A Method to Study de novo Formation of Chromatin Domains
Summary
This method is designed to follow formation of PRC2-mediated chromatin domains in cell lines, and the method can be adapted to many other systems.
Abstract
The organization and structure of chromatin domains are unique to individual cell lineages. Their misregulation might lead to a loss in cellular identity and/or disease. Despite tremendous efforts, our understanding of the formation and propagation of chromatin domains is still limited. Chromatin domains have been studied under steady-state conditions, which are not conducive to following the initial events during their establishment. Here, we present a method to inducibly reconstruct chromatin domains and follow their re-formation as a function of time. Although, first applied to the case of PRC2-mediated repressive chromatin domain formation, it could be easily adapted to other chromatin domains. The modification of and/or the combination of this method with genomics and imaging technologies will provide invaluable tools to study the establishment of chromatin domains in great detail. We believe that this method will revolutionize our understanding of how chromatin domains form and interact with each other.
Introduction
Eukaryotic genomes are highly organized and changes in the chromatin accessibility directly controls gene transcription1. The genome contains distinct types of chromatin domains, which correlate with transcriptional activity and replication timing2,3. These chromatin domains range in size from a few kilobases (kb) to more than 100 kb and are characterized by an enrichment in distinct histone modifications4. The central questions are: how are these domains formed and how are they propagated?
One of the most well-characterized chromatin domains is fostered through the activity of the Polycomb repressive complex 2 (PRC2). PRC2 is a multi-subunit complex composed of a subset of the Polycomb Group (PcG) of proteins5,6, and catalyzes the mono-, di- and trimethylation of lysine 27 of histone H3 (H3K27me1/me2/me3)7,8,9,10. H3K27me2/me3 are associated with a repressive chromatin state, but the function of H3K27me1 is unclear6,11. One of the core components of PRC2, embryonic ectoderm development (EED), binds to the end product of PRC2 catalysis, H3K27me3, through its aromatic cage and this feature results in the allosteric stimulation of PRC212,13. The PRC2 enzymatic activity is crucial for preserving cellular identity during development as the inappropriate expression of certain developmental genes that are contraindicated for a specific lineage, would be detrimental5,6. Hence, unraveling the mechanisms by which PRC2 fosters the formation of repressive chromatin domains in mammals is of fundamental importance to understanding cellular identity.
All of the past experimental systems designed to investigate chromatin domain formation including PRC2-mediated chromatin domains, were performed under steady-state conditions, which are unable to track the unfolding events of chromatin domain formation in cells. Here, we present a detailed protocol to generate an inducible cellular system which monitors the initial recruitment and propagation of chromatin domains. Specifically, we focus on tracking the formation of PRC2-mediated repressive chromatin domains that comprise H3K27me2/3. This system that can capture the mechanistic details of chromatin domain formation, could be adapted to incorporate other chromatin domains, such as the widely studied domains comprising either H2AK119ub or H3K9me. In combination with genomics and imaging technologies, this approach has the potential to successfully address various, key questions in chromatin biology.
Protocol
Generation of inducible EED rescue mESCs
1. Cell culture
- Use feeder-free C57BL/6 mouse embryonic stem cells (mESCs) possessing a stably integrated CreERT2 transgene, which can translocate to the nucleus upon administration of 4-Hydroxytamoxifen (4-OHT)13.
- Grow mESCs in conventional ESC medium14,15, supplemented with 1000 U/mL LIF, 1 μM ERK inhibitor PD0325901 and 3 μM GSK3 inhibitor CHIR99021. For conventional ESC medium, use knockout DMEM containing 15% fetal bovine serum (FBS), 2 mM L-glutamine, 1X penicillin/streptomycin and 0.1 mM 2-mercaptoethanol.
- Use plates coated with 0.1% gelatin solution for culturing mESCs.
2. Generation of clonal EED knockout (KO) mESCs
- Design guide RNAs (gRNAs) to delete Exon 10 and Exon 11 of endogenous copy of EED in mESCs using the CRISPR design tool in Benchling16. EED-KO-gRNA-1 targets the intron immediately upstream of exon 10 and EED-KO-gRNA-2 targets the intron immediately downstream of Exon 11. The simultaneous application of these gRNAs deletes both Exon 10 and Exon 11 by non-homologous end joining (NHEJ).
