An Intestinal Gut Organ Culture System for Analyzing Host-Microbiota Interactions
Summary
This article presents a unique method for analyzing host-microbiome interactions using a novel gut organ culture system for ex vivo experiments.
Abstract
The structure of the gut tissue facilitates close and mutualistic interactions between the host and the gut microbiota. These cross-talks are crucial for maintaining local and systemic homeostasis; changes to gut microbiota composition (dysbiosis) associate with a wide array of human diseases. Methods for dissecting host-microbiota interactions encompass an inherent tradeoff among preservation of physiological tissue structure (when using in vivo animal models) and the level of control over the experiment factors (as in simple in vitro cell culture systems). To address this tradeoff, Yissachar et al. recently developed an intestinal organ culture system. The system preserves a naive colon tissue construction and cellular mechanisms and it also permits tight experimental control, facilitating experimentations that cannot be readily performed in vivo. It is optimal for dissecting short-term responses of various gut components (such as epithelial, immunological and neuronal elements) to luminal perturbations (including anaerobic or aerobic microbes, whole microbiota samples from mice or humans, drugs and metabolites). Here, we present a detailed description of an optimized protocol for organ culture of multiple gut fragments using a custom-made gut culture device. Host responses to luminal perturbations can be visualized by immunofluorescence staining of tissue sections or whole-mount tissue fragments, fluorescence in-situ hybridization (FISH), or time-lapse imaging. This system supports a wide array of readouts, including next-generation sequencing, flow cytometry, and various cellular and biochemical assays. Overall, this three-dimensional organ culture system supports the culture of large, intact intestinal tissues and has broad applications for high-resolution analysis and visualization of host-microbiota interactions in the local gut environment.
Introduction
The intestine is a highly complex organ containing a wide range of cell types (epithelial cells, immune system cells, neurons, and more) organized in a particular structure that allows cells to interact and communicate with one another and with the luminal content (microbiota, food, etc.)1. Currently, the research toolbox available for analyzing host-microbiota interactions includes in vitro cell cultures and in vivo animal models2. In vivo animal models provide a physiological tissue construction3 but with poor experimental control and limited ability to manipulate the experiment conditions. In vitro culture systems, on the other hand, use primary cells or cell lines that can be supplemented with microbes4, offering tight control over the experiment parameters but lack the cellular complexity and the tissue architecture. Modern in vitro assays allow the advanced use of healthy and pathological human tissue samples, like epithelial organoids derived from mouse or human sources5,6, and samples that mimic the mucosal microenvironment7. Another example is the 'gut on a chip' assay, which includes the human colonic epithelial cell line (Caco2), extracellular matrix and microfluidic channels to mimic the physiological condition of the gut invariant8. However, as advanced and innovative as in vitro samples may be, they do not maintain a normal tissue architecture or naïve cellular composition.
To address that, Yissachar et al. recently developed an ex vivo organ culture system9 (Figure 1) that maintains intact gut fragments ex vivo, benefiting from the advantages of both in vivo and in vitro models. This ex vivo gut organ culture system is based on a custom-made culture device that supports a multiplexed culture of six colon tissues, allowing examining experimental inputs under comparable conditions while controlling the system's inputs and outputs. Recent works have demonstrated that this system is valuable for analyzing intestinal responses to individual gut bacteria9, whole human microbiota samples10 and microbial metabolites11. This system allows, for the first time, the study of these early host-microbiota interactions with a high level of control over the host, microbial and environmental components. Furthermore, it allows monitoring and manipulating the system throughout the experiment, in real-time.
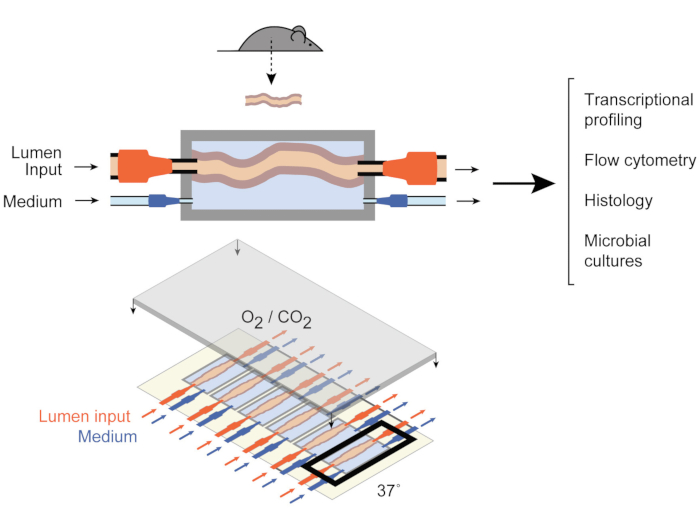
Figure 1: Schematics of the gut culture device. A whole intestinal tissue fragment is attached to the output and input ports of the chamber (top), with pumps regulating the medium flow inside the lumen and in the external medium chamber. The entire device (bottom) contains 6 such chambers. This figure has been modified from Yissachar et al. 2017. Please click here to view a larger version of this figure.
