Uptake of Fluorescent Labeled Small Extracellular Vesicles In Vitro and in Spinal Cord
Summary
We describe a protocol to label macrophage-derived small extracellular vesicles with PKH dyes and observe their uptake in vitro and in the spinal cord after intrathecal delivery.
Abstract
Small extracellular vesicles (sEVs) are 50-150 nm vesicles secreted by all cells and present in bodily fluids. sEVs transfer biomolecules such as RNA, proteins, and lipids from donor to acceptor cells, making them key signaling mediators between cells. In the central nervous system (CNS), sEVs can mediate intercellular signaling, including neuroimmune interactions. sEV functions can be studied by tracking the uptake of labeled sEVs in recipient cells both in vitro and in vivo. This paper describes the labeling of sEVs from the conditioned media of RAW 264.7 macrophage cells using a PKH membrane dye. It shows the uptake of different concentrations of labeled sEVs at multiple time points by Neuro-2a cells and primary astrocytes in vitro. Also shown is the uptake of sEVs delivered intrathecally in mouse spinal cord neurons, astrocytes, and microglia visualized by confocal microscopy. The representative results demonstrate time-dependent variation in the uptake of sEVs by different cells, which can help confirm successful sEVs delivery into the spinal cord.
Introduction
Small extracellular vesicles (sEVs) are nanosized, membrane-derived vesicles with a size range of 50-150 nm. They originate from multi-vesicular bodies (MVBs) and are released from cells upon fusion of the MVBs with the plasma membrane. sEVs contain miRNAs, mRNAs, proteins, and bioactive lipids, and these molecules are transferred between cells in the form of cell-to-cell communication. sEVs can be internalized by recipient cells by a variety of endocytic pathways, and this capture of sEVs by recipient cells is mediated by the recognition of surface molecules on both EVs and the target cells1.
sEVs have gained interest due to their capacity to trigger molecular and phenotypic changes in acceptor cells, their utility as a therapeutic agent, and their potential as carriers for cargo molecules or pharmacological agents. Due to their small size, the imaging and tracking of sEVs can be challenging, especially for in vivo studies and clinical settings. Therefore, many methods have been developed to label and image sEVs to assist their biodistribution and tracking in vitro and in vivo2.
The most common technique to study sEV biodistribution and target cell interactions involves labeling them with fluorescent dye molecules3,4,5,6,7. EVs were initially labeled with cell membrane dyes that were commonly used to image cells. These fluorescent dyes generally stain the lipid bilayer or proteins of interest on sEVs. Several lipophilic dyes display a strong fluorescent signal when incorporated into the cytosol, including DiR (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide), DiL (1, 1′-dioctadecyl-3, 3, 3′, 3′-tetramethyl indocarbocyanine perchlorate), and DiD (1, 1′-dioctadecyl-3, 3, 3′, 3′-tetramethyl indocarbocyanine 4-chlorobenzenesulfonate salt)8,9,10,11.
Other lipophilic dyes, such as PKH67 and PKH26, have a highly fluorescent polar head group and a long aliphatic hydrocarbon tail that readily intercalates into any lipid structure and leads to long-term dye retention and stable fluorescence12. PKH dyes can also label EVs, which allows the study of EV properties in vivo13. Many other dyes have been used to observe exosomes using fluorescence microscopy and flow cytometry, including lipid-labeling dyes14 and cell-permeable dyes such as carboxyfluorescein diacetate succinimidyl ester (CFDA-SE)15,16 and calcein acetoxymethyl (AM) ester17.
Studies of sEV-mediated crosstalk between different cells in the CNS have provided important insights on the pathogenesis of neuroinflammatory and neurodegenerative diseases18. For example, sEVs from neurons can spread beta-amyloid peptides and phosphorylated tau proteins and aid in the pathogenesis of Alzheimer's disease19. Additionally, EVs derived from erythrocytes contain large amounts of alpha-synuclein and can cross the blood-brain barrier and contribute to Parkinson's pathology20. The ability of sEVs to cross physiological barriers21 and transfer their biomolecules to target cells makes them convenient tools to deliver therapeutic drugs to the CNS22.
Visualizing sEV uptake by myriad CNS cells in the spinal cord will enable both mechanistic studies and the evaluation of the therapeutic benefits of exogenously administered sEVs from various cellular sources. This paper describes the methodology to label sEVs derived from macrophages and image their uptake in vitro and in vivo in the lumbar spinal cord by neurons, microglia, and astrocytes to qualitatively confirm sEV delivery by visualization.
Protocol
NOTE: All procedures were performed in compliance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care & Use Committee of Drexel University College of Medicine. Timed-pregnant CD-1 mice were used for astrocytic culture, and all dams were received 15 days after impregnation. Ten-twelve weeks old C57BL/6 mice were used for in vivo uptake experiments.
1. Isolation of sEVs from RAW 264.7 macrophage cells
- Culture RAW 264.7 cells in 75 cm2 flasks in DMEM exosome-depleted medium containing 10% exosome-depleted fetal bovine serum (FBS) and 1% penicillin-streptomycin (pen-strep) for 24-48 h.
- Collect 300 mL of conditioned medium and centrifuge at 300 × g for 10 min at 4 °C.
- Collect the supernatant and centrifuge at 2,000 × g for 20 min at 4 °C.
- Transfer the supernatant to centrifuge tubes, centrifuge for 35 min at 12,000 × g at 4 °C.
- Collect the supernatant and filter through a 0.22 µm syringe filter.
- Transfer to ultracentrifuge tubes and centrifuge for 80 min at 110,000 × g at 4 °C.
- Store the supernatant (exosome-depleted medium), resuspend the pellet in 2 mL of 1x phosphate-buffered saline (PBS), and centrifuge for 1 h at 110,000 × g at 4 °C.
- Resuspend the pellet in 100 µL of PBS for further characterization using nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) or in radioimmunoprecipitation assay (RIPA) buffer for western blotting.
2. Characterization of sEVs
- Nanoparticle tracking analysis (NTA)
NOTE: The size distribution and particle number/concentration of the purified sEVs from RAW 264.7 cells were measured by NTA.- Dilute the sEVs in filtered PBS to obtain 20-60 vesicles per field of view for optimal tracking.