- Clone gRNAs into pSpCas9(BB)-2A-GFP (PX458) using instructions in Ran et al., 201317.
- In a 6-well plate format, transfect 2 x 105 mESCs with 1 μg of each EED-KO-gRNA-1 and EED-KO-gRNA-2 (see Table of Materials) using transfection reagent by following the manufacturer's instructions. Change the media 24 h after transfection.
- Two days after the transfection, isolate GFP positive cells using Fluorescence-activated cell sorting (FACS). Expect the transfection efficiency to vary around 10-30 %. Sort around 5 x 105 cells to capture sufficient GFP positive cells for plating.
- Plate the isolated GFP positive mESCs into 15 cm plates pre-coated with 0.1% gelatin (10-20 x103 cells per plate) for colony picking.
- Grow the cells in ESC medium for about one week until single colonies are visible. Change the media every 2 days.
- Pick a minimum of 48 colonies using 20 μL micropipette tip. Scrape over the colony while aspirating into the micropipette tip. Do not break up the colony into single cells. Transfer the colony into an accutase-containing (20 μL) 96 well plate.
- Incubate the colonies for 10 min at 37 °C until all cells are dissociated.
- Add 200 μL of ESC media into each well using multichannel pipette.
- Mix well and plate the cells into two separate 96 well plates using multichannel pipette.
- Use one of the 96 well plates for genotyping and keep the other growing until genotyping is concluded. Use DNA extraction solution to extract DNA from the 96 well plate by following the manufacturer's instructions.
- Use genotyping primers Gnt_EED-KO-up and Gnt_EED-KO_down, which span the deleted site and perform genotyping PCR with Taq DNA polymerase following the manufacturer’s instructions.
NOTE: Other types of DNA polymerases can also be used for genotyping, however, Taq DNA polymerase offers convenience as the PCR reaction can directly be loaded on a gel when its colored reaction buffer is used.- Observe a DNA product of lower molecular weight in cells with a homozygous deletion relative to the wild-type (WT) case.
NOTE: To generate PRC2 null cells, homozygous deletion of exons 10 and 11 is necessary to destabilize and degrade EED, an essential subunit of core PRC213. The CRISPR targeting efficiency is around 10 %.
- Observe a DNA product of lower molecular weight in cells with a homozygous deletion relative to the wild-type (WT) case.
- Validate the deletion of EED exon 10 and 11 and the loss of EED protein by Sanger sequencing and Western blotting, respectively.
- Confirm the depletion of EED and H3K27me2/me3 from chromatin in EED KO cells by ChIP-seq using antibodies against H3K27me2/me3 and EED. Use the protocol given in Oksuz et al., 201715 for ChIP-seq experiments.
3. Engineering the EED knockout mESCs to harbor Cre-ERT2 based inducible EED expression
- Design gRNA (EED-gRNA-inducible) to introduce a cut within the intron following exon 9 of EED using the CRISPR design tool in Benchling16 (see Table of Materials).
- Clone gRNAs into pSpCas9(BB)-2A-GFP (PX458) using instructions in Ran et al., 201317.
- Design a donor template DNA that comprises EED cDNA sequence after exon 9 and a C-terminal Flag-HA tag upstream of a T2A-GFP sequence, all in reverse orientation with respect to the endogenous gene sequence.
- Flank the cassette with a splice-acceptor and a polyadenylation sequence nested between heterologous inverted loxP sites (lox66 and lox71)18.
- Include at least 500 bp of homology arms from each end. Split the donor template into 2 segments of gBlocks gene fragments (see gBlock-1, gBlock-2-WT "https://benchling.com/s/seq-l2LLlWNEnLrfGXcbdCxI" and gBlock-2-cage-mutant "https://benchling.com/s/seq-n8eiZCB2XAkOuzzpv6qM") and assemble them into PCR Blunt vector using Gibson cloning following the manufacturer's instructions.