Protocol
This protocol follows the animal care guidelines approved by the ethics committee for animal welfare.
1. Experiment preparation
- Fabrication of the gut organ culture device (3 days)
- Using a 3D printer, print the reusable plastic molds for the organ culture device (the device has 6 wells, with 24 small and large holes, and for the device cover lid) (3D files attached).
NOTE: These plastic molds may be used for fabrication of numerous devices. - Insert the blunt-end needles (22 G & 18 G) to the appropriate position within the device mold and cast approximately 20 g of polydimethylsiloxane (PDMS) mix (1:10 weight ratio, base to catalyst) for one set of the device and lid.
- Place the molds in a vacuum chamber for 30 min, to remove air bubbles from the PDMS mix.
- Incubate the molds at 55 °C overnight, to complete PDMS polymerization.
- When the PDMS is set, pull out the needles from the mold and carefully release the culture device and lid from the plastic molds.
- Remove PDMS residues from the well outline using a surgical blade. Attach the PDMS device and the device cover onto a cover glass (75 mm x 50 mm micro slides) using non-toxic silicon adhesive and leave the parts to set overnight (apply the glue to the smooth side of the device).
- Insert twelve 22 G needles for the lumen and twelve 18 G needles for the well. Fix all the needles in place using silicone and let it set overnight. Insert two 18 G needles into the cover lid for proper air flow in and out of the gut organ culture device.
- Check all the needles for leaks using a water-filled syringe. Check that there is no leaking from the wells by filling the wells with water.
- Place two surgical knots on each 22 G needle that will be connected to the colon. Place the device and the lid in an autoclave paper bag and sterilize in an autoclave.
- Using a 3D printer, print the reusable plastic molds for the organ culture device (the device has 6 wells, with 24 small and large holes, and for the device cover lid) (3D files attached).
- Culture medium (0.5 h)
- In a biological hood, mix the following (in 50 mL tube): 37 mL of Iscove's Modified Dulbecco's Medium (IMDM), 10 mL of KSR serum replacement, 1 mL of B27 supplement, 0.5 mL of N2 supplement, 0.5 mL of 1 M HEPES buffer, and 0.5 mL of non-essential amino acids.
- Filter, and store the complete medium at 4 °C.
- Tubing and surgical tool preparation
- Cut the appropriate length of tubes for the input lumen, input well, output lumen, and output well (12 short tubes and 12 long tubes). Connect an appropriate adaptor to each side of the tube.
- Prepare the surgical tools: straight scissors, 4x thin forceps, and 2x sharp forceps.
- Place the tubes and the surgical tools in an autoclave paper bag and sterilize them using an autoclave.
- Prepare the luminal input (desired stimulation – bacteria, stool, drugs, etc.).
- Before the experiment, determine the bacterial load of the bacterial culture, by serial dilutions12, and culture under aerobic and/or anaerobic conditions.
- After calculating the bacterial load, dilute the bacterial cultures in sterile tissue culture medium to obtain the required bacterial concentration. For fecal samples, filter using a 100 µm strainer.
NOTE: For nonbacterial stimulation (drugs, metabolites, etc.), dilute the substance to the required concentration using the gut culture medium.
2. Experiment setup preparation
- Inside the organ culture incubator, turn on the heater unit, and set it to 37 °C.
- Set up the pumps as well as the input and output syringes.
- Tissue culture medium input
- In a biological or laminar flow hood, fill the input well syringes with complete culture medium. The final volume depends on the experiment's duration and the flow rate; usually 1 mL/h plus additional medium for purge.
- Connect the tubes to the syringes using the Luer-lock adaptor. Place the filled syringes in the syringe pumps.
- Purge the input syringes. Make sure that the well medium flows out from all tubes into a waste glass.
- Luminal input
- Fill the luminal input syringes with stimulation treatment (bacteria, drugs, etc.). The volume depends on the duration of the experiment and the flow rate (usually 30 µL/h plus additional medium for purge).
- Connect the tubes to the syringes using the Luer-lock adaptor. Place the filled syringes in the syringe pumps.