- Introduce the diluted sample into a flow cell using a syringe pump with a constant flow rate.
- Take 3-5 videos of 30 s each. Set the shutter speed and gain, and manually focus the camera settings for the maximum number of vesicles to be visible and capable of being tracked and analyzed.
- Advance the samples between each recording to perform replicate measurements. Optimize the NTA post-acquisition settings and keep the settings constant between the samples.
- Analyze each video using the NTA software to obtain the average size and concentration of the vesicles.
- Carry out all NTA measurements with identical system settings for consistency.
- Western blot
- Quantify the total protein amounts in sEVs, cell lysates, and exosome-depleted media using a protein assay kit following the manufacturer's instructions.
- For cell lysate preparation, culture the RAW 264.7 cells in 75 cm2 flasks until 80-90% confluent. Detach the cells with 0.25% trypsin for 10-15 min, neutralize the trypsin with culture media, and pellet the cells by spinning at 400 × g for 5 min. Resuspend the cells in fresh growth medium.
- Count the cells using a hemocytometer and transfer 1 × 106 cells to another tube. Wash the cells with PBS twice using the same centrifugation conditions as above and add 50 µL of lysis buffer (RIPA buffer with protease inhibitor cocktail added) to the cell pellet from the final spin.
- Vortex the cells and keep them on ice for 20 min. Subject the mixture to centrifugation at 10,000 × g for 30 min at 4 °C, collect the supernatant (i.e., the lysate) in fresh microcentrifuge tubes, and keep at −80 °C until use.
- Concentrate 2 mL of exosome-depleted media to 100 µL using 3 kDa-cutoff centrifugal filters before quantifying the amount of protein. Mix the sEVs with lysis buffer in a 1:1 ratio, vortex for 30 s, and incubate on ice for 15 min to quantify the amount of protein.
- Mix equal amounts of protein (2 µg) of the sEVs, RAW 264.7 cell lysate, and exosome-depleted media with reducing sample buffer.
- Denature the samples at 95 °C for 5 min, keep them on ice for 5 min, and spin for 2 min at 10,000 × g. Load the samples on a 12% Tris-glycine protein gel and run the gel at 125 V for 45 min.
- Transfer the protein onto a polyvinylidene difluoride (PVDF) membrane at 25 V for 2 h.
- Following the transfer, block the PVDF membranes with blocking buffer (see the Table of Materials) for 1 h at room temperature.
- Incubate the blot with primary antibodies on a shaker overnight at 4 °C.
NOTE: Primary antibodies used were anti-CD81 (1:1,000), anti-alpha-1,3/1,6-mannosyltransferase (ALG-2)-interacting protein X (Alix) (1:1,000), anti-Calnexin (1:1,000), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000). - Wash the blots 3 x 15 min with 1x Tris-buffered saline, 0.1% Tween 20 (TBST), and incubate at room temperature with goat anti-mouse IgG-horseradish peroxidase (HRP)- or donkey anti-rabbit IgG-HRP-conjugated secondary antibodies (1:10,000) for 1 h on the shaker.
- Wash the blots 3 x 15 min with 1x TBST, and detect the proteins using an HRP substrate.
- Analyze the blots by enhanced chemiluminescence using a western blot imager.
- Transmission electron microscopy (TEM)
- Fix sEVs by resuspending them in 2% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB); vortex for 2 x 15 s.
- Place a drop of 10 µL of sEV suspension on clean parafilm. Float the carbon-coated formvar grid on the drop with their coated side facing the suspension. Let the membranes absorb for 20 min in a dry environment.
- Place the grids (membrane side down) on a drop of PB to wash for 3 x 2 min.
- Transfer the grids to 50 µL of 1% glutaraldehyde for 5 min.
- Wash the grids with 100 µL of distilled water for 8 x 2 min.
- Contrast the sample by placing the grids on a drop of 1% uranyl acetate for 2 min.
- Embed the sample with 50 µL of 0.2% uranyl acetate with 2% methylcellulose solution for 10 min on a parafilm-covered ice dish.
- Use stainless steel loops to hold the grids and remove excess fluid with filter paper.
- Air-dry the grid for 10 min while still on the loop.
- Observe under a transmission electron microscope at 80 kV.
3. Labeling of sEVs
- Dilute 20 µg of sEVs in 1 mL of diluent buffer or the same volume of PBS in 1 mL of diluent buffer for dye control.
- Dilute 3 µL of PKH67 or PKH26 dye in 1 mL of diluent buffer and mix by pipetting.
- Add diluted PKH dye to the diluted sEVs and mix by pipetting. Incubate for 5 min in the dark at room temperature. For a dye control, mix the diluted dye with diluted PBS from step 3.1.
- Add 2 mL of 1% bovine serum albumin (BSA) in PBS to the tube with the dye and the sEV mix and to the dye control tube to absorb excess dye.
- Centrifuge for 1 h at 110,000 × g at 4 °C. Discard the supernatant, resuspend the pellet in 2 mL of PBS, and centrifuge for 1 h at 110,000 × g at 4 °C. Repeat the wash with PBS and resuspend the labeled sEVs or the dye control in an equal volume of PBS.
- Quantify the amount of total protein by the Bradford method.
4. Uptake of sEVs by Neuro-2a cells
- Place 18-mm coverslips in a 12-well plate and plate 10 × 104 Neuro-2a cells in each well in a total of 1 mL of complete DMEM medium containing 10% FBS and 1% pen-strep.
- Change the medium to DMEM exosome-depleted medium when the cell confluency is 80-90%. Add 1, 5, or 10 µg of labeled sEVs in each well for 1, 4, and 24 h for dose- and time-dependent uptake, or add an equal volume of dye control.
5. Primary astrocytic cultures
- Anesthetize 4 postnatal pups 4 days after birth by inducing hypothermia.
- Collect the brains in a 60-mm Petri dish containing ice-cold Hank's Balanced Salt Solution (HBSS) supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES).
- Dissect both cortical lobes and remove the meninges. Mince the tissues with a sterilized blade.
- Transfer the tissues to a 15 mL conical tube containing papain/deoxyribonuclease I dissociation buffer and incubate for 20 min at 37 °C. Swirl every 5 min.