NOTE: gblock-2 contains an aromatic residue (Y365) within the cage of EED that is important for interaction with H3K27me312. gBlock-2-Wt contains wild type residue, whereas gblock-2-cage-mutant contains the cage-mutant of EED (Y365A), incapable of binding to H3K27me3. Inducible WT EED rescue is denoted as i-WT-r and inducible cage-mutant EED rescue is denoted as i-MT-r for convenience.
- In a 6-well plate format, transfect 2 x 105 mESCs with 1 μg of the gRNA (EED-gRNA-inducible) and 1 μg of the Donor templates (i-WT-r or i-MT-r) using transfection reagent and isolate GFP positive cells using FACS.
- Follow steps 2.3-2.11 to isolate individual colonies ready for genotyping for successful targeting.
- Use genotyping primers Inducible_Genotype-FW-1 and Inducible_Genotype-REV-1, which span the inserted cassette and perform genotyping PCR using Taq DNA polymerase.
NOTE: The targeting efficiency for CRISPR is around 10 %.- Confirm correct integration of the cassette by PCR using Inducible_Genotype-FW-2 and Inducible_Genotype-REV-2 primers, which are outside of the homology arms.
NOTE: Homozygous integration of the cassette is necessary to accomplish efficient rescue of EED. Note that cells with homozygous integration will produce a single large DNA product as compared to WT EED, which will produce a short product.
- Confirm correct integration of the cassette by PCR using Inducible_Genotype-FW-2 and Inducible_Genotype-REV-2 primers, which are outside of the homology arms.
- Confirm the integration of the cassette by Sanger sequencing.
- Confirm flipping of the cassette and the expression of EED and other PRC2 core components (e.g., EZH2 and SUZ12) upon 4-OHT administration by Western blotting.
- Confirm the expression of T2A-GFP and percentage of flip by flow cytometry. Note that expression of GFP is indicative of flipping of the cassette and the expression of EED.
4. Following nucleation and spreading of PRC2 activity on chromatin
- Confirm that i-WT-r and i-MT-r mESCs do not have leaky expression of GFP by flow cytometry. In the case of leaky expression of GFP, isolate GFP-negative cells by FACS before starting the experiment.
- Expand the i-WT-r and i-MT-r mESCs into five 15 cm plates (5x 106 cells per plate).
- Induce expression of WT or cage-mutant EED by administration of 0.5 μM 4-OHT for 0 h, 12 h, 24 h, 36 h and 8 days (one 15 cm plate per condition). Change the media after 12 h for treatments longer than 12 h. Isolate the successfully recombined cells by FACS using GFP. Adjust the time of the treatments such that all of the conditions are collected at once.
- Perform ChIP-seq for H3K27me2, H3K27me3 to investigate their temporal deposition to chromatin in response to re-expression of WT or cage-mutant EED. Use ChIP-seq protocol including library preparation detailed in Oksuz et al., 201715.
- Use spike-in control in each sample to allow for quantitative comparison among different time points19.
- For spike-in control, use chromatin from Drosophila melanogaster (in a 1:50 ratio to the mESC-derived chromatin) as well as Drosophila specific H2Av antibody (1 μL of H2Av antibody per 4 μg of Drosophila chromatin according to manufacturer’s instructions) in each sample.
- Prepare the chromatin from Drosophila in a manner similar to that from mESCs using the protocol detailed in Oksuz et al., 201715.
- Map the sequence reads for ChIP-seq to mm10 genome with Bowtie 2 using default parameters20.
- Normalize the mouse ChIP-seq reads to spike-in Drosophila read counts21. Calculate the spike-in normalization factor using the following formula: 1 x 106/unique Drosophila read counts. Expect to get around 1 x 106 Drosophila read counts and 20 x 106 mouse read counts per experiment.
- Do not include the Drosophila chromatin when sequencing the input samples.
