (HCC). The upper panel depicts the spectra from a human liver tissue where a neoplastic lesion was not present, but from a patient with chronic hepatitis and liver cirrhosis. The lower panel shows the averaged spectra of HCC from the same patient. At a glance, while these two have very similar spectral patterns, there are many small differences in spectral pattern. The abscissa shows the mass-to-charge ratio (m/z) and the ordinate depicts the relative intensity of each spectrum. Acquisition of spectra was performed using the positive ion mode after extracting the tissue, as depicted in Figure 2C. Please click here to view a larger version of this figure.
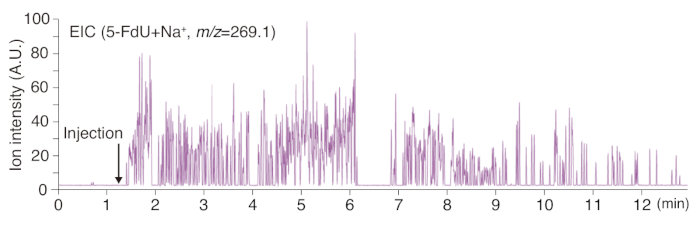
Figure 6: Real-time measurements of drug metabolism. Metabolic changes in 5-FdU injected intravenously into the tail vessel of a mouse. Very rapid and sensitive detection of 5-FdU with sodium adduct was achieved in the liver in situ. Because of unstable ionization during measurement, cessation of the signal can be noted at around 6-7 s. Please click here to view a larger version of this figure.
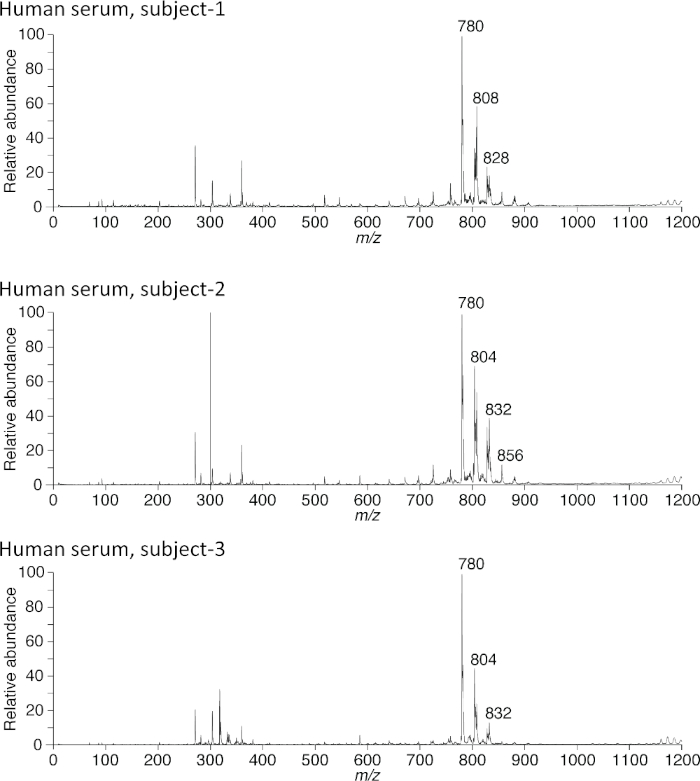
Figure 7: Examples of data from three individuals. Human serum from three individuals was analyzed using PESI-MS. There were clear differences in the spectral patterns among the individuals. Data were obtained using the positive ion mode. Please click here to view a larger version of this figure.
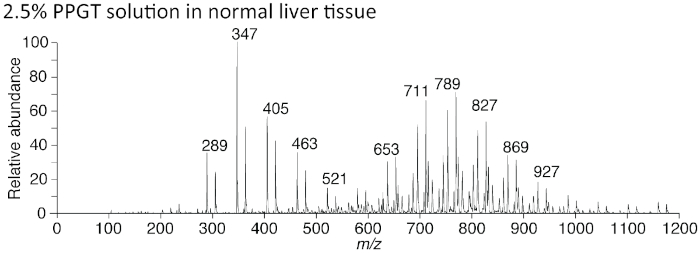
Figure 8: Polymer compounds interfere with precise data acquisition. Because of the periodical emergence of spectral peaks overlaid on the original specimen, it becomes hard to interpret the data precisely. In this case, the spectra derived from polypropylene triol glycerol (PPGT) appear as a noisy overlay. Those compounds, usually used for calibration, mask the original spectra from the liver. Please click here to view a larger version of this figure.
Discussion
Although PESI is a derivative of ESI for mass spectrometry4, it is most advantageous for monitoring real-time metabolomics, as well as for analyzing biochemical reactions without performing any complex or time-consuming pretreatments5,14,15,17. It is an easy and instantaneous mass spectrometry technique that can be applied to the integrated state of living organisms. Since it does not require complex cascades of sample pretreatments, there is a much greater possibility to assess the molecular composition of the entire specimen, because we avoid any possible loss of specimen that can occur in conventional ESI, which requires complex steps of sample pretreatments using large amounts of organic solvents. Therefore, PESI allows us to obtain much more information from the specimen if all conditions are appropriately controlled.