NOTE: For 4 mouse cortices, 9 mL of 7.5 U/mL papain in HBSS is activated at 37 °C for at least 30 min, filtered through a 0.22 µm syringe filter, and mixed with deoxyribonuclease I to a final concentration of 0.1 mg/mL. - Aspirate the supernatant and add 5 mL of complete DMEM to inactivate the enzyme activity. Carefully triturate to dissociate the tissues with a 5 mL glass serological pipette and a flame-polished Pasteur pipette.
- Pass the cell suspension through a 40 µm cell strainer and centrifuge the cells at 250 × g for 5 min at 4 °C. Aspirate the medium and seed the cells in 10 mL of complete DMEM in a 75 cm2 flask. Replace the supernatant medium with 15 mL of fresh DMEM medium 4 h after plating.
- After 14 days in vitro, transfer the flask to an orbital shaker to detach the microglia and oligodendrocytes at 320 rpm for 6 h.
- Trypsinize the remaining astrocytes using 5 mL of the cell dissociation enzyme (Table of Materials) for 10 min at 37 °C. Add 5 mL of complete DMEM to inactivate the enzymatic action and pellet the cells at 250 × g for 5 min at 4 °C.
- Resuspend the cells in complete DMEM. Seed 5 × 104 cells on 12 mm #1.5 coverslips in a 24-well plate.
6. Uptake of sEVs by astrocytes
- When astrocytes reach 80-90% confluency, change the medium to DMEM exosome-depleted medium.
- Add 1 µg of unlabeled, labeled sEVs, an equal volume of dye control, or PBS to the cells. Use the cells for staining 1 h and 24 h after sEV treatment.
7. Immunofluorescence
- Rinse the cells with PBS 3x and fix them with 4% PFA in PB for 10 min at room temperature.
- Wash the fixed cells 3 x 5 min with PB and permeabilize them using 0.1% Triton X-100 in PB for 10-15 min and wash with PB 3 x 5 min.
- Block the cells with 5% normal goat serum (NGS) in PB for 1 h at room temperature.
- Incubate the cells with primary antibodies: microtubule-associated protein 2 (MAP2A, 1:500) for Neuro-2a cells or glial fibrillary acidic protein (GFAP, 1:500) for primary astrocytes in fresh 5% NGS/PB overnight at 4 °C with gentle shaking.
- Wash 3 x 10 min with PB and add fluorophore-conjugated secondary antibodies (Goat Anti-Mouse IgG1, Alexa Fluor 594; or Goat Anti-Mouse IgG H&L, Alexa Fluor 488) in 5% NGS and incubate for 2 h at room temperature on a rocker.
- Wash 3 x 10 min with PB and incubate with 1 µg/mL of nuclear stain 4',6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature. Wash the cells again 3x with PB.
- Mount the coverslips on #1 slides using an antifade mounting medium. Let them dry overnight in the dark, and store the prepared glass slides at 4 °C until imaging on a confocal microscope.
8. In vivo uptake of sEVs
- Perform intrathecal injection of 5 µg of unlabeled or labeled sEVs resuspended in 10 µL of PBS, or equal volume (10 µL) of dye control (as prepared in section 3) into C57BL/6 mice.
- After 6 and 18 h post-injection of sEVs, deeply anesthetize mice by intraperitoneal injection of 100 mg/kg body weight of ketamine and 10 mg/kg body weight of xylazine.
- Perform intracardial perfusion of mice with 0.9% saline to flush out blood, followed by freshly made ice-cold 4% PFA/PB.
- Dissect the spinal cord and fix in 4% PFA/PB at 4 °C for 24 h. Cryoprotect the tissues in 30% sucrose in PB at 4 °C for 24 h or until the tissues sink. Store the tissues at 4 °C until immunohistochemistry.
9. Immunohistochemistry
- Embed L4-L5 spinal cord in O.C.T compound. Freeze on dry ice until completely solidified.
- Section the tissues at 30 µm (cross-sectionally for the spinal cord) using a cryostat, and collect the sections in a 24-well plate containing PB. Wash the sections 3 x 5 min with 0.3% Triton in PB.
- Block non-specific binding sites with 5% NGS in 0.3% Triton/PB for 2 h at room temperature.
- Dilute primary antibodies: Anti-MAP2A (1:500), GFAP (1:1,000), Iba1 for microglia (1:2,000) with 5% NGS in 0.3% Triton/PB, and incubate the sections overnight at 4 °C on a shaker.
- Wash the sections 3 x 5 min with 0.3% Triton/PB, and add secondary antibodies (Donkey Anti-Rabbit IgG Alexa Fluor 488, 1:500, or Goat Anti-Mouse IgG Alexa Fluor 488, 1:500) in 5% NGS/PB for 2 h at room temperature.
- Wash 3 x 5 min with PB, and incubate the sections in 1 µg/mL of DAPI for 10 min at room temperature. Wash the sections 3 x 5 min with PB.
- Mount the sections on a clean adhesive slide (Table of Materials) with a fine paintbrush under a light microscope.
- Wet the coverslip with mounting medium. Cure overnight in the dark at room temperature.
- Image under a confocal microscope with the respective lasers.
Representative Results
After the isolation of sEVs from RAW 264.7 conditioned media via centrifugation, NTA was used to determine the concentration and size distribution of the purified sEVs. The average mean size of RAW 264.7-derived sEVs was 140 nm, and the peak particle size was 121.8 nm, confirming that most detectable particles in the light scattering measurement fell within the size range of exosomes or sEVs at 50-150 nm (Figure 1A). As suggested in the minimal information for studies of extracellular vesicles 2018 (MISEV2018)23, we analyzed a set of proteins that should be present or excluded from distinct EV populations. Western blotting of sEVs, cell lysate, and exo-depleted media demonstrated that sEV-derived protein samples contained the sEV marker proteins Alix, CD81, and GAPDH. The cell lysate fraction was enriched with the endoplasmic reticulum resident protein, calnexin, which was absent in the sEVs. Thus, calnexin served as a negative marker for cellular contamination (Figure 1B).