- Use genomecov tool from bedtools to convert bam file into bedgraph22. Next, convert the bedgraph into bigwig file using bedGraphToBigWig tool from USCS23,24. See the following script:
gen="path to mm10 genome"
chr="path to mm10 chromosome sizes"
inp_bam="path to input bam file "
multiply="calculate the spike-in normalization factor"
bedtools genomecov -bg -scale $multiply -ibam $inp_bam -g $gen > output.bedGraph
sort -k1,1 -k2,2n output.bedGraph > output_sorted.bedGraph
bedGraphToBigWig output_sorted.bedGraph $chr output.bw - Visualize the ChIP-seq read densities by uploading the bigwig files on the USCS genome browser24.
5. Monitoring emergence and growth of the H3K27me3 foci in the mESCs nuclei
- Plate 1x 104 mESCs into 8-well chamber slides pre-coated with 0.1% gelatin.
- Next day, start performing consecutive 4-OHT (0.5 μM) inductions to collect the cells expressing EED for indicated time points (0 h, 12 h, 24 h and 36 h). Change the media after 12 h for treatments longer than 12 h.
- Fix the mESCs with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature (RT) for 10 min.
- Permeabilize the cells with PBS/0.25% Triton X-100 at RT for 30 min.
- Block the cells with PBS/5% donkey serum/0.1% Triton X-100 (blocking buffer) at RT for 30 min.
- Dilute H3K27me3 primary antibody 1:500 in the blocking buffer and add onto the cells.
- Incubate the cells with primary antibody overnight at 4 °C.
- Next day, perform 3 washes with PBS/0.1% Triton X-100.
- Dilute secondary antibody (Alexa fluor 595) 1:1000 in the blocking buffer and add onto cells.
- Perform all incubations in dark in the following steps as secondary antibodies conjugated with Alexa Fluor are light sensitive.
- Incubate the secondary antibody for 2 h at RT.
- Perform 3 washes with PBS/0.1% Triton X-100.
- Dilute DAPI (1 mg/mL) 1:4000 with PBS/0.1% Triton X-100 and add onto cells.
- Incubate the cells with DAPI for 10 min at RT.
- Mount the cells with Aqua mount.
- Image the cells with confocal microscopy at 63X magnification.
- Process and pseudo-color the images using a distribution of ImageJ, Fiji25.
Representative Results
A general scheme of the conditional rescue system
Figure 1 shows the targeting scheme to conditionally rescue EED KO cells with either WT or cage-mutant (Y365A) EED that is expressed from the endogenous EED locus. After knocking out EED, a core subunit of PRC2 that is essential for its stability and enzymatic activity, a cassette within the intron following exon 9 of EED is introduced (Figure 1). The cassette consists of the remaining 3' cDNA sequence of EED, in reverse orientation with respect to the endogenous gene sequence. The cassette is flanked by heterologous inverted loxP sites (lox66 and lox71)18. The cells are propagated until the complete loss of H3K27me2/3 is observed. Upon activation of Cre recombinase expression by the addition of 4-OHT, the cassette is inverted such that exon 9 is spliced into the cassette using the splice acceptor sequence (Figure 1). With this system, EED KO cells can now be rescued by either WT or cage-mutant versions of EED both of which have a C-terminal Flag-HA tag. The downstream T2A-GFP provides a marker to select for cells that undergo a successful inversion event. The polyA signal prevents transcription of downstream sequences. With this system, the kinetics of PRC2 recruitment and the formation of H3K27me domains can now be followed.
Tracking the temporal deposition of PRC2-mediated H3K27me2/3
To follow the temporal dynamics of PRC2-mediated chromatin domain establishment, the WT or cage-mutant version of EED is re-expressed in the background of EED KO cells, upon 4-OHT treatment (Figure 2A,B). To follow deposition of H3K27me2/3 marks, ChIP-seq of H3K27me2/me3 are performed after re-expression of WT or cage-mutant EED for the indicated time points. The emergence of H3K27me3 is observed at 12 h after WT EED expression at discrete regions that are denoted as "nucleation sites" (Figure 2C), and then at regions distant from the initial nucleation sites by 24 h13. Eventually, the distribution of H3K27me3 nearly approximates that of the levels seen in WT parental cells at 36 h after WT EED expression. The temporally established downstream sites of H3K27me3 are termed "spreading sites"13. This system can also track the temporal deposition of H3K27me2, which appears to precede H3K27me3 deposition (Figure 2C,D). Lastly, re-expression of cage-mutant EED exhibits different dynamics relative to WT EED (Figure 2C,D). In this case, deposition of H3K27me3 is very inefficient and instead, H3K27me2 becomes apparent and more concentrated at the nucleation sites, indicating that the cage-mutant EED is unable to spread the modification to neighboring regions.