We first need to mention some of the technical aspects required to obtain good results when considering PESI-MS analysis. The most critical factor in PESI is the maintenance of ionization efficiency during analysis. To achieve this, it is advisable to adjust and optimize the distance between the tip of the needle and the sample surface, to generate a stable TIC by referring to the computer monitor during analysis. To do this, it is necessary to change the depth of the needle in a stepwise manner to achieve maximal TIC. Because the duration of ionization is relatively short compared to ESI, PESI is not ideal for reproducibility or quantification. Second, because the shape of the needle tip plays a very important role in the ionization by forming an ideal Taylor cone that serves as the ion source, care must be taken not to deform the needle tip. Deformation of the tip may lead to lower ionization efficiency.
While PESI is advantageous because it can be applied directly to fresh samples (e.g. liver, kidney, and brain) and living organism without special treatment, it is relatively sensitive to other exogenous contaminants because it can ionize almost all components contained in a fluid21,22,23. That is, the downside of the advantage of skipping sample pretreatment means that PESI-MS is at a disadvantage in several situations that are often encountered in biological experiments. As explained in the protocol section, PESI is sensitive to contamination by RNA preservatives, aldehydes, and polymer compounds such as the cryogels often used for histological preparation. Blood clots can adhere to the PESI needle and often mask the intended spectra. Moreover, hemolysis of RBCs liberates hemoglobin, and the dissociated ferric ion gives rise to much stronger spectra than those derived from the intended analytes. Polymer compounds also give periodic spectral peaks that overlay the spectra derived from the intended analytes, making it much harder to interpret the real data because of the noise (Figure 8). To avoid any problems in the acquisition of mass spectra, it is advisable to follow our protocol, taking special note of references to the contamination. In this regard, the application of PESI is limited in some cases.
Another disadvantage of PESI lies in its relatively unstable ionization process, which requires us to adjust the depth of the probe needle to the analytes. In addition, this technique can cover a very restricted area in a single analysis. Because of the very small size of the needle tip that is directly applied to the tissue, PESI-MS analysis is not as comprehensive as the 2D analysis that can be achieved with a MALDI-TOF MS-based imaging mass spectrometry or DESI-based imaging7,8. PESI can, however, reconstruct an image of the hippocampal formation, although it takes several hours23 because the 2D scanning of a sample takes a long time, chiefly because of this technique's unstable ionization efficiency. Taking this into consideration, mass spectral imaging by PESI-MS is not a valuable application.
Regarding the electrochemical properties of PESI, the volume and concentration of the sample that adheres to the needle tip is critical18, as the suppression effect is somewhat attenuated by downsizing the electrospray size. Considering that PESI is miniaturization of ESI, samples must ideally be diluted to avoid forming a slurry on the tip. Therefore, the addition of approximately 30 µL of 50% ethanol is necessary to achieve efficient ionization when the direct tissue method is employed (Figure 2B). This maximizes the ionization efficiency and achieves almost complete ionization of the molecules attached to the tip of the needle.
Choosing the most appropriate method of data processing is very important when using this system24. Concerning the implementation of machine learning for any disease diagnosis, the construction of a database is necessary to establish a reference for classifying or categorizing diseases7. For example, we have used the support vector machine or logistic regression to make a diagnosis of primary liver malignancies25.
PESI-MS is a versatile and easy technique that has great potential for drug screening, doping tests, food safety tests of agricultural products26, and some environmental tests. Because this ionization unit is compatible with other spectrometers by preparing an attachment for each specific instrument, PESI can be applied to various purposes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Ayumi Iizuka for operating the PESI-MS and Kazuko Sawa-nobori for her secretarial assistance. We thank Bronwen Gardner, Ph.D., from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Materials
| 5-Fluoro-2'-deoxyuridine (5-FdU) | Sigma-Aldrich | F8791-25MG | 25mg |
| disposable biposy punch (Trepan) | kai Europa GmbH | BP-30F | bore size 3mm |
| ethanol | nacalai tesque | 14710-25 | extra pure reagent |
| LabSolutions | Shimadzu | ver. 5.96, Data analyzer | |
| micropestle | United Scientific Supplies | S13091 | |
| microtube | Treff | 982855 | 0.5 mL clear |
| PESI-MS (Direct Probe Ionization-MS) | Shimadzu | DPiMS-2020 | Mass spectrometer equipped with PESI |
| PPGT solition | Shimadzu | ND | Attached to DPiMS-2020 |
References
- Fenn, J. B., Mann, M., Meng, C. K., Wong, S. F., Whitehouse, C. M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 246, 64-71 (1989).