We next performed dose-response and time-course experiments for sEV uptake in vitro. Neuro-2a cells were incubated with a single 1 µg dose of PKH67-labeled sEVs for 1, 4, and 24 h, following which the uptake of different concentrations of sEVs (1, 5, and 10 µg) was examined at 1 h. The results of the NTA indicated that 1 µg of protein on average was equal to ~1 x 109 particles. In parallel, PBS, unlabeled sEVs, and dye-alone controls were also tested. We observed that uptake of sEVs occurred at 1 h (Figure 2A) and for the 1, 5, and 10 µg sEVs (Figure 2B). Fluorescence could be detected at 4 h for 5 and 10 µg of sEVs (Figure 2C) post incubation. Next, uptake of PKH26-labeled sEVs by primary astrocytes was examined (Figure 3). Maximal fluorescence from sEV uptake in primary cortical astrocytes occurred at 24 h. Unlabeled sEVs did not show fluorescence, demonstrating that sEV autofluorescence does not significantly contribute to false positives (Supplemental Figure S1A).
Next, labeled sEVs were intrathecally injected into mice to assess the delivery and uptake of sEVs by different cells in the spinal cord using immunohistochemistry and confocal microscopy. We stained for MAP2 as a neuronal marker, GFAP as an astrocytic marker, and IBA1 as a microglial marker. Neurons (Figure 4), astrocytes (Figure 5), and microglial cells (Figure 6) all took up PKH26-labeled sEVs, and maximal sEV fluorescence was observed at 6 h post-injection. While the sEVs did not always colocalize with the cellular markers, we did not observe any differential uptake by CNS cells. Intrathecal injection with 5 µg of unlabeled RAW 264.7 sEVs or dye control did not show significant fluorescence (Supplemental Figure S1B). Fluorescent signals were observed in the meninges, both 6 h and 18 h after the injection of sEVs (Supplemental Figure S1C).
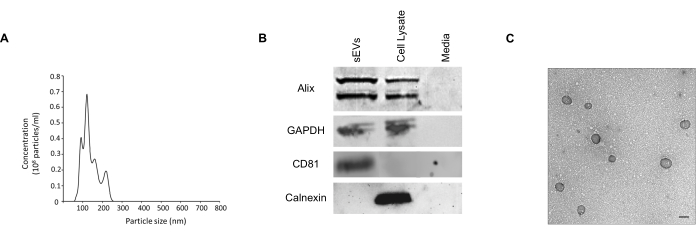
Figure 1: Characterization of purified RAW 264.7 sEVs. (A) Size and concentration of sEVs were determined using NanoSight NS300. The particles were tracked and sized based on Brownian motion and diffusion coefficient. The size distribution of sEVs is shown in nm. The concentration of sEVs was expressed as particles/mL. (B) Western blot of proteins derived from purified sEVs, cell lysate, and exosome-depleted media using sEV markers ALIX, GAPDH, and CD81. The endoplasmic reticulum protein marker, calnexin, serves as a control to monitor cellular contamination in sEV preparations. (C) Transmission electron microscopy demonstrated the size and morphology of sEVs. Scale bar = 100 nm. Abbreviations: sEVs = small extracellular vesicles; ALIX = Alpha-1,3/1,6-Mannosyltransferase (ALG-2)-interacting protein X; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; CD81 = cluster of differentiation 81. Please click here to view a larger version of this figure.
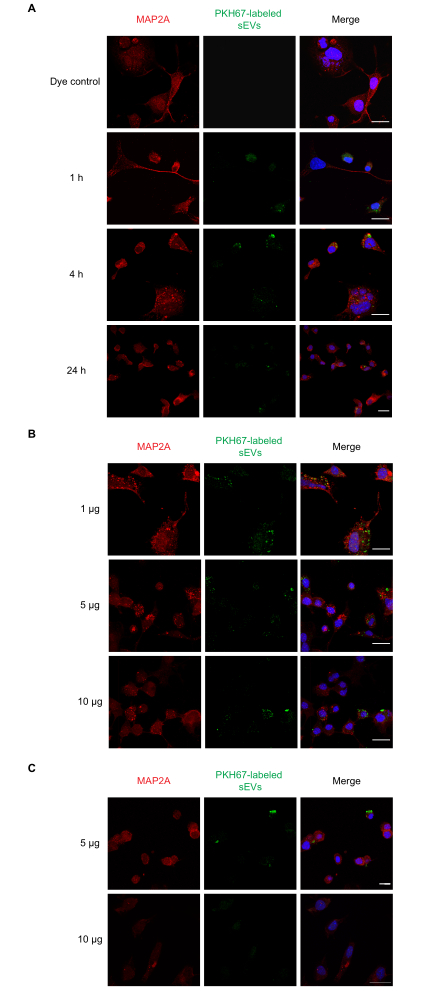
Figure 2: Uptake of labeled RAW 264.7 sEVs by Neuro-2a cells. (A) PKH67-labeled sEVs (1 µg) were added to the cultured Neuro-2a cells for 1, 4, or 24 h. sEV uptake was observed at all time points with confocal microscopy. (B) PKH67-labeled sEVs (1, 5, or 10 µg) were added to Neuro-2a cells for 1 h. (C) PKH67-labeled sEVs (5 or 10 µg) were added to Neuro-2a cells for 4 h. sEV uptake was observed in all dosage groups with confocal microscopy. Negative control groups treated with PKH dye alone did not show sEV staining (Supplemental Figure S1). Neuro-2a cells were immunostained with MAP2A (probed with Alexa Fluor 594, shown in red), while cell nuclei were stained with DAPI (shown in blue) and sEVs with PKH67 (shown in green). Scale bar = 50 µm. Abbreviations: sEVs = small extracellular vesicles; MAP2A = microtubule-associated protein 2A; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