The emergence of H3K27me3 foci and their growth can also be visualized by microscopy (Figure 2E)13. Before the induction of WT EED expression, H3K27me3 staining is not apparent. However, 12 h of WT EED expression shows evidence of H3K27me3 foci formation. These foci increase in number and size by 24 h, and eventually spread to large regions of the nucleus by 36 h of WT EED expression. This system gives evidence of the de novo formation of PRC2-mediated chromatin domains as monitored by ChIP-seq and immunofluorescence (Figure 2C-E)13.
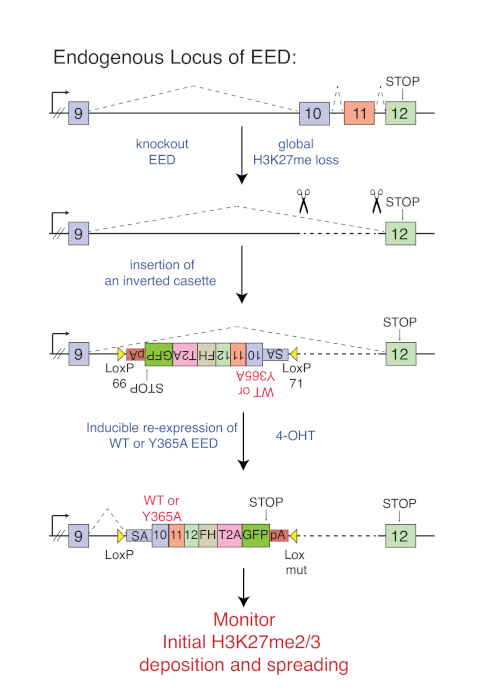
Figure 1: Targeting scheme to conditionally rescue EED KO mESCs either with WT or cage-mt EED (Y365A). Deletion of exon 10 and 11 causes destabilization and degradation of EED and global loss of H3K27me2/me3. A cassette in the intron between exon 9 and 10 is inserted in the reverse direction as indicated. Addition of 4-OHT inverts the cassette such that endogenous exon 9 splices into the cage-mutant or wild type cDNA of the cassette allowing inducible re-expression of WT or cage-mutant EED. This figure has been modified from Oksuz et al., 201813. Please click here to view a larger version of this figure.
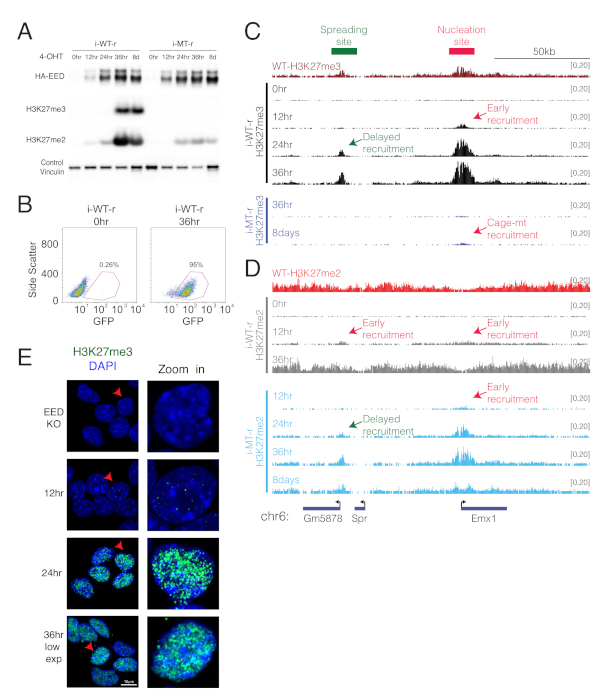
Figure 2: Validations and representative applications of inducible EED rescue systems (i-WT-r and i-MT-r). (A). i-WT-r and i-MT-r systems are validated by Western blot using indicated antibodies on whole extracts after 4-OHT treatment to induce expression of WT (i-WT-r) or cage-mutant (i-MT-r) EED, at the time point indicated. (B). Flow cytometry analysis of GFP in i-WT-r cells before (0 h) and after (36 h) 4-OHT treatment for confirming the efficiency of the expression of T2A-GFP, which is indicative of successful flip of the inverted cassette. (C, D). ChIP-seq tracks for H3K27me3 (C) and H3K27me2 (D) near the Emx1 gene in the WT, or in i-WT-r or in i-MT-r cells, for the indicated times of 4-OHT treatment. Early and delayed sites for deposition of the histone marks are indicated. (E). Immunofluorescence using H3K27me3 antibody at 0, 12, 24 and 36 h after rescue of WT EED expression in i-WT-r mESCs. Staining at 36 h is shown at lower exposures. The rightmost panel is a zoomed-in image of the cell labeled with a red arrow. This figure has been modified from Oksuz et al., 201813. Please click here to view a larger version of this figure.