- Karas, M., Bachman, D., Bahr, U., Hillenkamp, F. Matrix-Assisted Ultraviolet Laser Desorption of Non-Volatile Compounds. International Journal of Mass Spectrometry and Ion Processes. 78, 53-68 (1987).
- Tanaka, K., et al. Protein and polymer analyses up to m/z 100000 by laser ionization time-of flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2, 151-153 (1988).
- Hiraoka, K., Nishidate, K., Mori, K., Asakawa, D., Suzuki, S. Development of probe electrospray using a solid needle. Rapid Communications in Mass Spectrometry. 21, 3139-3144 (2007).
- Yoshimura, K., Chen, L. C., Yu, Z., Hiraoka, K., Takeda, S. Real time analysis of living animals by electrospray ionization mass spectrometry. Analytical Biochemistry. 417, 195-201 (2011).
- Balog, J., et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Science Translational Medicine. 5, 194ra93 (2013).
- Boughton, B. A., Hamilton, B. Spatial metabolite profiling by matrix-assisted laser desorption ionization mass spectrometry imaging. Advances in Experimental Medicine and Biology. 965, 291-321 (2017).
- Shimma, S., Sugiura, Y., Hayasaka, T., Hoshikawa, Y., Noda, T., Setou, M. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 855, 98-103 (2017).
- Sugiyama, E., Setou, M. Visualization of brain gangliosides using MALDI imaging mass spectrometry. Methods in Molecular Biology. 1804, 223-229 (2018).
- Zhang, J., et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Science Translational Medicine. 9, 406 (2017).
- Pirro, V., Jarmusch, A. K., Vincenti, M., Cooks, R. G. Direct drug analysis from oral fluid using swab touch spray mass spectrometry. Analytica Chimca Acta. 861, 47-54 (2015).
- Chen, L. C., et al. Characterization of probe electrospray generated from a solid needle. Journal of Physical Chemistry. B. 112, 11164-11170 (2008).
- Mandal, M. K., Chen, L. C., Hiraoka, K. Sequential and exhaustive ionization of analytes with different surface activity by probe electrospray ionization. Journal of the American Society for Mass Spectrometry. 22, 1493-1500 (2011).
- Yoshimura, K., Chen, C. L., Asakawa, D., Hiraoka, K., Takeda, S. Physical properties of the probe electrospray ionization (PESI) needle applied to the biological samples. Journal of Mass Spectrometry. 44, 978-985 (2009).
- Yoshimura, K., et al. Analysis of renal cell carcinoma as a first step for mass spectrometry-based diagnostics. Journal of the American Society for Mass Spectrometry. 23, 1741-1749 (2012).
- Ashizawa, K., et al. Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncology. 75, 111-119 (2017).
- Johno, H., et al. Detection of potential new biomarkers of atherosclerosis by probe electrospray ionization mass spectrometry. Metabolomics. 14, 38 (2018).
- Zaitsu, K., et al. Intact endogenous metabolite analysis of mice liver by probe electrospray ionization/triple quadrupole tandem mass spectrometry and its preliminary application to in vivo real-time analysis. Analytical Chemistry. 88, 3556-3561 (2016).
- Yoshimura, K., et al. Real time diagnosis of chemically induced hepatocellular carcinoma using a novel mass spectrometry-based technique. Analytical Biochemistry. 441, 32-37 (2013).
- Nakagawa, H., et al. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers. 10, 447-461 (2018).
- Mandal, M. K., Chen, L. C., Hashimoto, Y., Yu, Z., Hiraoka, K. Detection of biomolecules from solutions with high concentration of salts using probe electrospray and nano-electrospray ionization mass spectrometry. Analytical Methods. 2, 1905-1912 (2010).
- Yoshimura, K., Chen, L. C., Johno, H., Nakajima, M., Hiraoka, K., Takeda, S. Development of non-proximate probe electrospray ionization for real-time analysis of living animal. Mass Spectrometry. 3, S0048 (2014).
- Chen, L. C., et al. Ambient imaging mass spectrometry by electrospray ionization using solid needle as sampling probe. Journal of Mass Spectrometry. 44, 1469-1477 (2009).
- Yoshimura, K., Chen, C. L., Asakawa, D., Hiraoka, K., Takeda, S. Physical properties of the probe electrospray ionization (PESI) needle applied to the biological samples. Journal of Mass Spectrometry. 44, 978-985 (2009).
- Takeda, S., Yoshimura, K., Hiraoka, K. Innovations in analytical oncology – Status quo of mass spectrometry-based diagnostics for malignant tumor. Journal of Analytical Oncology. 1, 74-80 (2012).
- Hiraoka, K., et al. Component profiling in agricultural applications using an adjustable acupuncture needle for sheath-flow probe electrospray ionization/mass spectrometry. Journal of Agricultural and Food Chemistry. 67, 3275-3283 (2019).