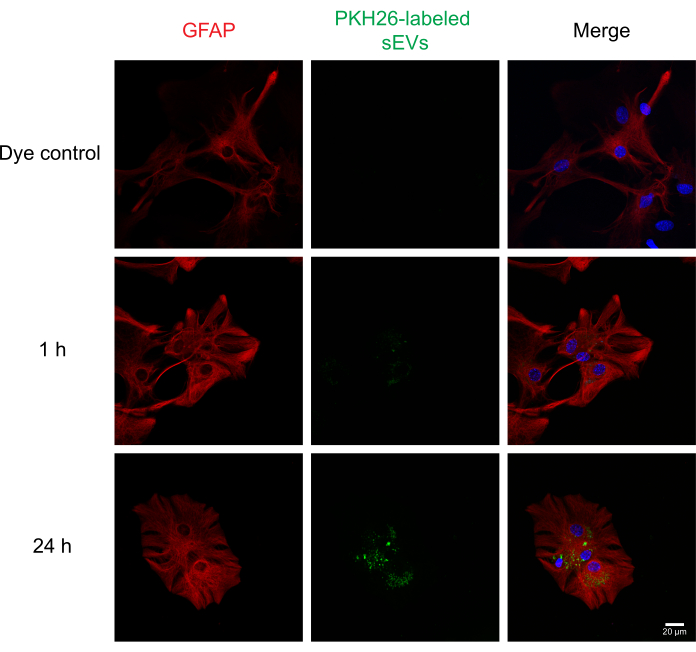
Figure 3: Uptake of PKH26-labeled RAW 264.7 sEVs by primary mouse cortical astrocytes. One µg of sEVs was labeled with PKH26 dye and added to the primary astrocyte culture medium. Uptake of sEVs was observed at 1 and 24 h post-addition using a confocal laser scanning microscope. Astrocytes were stained with GFAP (probed with Alexa Fluor 488, shown in red), while cell nuclei were counterstained with DAPI (shown in blue), and sEVs were previously stained with PKH26 (shown in green). Scale bar = 20 µm. PKH26 dye alone served as a negative control for sEV staining. Abbreviations: sEVs = small extracellular vesicles; GFAP = glial fibrillary acidic protein; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
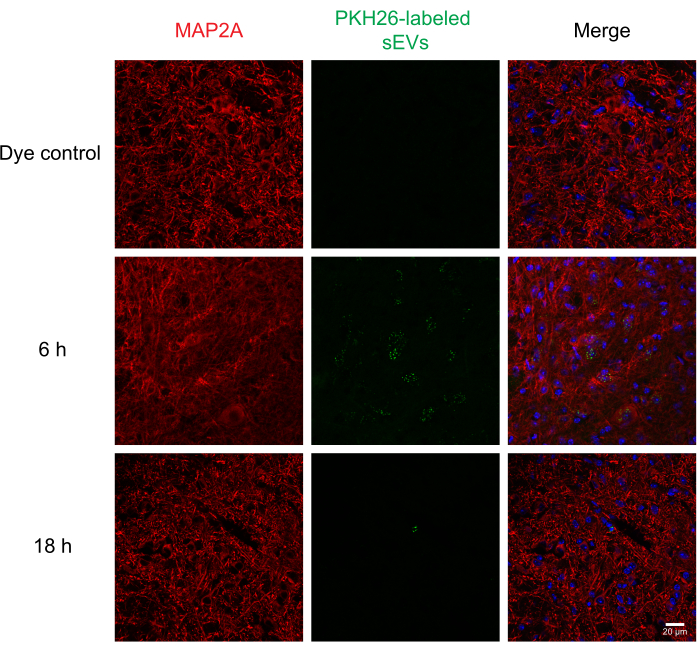
Figure 4: Uptake of RAW 264.7 sEVsin neurons. PKH26-labeled sEVs were injected intrathecally in mice; 6 and 18 h later, the mice were perfused with 4% PFA, and the spinal cord was isolated and sectioned at 30 µm. Spinal cord sections were immunostained with a cell marker (probed with Alexa Fluor 488, shown in red) and DAPI nuclear counterstain (shown in blue), while sEVs were previously labeled with PKH26 (shown in green). Spinal cord sections were immunostained for MAP2A to visualize the neurons (red). Confocal microscopy shows sEVs in MAP2A-positive neurons at different time points. The negative control, PKH26 dye-alone group, did not show sEV staining. Scale bar = 20 µm. Abbreviations: sEVs = small extracellular vesicles; PFA = paraformaldehyde; MAP2A = microtubule-associated protein 2A; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
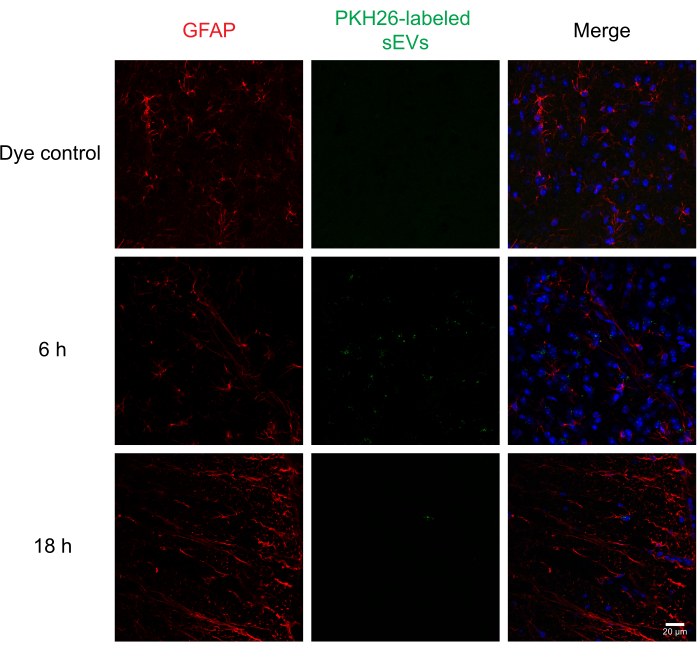
Figure 5: Uptake of RAW 264.7 sEVs in astrocytes. PKH26-labeled sEVs were injected intrathecally in mice; 6 and 18 h later, the mice were perfused with 4% PFA, and the spinal cord was isolated and sectioned at 30 µm. Spinal cord sections were immunostained with a cell marker (probed with Alexa Fluor 488, shown in red) and DAPI nuclear counterstain (shown in blue), while sEVs were previously labeled with PKH26 (shown in green). Spinal cord sections were immunostained for GFAP to visualize the astrocytes (red). Confocal microscopy shows sEVs in GFAP-positive astrocytes at different time points. The negative control, PKH26 dye-alone group, did not show sEV staining. Scale bar = 20 µm. Abbreviations: sEVs = small extracellular vesicles; PFA = paraformaldehyde; GFAP = glial fibrillary acidic protein; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
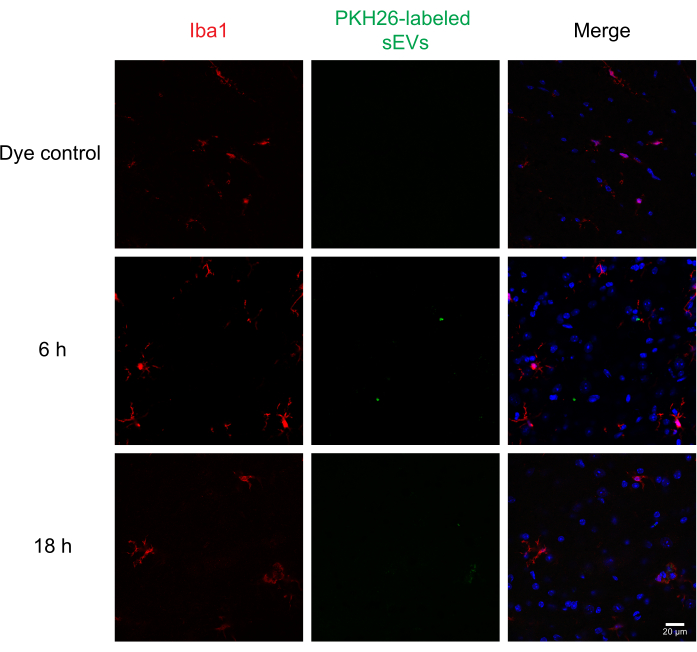
Figure 6: Uptake of RAW 264.7 sEVs in microglia. PKH26-labeled sEVs were injected intrathecally in mice; 6 and 18 h later, the mice were perfused with 4% PFA, and the spinal cord was isolated and sectioned at 30 µm. Spinal cord sections were immunostained with a cell marker (probed with Alexa Fluor 488, shown in red) and DAPI nuclear counterstain (shown in blue), while sEVs were previously labeled with PKH26 (shown in green). Spinal cord sections were immunostained for IBA1 to visualize the microglia (red). Confocal microscopy shows sEVs in IBA1-positive microglia at different time points. The negative control, PKH26 dye-alone group, did not show sEV staining. Scale bar = 20 µm. Abbreviations: sEVs = small extracellular vesicles; PFA = paraformaldehyde; IBA1 = ionized calcium-binding adaptor molecule 1; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