Discussion
A powerful approach towards understanding the mechanistic details during the formation of a given chromatin domain, is to first disrupt the domain and then track its reconstruction in progress within cells. The process can be paused at any time during the reconstruction to analyze in detail the events in progress. Previous studies on chromatin domains were unable to resolve such events as they were performed under steady-state conditions (e.g., comparing wild-type and gene knockout). Here, we outline a system to assess the dynamic formation of chromatin domains, as highlighted by the recruitment and spreading of PRC2-mediated repressive domains in cells.
The most critical step for this inducible system is the accurate design of DNA constructs to re-express the desired proteins (or RNA) after their respective deletion from the cells. Various mutations, tags, and fluorescent markers could be introduced within these constructs, depending on the downstream applications. For example, instead of using self-cleaving T2A peptide immediately before GFP to disconnect it from the protein of interest, various fluorescent fusion proteins could be generated for high resolution imaging and/or single-particle tracking experiments to monitor the initial events of chromatin domain formation in living cells.
Although this method could be modified in many ways to provide insights into the formation of chromatin domains, it does have some limitations. First, it requires a cell line that harbors the Cre-ERT2 gene. Second, optimizations are necessary to determine the time points after 4-OHT treatment. Third, this system is not reversible so as to monitor the events during deconstruction of a chromatin domain. Degron-based methods could be used as an alternative to the method described here26,27. These systems enable reversible and rapid degradation of proteins, but usually require costly molecules for protein destabilization, such as auxin or shield26,27. Furthermore, these degron-based methods have limitations. They could only be used to re-express the WT version of the protein after its degradation. In contrast, the system described herein allow conditional expression of the mutant version of the protein of interest in addition to its WT counterpart. Combining such a degron system with the method described here would be a powerful means to reversibly modulate formation of chromatin domains. An alternative method for following the formation of chromatin domains entails the use of specific inhibitors to deplete a defined chromatin modification from the chromatin and then washout the inhibitor to follow de novo deposition of the modification. For example, treatment with EZH2 inhibitor and subsequent washout is used for tracking the re-formation of H3K27me domains28. However, EZH2 inhibitor is not effective in completely depleting H3K27me, even 7 days after treatment. In this case, the presence of pre-existing H3K27me might recruit and activate PRC212, thereby complicating interpretation of the results. As well, the use of the inhibitor and washout strategy is limited due to the absence of specific and potent inhibitors for many chromatin domains.
In addition to the applicability of this method to cellular systems, it can be used to generate inducible mutations/tagging on desired proteins in animals. In some cases, mutating a protein or inserting a tag could be lethal during development. This method bypasses this lethality and provides a temporal and spatial control of the mutation/tagging of proteins by coupling with tissue specific Cre-ERT2 mouse strains. These types of experiments would be valuable to determine the effect of a mutation in a specific tissue and/or at a specific stage of development. Importantly, it allows for the isolation of cells that undergo a successful recombination via the reporter within the cassette. This facilitates biochemical analyses, such as affinity purification from specific tissues, to isolate the tissue-specific interactome for a given protein. This system could be geared to monitor dynamic changes in any given cellular process, and hence is not limited to tracking chromatin domain formation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Drs. L. Vales, D. Ozata and H. Mou for revision of the manuscript. The D.R. Lab is supported by the Howard Hughes Medical Institute and the National Institutes of Health (R01CA199652 and R01NS100897).