Supplemental Figure S1: Uptake of labeled RAW 264.7 sEVs by primary mouse cortical astrocytes and in spinal cord. (A) Controls for the uptake of PKH26-labeled RAW 264.7 sEVs by primary mouse cortical astrocytes. One µg of unlabeled sEVs resuspended in PBS or an equal volume of PBS was added in parallel to the culture medium of astrocytes. No fluorescence was observed at 1 h for PBS and the unlabeled control using a confocal laser scanning microscope. Astrocytes were stained with GFAP (probed with Alexa Fluor 488, shown in red), while the nuclei were counterstained with DAPI (blue), and unlabeled sEVs were visualized under the same Alexa Fluor 546 channel as PKH26-labeled sEVs. Scale bar = 50 µm. (B) Controls for the uptake of PKH26-labeled RAW 264.7 sEVs by mouse spinal cord in vivo. Five µg of unlabeled sEVs or dye control were injected intrathecally in mice. Again, fluorescent signals were not observed for unlabeled sEVs or dye-alone control using a confocal laser scanning microscope. Astrocytes were stained with GFAP (probed with Alexa Fluor 488, shown in red), while nuclei were counterstained with DAPI (blue), and unlabeled sEVs were visualized under the same Alexa Fluor 546 channel as PKH26-labeled sEVs. Scale bar = 50 µm. (C)Representative images reveal the presence of RAW 264.7 sEVs in mouse spinal meninges 6 h and 18 h after intrathecal delivery. Five µg of sEVs were labeled with PKH26 dye (shown in green), and the nuclei were counterstained with DAPI (shown in blue). Asterisks indicate the anterior spinal artery. Scale bar = 50 µm. Abbreviations: sEVs = small extracellular vesicles; PBS = phosphate-buffered saline; GFAP = glial fibrillary acidic protein; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to download this File.
Discussion
In this protocol, we showed the labeling of sEVs with PKH dyes and the visualization of their uptake in the spinal cord. PKH lipophilic fluorescent dyes are widely used for labeling cells by flow cytometry and fluorescent microscopy3,5,6,12,24,25. Due to their relatively long half-life and low cytotoxicity, PKH dyes can be used for a wide range of in vivo and in vitro cell-tracking studies26,27. Although excellent membrane retention and biochemical stability are advantageous, the intercalation of fluorescent probes with lipoprotein contaminants purified with sEVs can compromise the interpretation of sEV internalization and functional studies. Thus, the purification and labeling of sEVs are critical steps in the protocol because the persistence of the dyes with contaminants can lead to misinterpretation of the in vivo distribution28. The inclusion of controls is critical to avoid false-positive fluorescence signals due to non-specific labeling of particles and the long half-life of these dyes.
Aggregation and micelle formation of lipophilic dyes may also yield false signals. We addressed the problem of free or unbound dye by including a dye-alone control and visualizing the EV uptake at earlier time points. An important limitation reported for PKH labeling is that numerous PKH26 nanoparticles are formed during PKH26 dye labeling of sEVs. Although not included in this protocol, it is reported that PKH26 nanoparticles can be removed by a sucrose gradient29. Another study evaluated the effect of PKH labeling on the size of sEVs by NTA and reported an increase in size following PKH labeling30. Nevertheless, PKH dyes serve as a pragmatic and valuable tracer to show where the sEVs have traversed. Another limitation of this study is that we did not quantify sEVs as this protocol focuses on the confirmation of cellular uptake after intrathecal delivery. Novel cyanine-based membrane probes have been developed recently for highly sensitive fluorescence imaging of sEVs without altering the size or generating artifacts, such as the formation of PKH nanoparticles31, and will undoubtedly improve future labeling studies.
Although macrophages play important roles in neuroinflammation, they also exert neuroprotective functions by delivering their cargo via exosomes32. Our studies show that labeled macrophage-derived sEVs are taken up by Neuro-2a cells, primary astrocytes, and in the lumbar spinal cord after intrathecal administration. The results indicate that a longer incubation time can lead to lower sEV signal intensity, which could be attributed to the degradation of sEVs or cell division by Neuro-2a cells in culture33,34. Although low-throughput, this protocol for visualizing labeled sEVs in the spinal cord can be used for initial validation studies that confirm sEV uptake before investigating the functional impact of sEVs delivered intrathecally. As we observed generally similar sEV uptake in several CNS cell types, the uptake process appears to be non-selective. If autofluorescence is an issue in imaging, unlabeled sEVs can be used as an additional control to negate sEV autofluorescence during imaging of tissues and cultures. Although the dose and the route of administration of sEVs can influence the pattern of biodistribution11, this protocol is not optimized for the quantitative analysis of sEV uptake. Several different approaches and various imaging strategies are being employed to investigate sEVs, and these are being continually refined and optimized for in vivo tracking of sEVs2.
This protocol is meant to be just one approach to confirm sEV uptake. As with all protocols, cross-validation using multimodal approaches can be beneficial. Specifically, the uptake of sEVs can be confirmed by investigating biomolecular cargo transfer to recipient cells and tissues. If the investigator knows the miRNA composition of delivered sEVs, an alternate approach to confirm sEV transfer would be to check for miRNA changes in the recipient cells or determine the changes in expression levels of target genes for the miRNAs transferred. PBS-treated samples can be used as a control for this approach. Overall, these results support the concept that macrophage-derived sEVs are taken up by CNS cells in vitro and in vivo. This protocol can be used to investigate the role of sEVs in spinal disorders, pain, and inflammation and to determine whether sEVs can be developed as cellular vehicles for the delivery of therapeutic small molecules, RNA, and proteins.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was supported by grants from NIH NINDS R01NS102836 and the Pennsylvania Department of Health Commonwealth Universal Research Enhancement (CURE) awarded to Seena K. Ajit. We thank Dr. Bradley Nash for critical reading of the manuscript.
Materials
| Amicon Ultra 0.5 mL centrifugal filters | MilliporeSigma | Z677094 | |
| Anti-Alix Antibody | Abcam | ab186429 | 1:1000 |
| Anti-Calnexin Antibody | Abcam | Ab10286 | 1:1000 |
| Anti-CD81 Antibody | Santa Cruz Biotechnology | sc-166029 | 1:1000 |
| Anti-GAPDH Monoclonal Antibody (14C10) | Cell Signaling Technology | 2118 | 1:1000 |
| Anti-Glial Fibrillary Acidic Protein Antibody | Sigma-Aldrich | MAB360 | 1:500 for IF; 1:1000 for IHC |
| Anti-Iba1 Antibody | Wako | 019-19741 | 1:2000 |
| Anti-MAP2A Antibody | Sigma-Aldrich | MAB378 | 1:500 |
| Bovine Serum Albumin (BSA) | VWR | 0332 | |
| Cell Strainer, 40 μm | VWR | 15-1040-1 | |
| Centrifuge Tubes | Thermo Scientific | 3118-0050 | 12,000 x g |
| Coverslip, 12-mm, #1.5 | Electron Microscopy Sciences | 72230-01 | |
| Coverslip, 18-mm, #1.5 | Electron Microscopy Sciences | 72222-01 | |
| DAPI | Sigma-Aldrich | D9542-1MG | 1 µg/mL |
| DC Protein Assay | Bio-Rad | 500-0116 | |
| Deoxyribonuclease I (DNAse I) | MilliporeSigma | D4513-1VL | |
| Donkey Anti-Rabbit IgG H&L (HRP) | Abcam | ab16284 | 1:10000 |
| Donkey Anti-Rabbit IgG H&L, Alexa Fluor 488 | Invitrogen | A-21206 | 1:500 |
| Double Frosted Microscope Slides, #1 | Thermo Scientific | 12-552-5 | |
| DPBS without Calcium and Magnesium | Corning | 21-031-CV | |
| Dulbecco's Modified Eagle Medium (DMEM) | Corning | 10-013-CV | |
| Exosome-Depleted Fetal Bovine Serum | Gibco | A27208-01 | |
| Fetal Bovine Serum (FBS) | Corning | 35-011-CV | |
| FluorChem M imaging system | ProteinSimple | ||
| FV3000 Confocal Microscope | Olympus | ||
| Goat Anti-Mouse IgG H&L (HRP) | Abcam | ab6789 | 1:10000 |
| Goat Anti-Mouse IgG H&L, Alexa Fluor 488 | Invitrogen | A-11001 | 1:500 |
| Goat Anti-Mouse IgG1, Alexa Fluor 594 | Invitrogen | A-21125 | |
| Hank's Balanced Salt Solution (HBSS) | VWR | 02-0121 | |
| HEPES | Gibco | 15630080 | |
| HRP Substrate | Thermo Scientific | 34094 | |
| Intercept blocking buffer, TBS | LI-COR Biosciences | 927-60001 | |
| Laemmli SDS Sample Buffer | Alfa Aesar | AAJ61337AC | |
| Micro Cover Glass, #1 | VWR | 48404-454 | |
| Microm HM550 | Thermo Scientific | ||
| NanoSight NS300 system | Malvern Panalytical | ||
| NanoSight NTA 3.2 software | Malvern Panalytical | ||
| Neuro-2a Cell Line | ATCC | CCL-131 | |
| Normal Goat Serum | Vector Laboratories | S-1000 | |
| O.C.T Compound | Sakura Finetek | 4583 | |
| Papain | Worthington Biochemical Corporation | NC9597281 | |
| Paraformaldehyde | Electron Microscopy Sciences | 19210 | |
| Penicillin-Streptomycin | Gibco | 15140122 | |
| PKH26 | Sigma-Aldrich | MINI26-1KT | |
| PKH67 | Sigma-Aldrich | MINI67-1KT | |
| Protease Inhibitor Cocktail | Thermo Scientific | 1862209 | |
| PVDF Transfer Membrane | MDI | SVFX8302XXXX101 | |
| RAW 267.4 Cell Line | ATCC | TIB-71 | |
| RIPA Buffer | Sigma-Aldrich | R0278 | |
| Sodium Chloride | AMRESCO | 0241-2.5KG | |
| Superfrost Plus Gold Slides | Thermo Scientific | 15-188-48 | adhesive slides |
| T-75 Flasks | Corning | 431464U | |
| Tecnai 12 Digital Transmission Electron Microscope | FEI Company | ||
| TEM Grids | Electron Microscopy Sciences | FSF300-cu | |
| Tris-Glycine Protein Gel, 12% | Invitrogen | XP00120BOX | |
| Tris-Glycine SDS Running Buffer | Invitrogen | LC26755 | |
| Tris-Glycine Transfer Buffer | Invitrogen | LC3675 | |
| TrypLE Express | cell dissociation enzyme | ||
| Triton X-100 | Acros Organics | 327371000 | |
| Trypsin, 0.25% | Corning | 25-053-CL | |
| Tween 20 | |||
| Ultracentrifuge Tubes | Beckman | 344058 | 110,000 x g |
References
- Mulcahy, L. A., Pink, R. C., Carter, D. R. F. Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles. 3 (1), 24641 (2014).