Materials
| (Z)-4-Hydroxytamoxifen (5 mg) | Sigma | H7904-5MG | For induction of EED expression |
| 16% Paraformaldehyde aqueous solution (10×10 ml) | Electron Microscopy Sciences | 15710 | For immunofluorescence |
| 2-mercaptoethanol | LifeTechnologies | 21985-023 | For mESCs culture |
| 2% Gelatin Solution | Sigma | G1393-100ml | For mESCs culture |
| Accutase 500 ML | Innovative Cell Tech/FISHER | AT 104-500 | For mESCs culture |
| Alexa Fluor 594 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson immunoresaerch | 711-585-152 | For immunofluorescence |
| Aqua-Mount Mounting Medium | FISHER/VWR | 41799-008 | For immunofluorescence |
| CHAMBER SLD TC PRMA 8-CHM 16 PK | Fisher Sci | 177445PK | For immunofluorescence |
| DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) – 10 mg | Life Tech | D1306 | For immunofluorescence |
| ERK inhibitor, PD0325901 | Stemgent | 04-0006 | For mESCs culture |
| ESGRO Recombinant Mouse LIF Protein | Millipore/Fisher | ESG1107 | For mESCs culture |
| FBS Stem Cell Qualified | Atlanta | S10250 | For mESCs culture |
| Gibson Assembly Master Mix | NEB | E2611L | For Donor template cloning |
| GSK3 inhibitor, CHIR99021 | Stemgent | 04-0004 | For mESCs culture |
| H3K27me2 (D18C8) rabbit mAB | Cell Signaling | 9728S | Antibody for ChIP-seq |
| H3K27me3 | Cell Signaling | 9733S | Antibody for ChIP-seq |
| Histone H2Av antibody (pAb) | Active motif | 39715 | Spike-in control for ChIP-seq |
| Knockout DMEM | Invitrogen | 10829-018 | For mESCs culture |
| L-glutamine | Sigma | G7513 | For mESCs culture |
| Lipofectamine 2000 | LifeTech | 11668019 | For transfection |
| MangoTaq DNA Polymerase | Bioline | BIO-21079 | For Genotyping PCR |
| Normal donkey serum (10 mL) | Jackson ImmunoResearch | 017-000-121 | For immunofluorescence |
| Penicillin-Streptomycin | Sigma/Roche | P0781 | For mESCs culture |
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | 48138 | For gRNA cloning |
| QuickExtract DNA Extraction Solution | Lucigen | QE0905T | For Genotyping PCR |
| Triton X-100 | Sigma | T8787-250ML | |
| Zero Blunt PCR Cloning Kit | Thermo Fisher | K270020 | For Donor template cloning |
| Primers/gBlocks | |||
| EED-KO-gRNA-1 | Sequence: ctctggctactgtcaactag. gRNAs pairs to knockout EED in C57BL/6 ESCs for i-WT-r and i-MT-r systems. | ||
| EED-KO-gRNA-2 | Sequence: TAGGCTATGACGCAGCTCAG. gRNAs pairs to knockout EED in C57BL/6 ESCs for i-WT-r and i-MT-r systems. | ||
| EED-gRNA-inducible | Sequence: atggcaccccgaaattagaa. gRNA and Donor to generate i-WT-r system in the EED-KO background. | ||
| i-WT-r Donor | https://benchling.com/s/seq-l2LLlWNEnLrfGXcbdCxI. gRNA and Donor to generate i-WT-r system in the EED-KO background. | ||
| EED-gRNA-inducible | Sequence: atggcaccccgaaattagaa. gRNA and Donor to generate i-WT-r system in the EED-KO background. | ||
| i-MT-r Donor | https://benchling.com/s/seq-n8eiZCB2XAkOuzzpv6qM. gRNA and Donor to generate i-MT-r system in the EED-KO background. | ||
| Genotyping Primers | |||
| Gnt-EED-KO-FW-1 | Sequence: ctgtaggctgccatctgtga. Wild type allele will produce a product of 1.9 kb. Knockout allele will produce a product of 200 bp. | ||
| Gnt-EED-KO-REV-1 | Sequence: agccagggctacacagagaa. Wild type allele will produce a product of 1.9 kb. Knockout allele will produce a product of 200 bp. | ||
| Inducible_Genotype-FW-1 | Sequence: tgcagtgaaacaaatttggaa. When the casette is inserted, the primers will produce 1863 bp. The wild type allele will produce a product of ~200 bp. | ||
| Inducible_Genotype-REV-1 | Sequence: gagaggggtggcactgtaaa. When the casette is inserted, the primers will produce 1863 bp. The wild type allele will produce a product of ~200 bp. | ||