- Betzer, O., et al. Advances in imaging strategies for in vivo tracking of exosomes. Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology. 12 (2), 1594 (2020).
- Dehghani, M., Gaborski, T. R. Fluorescent labeling of extracellular vesicles. Methods in Enzymology. 645, 15-42 (2020).
- González, M. I., et al. Covalently labeled fluorescent exosomes for in vitro and in vivo applications. Biomedicines. 9 (1), 81 (2021).
- Chuo, S. T. -. Y., Chien, J. C. -. Y., Lai, C. P. -. K. Imaging extracellular vesicles: current and emerging methods. Journal of Biomedical Science. 25 (1), 91 (2018).
- vander Vlist, E. J., Nolte-‘tHoen, E. N. M., Stoorvogel, W., Arkesteijn, G. J. A., Wauben, M. H. M. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nature Protocols. 7 (7), 1311-1326 (2012).
- Hoshino, A., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 527 (7578), 329-335 (2015).
- Heinrich, L., et al. Confocal laser scanning microscopy using dialkylcarbocyanine dyes for cell tracing in hard and soft biomaterials. Journal of Biomedical Materials Research. Part B, Applied Biomaterials. 81 (1), 153-161 (2007).
- Haney, M. J., et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. Journal of Controlled Release. 207, 18-30 (2015).
- Grange, C., et al. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. International Journal of Molecular Medicine. 33 (5), 1055-1063 (2014).
- Wiklander, O. P., et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. Journal of Extracellular Vesicles. 4, 26316 (2015).
- Lai, C. P., et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications. 6 (1), 7029 (2015).
- Deddens, J. C., et al. Circulating extracellular vesicles contain miRNAs and are released as early biomarkers for cardiac injury. Journal of Cardiovascular Translational Research. 9 (4), 291-301 (2016).
- Montecalvo, A., et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 119 (3), 756-766 (2012).
- Escrevente, C., Keller, S., Altevogt, P., Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 11, 108 (2011).
- Cho, E., et al. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. Journal of Controlled Release. 279, 326-335 (2018).
- Mantel, P. Y., et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host & Microbe. 13 (5), 521-534 (2013).
- Porro, C., Trotta, T., Panaro, M. A. Microvesicles in the brain: Biomarker, messenger or mediator. Journal of Neuroimmunology. 288, 70-78 (2015).
- De Toro, J., Herschlik, L., Waldner, C., Mongini, C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Frontiers in Immunology. 6, 203 (2015).
- Matsumoto, J., et al. Transmission of alpha-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: another mechanism for initiation and progression of Parkinson’s disease. Acta Neuropathologica Communications. 5 (1), 71 (2017).
- Matsumoto, J., Stewart, T., Banks, W. A., Zhang, J. The transport mechanism of extracellular vesicles at the blood-brain barrier. Current Pharmaceutical Design. 23 (40), 6206-6214 (2017).
- Shaimardanova, A., et al. Extracellular vesicles in the diagnosis and treatment of central nervous system diseases. Neural Regeneration Research. 15 (4), 586-596 (2020).
- Théry, C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 7 (1), 1535750 (2018).
- Hoen, E. N. M. N. -. t., et al. Quantitative and qualitative flow cytometric analysis of nanosized cell-derived membrane vesicles. Nanomedicine: Nanotechnology, Biology and Medicine. 8 (5), 712-720 (2012).
- Gangadaran, P., Hong, C. M., Ahn, B. -. C. An update on in vivo imaging of extracellular vesicles as drug delivery vehicles. Frontiers in Pharmacology. 9, 169 (2018).
- Teare, G. F., Horan, P. K., Slezak, S. E., Smith, C., Hay, J. B. Long-term tracking of lymphocytes in vivo: the migration of PKH-labeled lymphocytes. Cellular Immunology. 134 (1), 157-170 (1991).
- Kuffler, D. P. Long-term survival and sprouting in culture by motoneurons isolated from the spinal cord of adult frogs. Journal of Comparative Neurology. 302 (4), 729-738 (1990).
- Takov, K., Yellon, D. M., Davidson, S. M. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. Journal of Extracellular Vesicles. 6 (1), 1388731 (2017).
- Pužar Dominkuš, P., et al. PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochimica et Biophysica Acta. Biomembranes. 1860 (6), 1350-1361 (2018).
- Dehghani, M., Gulvin, S. M., Flax, J., Gaborski, T. R. Systematic evaluation of PKH labelling on extracellular vesicle size by nanoparticle tracking analysis. Scientific Reports. 10 (1), 9533 (2020).
- Shimomura, T., et al. New lipophilic fluorescent dyes for labeling extracellular vesicles: characterization and monitoring of cellular uptake. Bioconjugate Chemistry. 32 (4), 680-684 (2021).
- Yuan, D., et al. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials. 142, 1-12 (2017).
- Polanco, J. C., Li, C., Durisic, N., Sullivan, R., Götz, J. Exosomes taken up by neurons hijack the endosomal pathway to spread to interconnected neurons. Acta Neuropathologica Communications. 6 (1), 10 (2018).
- Jurgielewicz, B. J., Yao, Y., Stice, S. L. Kinetics and specificity of HEK293T extracellular vesicle uptake using imaging flow cytometry. Nanoscale Research Letters. 15 (1), 170 (2020).