| Inducible_Genotype-FW-2 | Sequence: ccccctctttctccttttct. When the casette is inserted, the primers will produce 3200 bp. The wild type allele will produce a product of 1560 bp. | ||
| Inducible_Genotype-REV-2 | Sequence: atgcctgggtgaatgaaaaa. When the casette is inserted, the primers will produce 3200 bp. The wild type allele will produce a product of 1560 bp. |
References
- Bonev, B., Cavalli, G. Organization and function of the 3D genome. Nature Reviews Genetics. 17 (11), 661-678 (2016).
- Dixon, J. R., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485 (7398), 376-380 (2012).
- Pope, B. D., et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 515 (7527), 402-405 (2014).
- Carelli, F. N., Sharma, G., Broad Ahringer, J. Chromatin Domains: An Important Facet of Genome Regulation. Bioessays. 39 (12), (2017).
- Margueron, R., Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature. 469 (7330), 343-349 (2011).
- Holoch, D., Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends in Biochemical Sciences. 42 (7), 531-542 (2017).
- Cao, R., et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 298 (5595), 1039-1043 (2002).
- Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P., Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes and Development. 16 (22), 2893-2905 (2002).
- Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 111 (2), 185-196 (2002).
- Müller, J., et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 111 (2), 197-208 (2002).
- Ferrari, K. J., et al. Polycomb-Dependent H3K27me1 and H3K27me2 Regulate Active Transcription and Enhancer Fidelity. Molecular Cell. 53 (1), 49-62 (2014).
- Margueron, R., et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 461 (7265), 762-767 (2009).
- Oksuz, O., et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Molecular cell. 70 (6), 1149-1162 (2018).
- Tee, W. W., Shen, S. S., Oksuz, O., Narendra, V., Reinberg, D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 156 (4), 678-690 (2014).
- Oksuz, O., Tee, W. W. Probing chromatin modifications in response to ERK signaling. Methods in Molecular Biology. 1487, 289-301 (2017).
- Benchling for Academics. Benchling Available from: https://benchling.com (2018)
- Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8 (11), 2281-2308 (2013).
- Zhang, Z., Lutz, B. Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein. Nucleic acids research. 30 (17), 90 (2002).
- Orlando, D. A., et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell reports. 9 (3), 1163-1170 (2014).
- Langmead, B., Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 9 (4), 357-359 (2012).
- Descostes, N. ChIPSeqSpike: ChIP-Seq data scaling according to spike-in control. R package version 1.2.1. , (2019).
- Quinlan, A. R., Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26 (6), 841-842 (2010).
- Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S., Karolchik, D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 26 (17), 2204-2207 (2010).
- . UCSC Genome Browser Home Available from: https://genome.ucsc.edu (2019)
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 6 (12), 917-922 (2009).
- Banaszynski, L. A., Chen, L., Maynard-Smith, L. A., Ooi, A. G. L., Wandless, T. J. A Rapid, Reversible, and Tunable Method to Regulate Protein Function in Living Cells Using Synthetic Small Molecules. Cell. 126 (5), 995-1004 (2006).
- Højfeldt, J. W., et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nature Structural & Molecular Biology. 25 (3), 225-232 (2018).