- Purge input syringes. Make sure that the stimulation flow out of all the tubes into a waste glass. Be careful not to contaminate the different stimulations.
- Outputs
- Place the empty syringes in the output syringe pumps (set pump mode to 'withdrawal').
- Device setup
- Set the regulator that controls the gas mixture (95% O2 + 5% CO2) flow rate for a gentle, minimal flow.
- Examine the device in a laminar hood.
- Flush the needles of the device with sterile IMDM to wash the needles.
- Add 500 µL of sterile culture medium into each well of the device.
- Preparation for tissue dissection
- Put the sterile surgical tools inside the laminar hood.
- Fill a 10 mL syringe with sterile IMDM and connect a sterile (autoclaved) 22 G blunt-end needle for flushing of the colon.
3. Organ cultures
- Mice sacrifice and tissue dissection
- In a laminar hood, sacrifice 12-14 day old mice by decapitation. Spray the mice with 70% ethanol and place the mice on a plastic plate.
- Using sharp scissors and forceps, dissect the mouse and take out the digestive tract from the stomach to the anus by cutting all the fat and connective tissues. Cut the colon and place it on a new plate.
NOTE: Minimize the contact with the colon tissue. Do not touch the middle part of the colon tissue. Hold the tissue gently and only at the edges of the tissue.
- Colon flush and wash
- Under a dissection microscope, gently flush the colon content with sterile IMDM (into the proximal side) with the prepared 10 mL syringe (step 2.7.2). After removing the feces from the intestinal tissue, place the colon in a new 6 well plate filled with 0.5 mL of sterile IMDM.
- Connecting the colon to the device
- Take the tissue and carefully connect it onto the 22 G needle and make a tight tie with the two threads. At this point, it is imperative to maintain the correct orientation of the colon to the lumen flow (proximal = input, distal = output). Repeat steps 3.2-3.3 for all 6 tissues.
- Verify that the input needle Luer locks are empty from the medium. If not, empty them. Add stimulations to the input needle Luer lock (to avoid entry of air bubbles into the lumen). Repeat this step for each colon with the appropriate stimulation.
- Check that all the tissues are connected and place the cover lid on top of the device.
- Connecting the organ culture device to the pumps
- Place the device into the pre-heated temperature-controlled chamber (37 °C).
- Connect gas flow
- Connect the gas adaptor to the cover lid using the appropriate input needle.
- Connect the input and output tubes to the device.
NOTE: Connect the proximal colon side to the input tubs.
- Purge the lumen with the stimulation inputs.
- Gently flow luminal stimulation through the gut and verify medium flow in the output tubes.
- Wash external medium.
- Wash the external medium 3 times (set the pumps at a rate of 600 µL/min for both input and output well). Each wash takes 1 min (starting by emptying the well).
- Start the experiment.
- Start the pumps at the following rates:
Flow rate: lumen: input- 30 µL/h, output- 35 µL/h
External medium: input- 1000 µL/h, output- 950 µL/h
NOTE: Experiment time can vary between 30 min to 24 h.
- Start the pumps at the following rates:
- End of experiment (up to 24 h for colon organ cultures)
- Disconnect all the tubes from the device.
- Disconnect the tissues from the needles and continue to the desired readouts.
Representative Results
The gut organ culture system maintains tissue viability ex vivo. The evaluation of the tissue viability was done throughout the culture period. Colon tissue fragments were incubated in the gut organ culture system and fixed following 2/12/24 h culture. The intestinal epithelial cell (IEC) layer integrity was validated by immunofluorescence staining using E-cadherin and cytokeratin-18 antibodies. Likewise, mucus-filled goblet cells in the colonic epithelium and mucus secretion within the lumen were detected as well as proliferating IEC in the colonic crypts, as indicated by Ki-67 staining (Figure 2). These results and others9 show that the gut culture system maintain intestinal function and structure ex vivo.
Rapid transcriptional host response was triggered by intestinal colonization with segmented filamentous bacteria (SFB). Colonization by two different immunomodulating gut microbes were used to examine the initial events induced in the gut. The primary microbe was segmented filamentous bacteria (SFB) that was previously shown to induce differentiation of Th17 cell in the small intestine13. SFB-negative SPF mice were sacrificed and segments of the small intestine (SI) (ileum) were dissected and connected to the gut organ culture system. As SFB is difficult to culture in vitro14, organ cultures were infused with a suspension of fecal pellets from SFB monocolonized mice15 or fecal pellets from GF mice as control. SFB induced Th17 through adherence to the intestinal epithelium13,16. Thus, the evaluation of the spatial localization of SFB early after ex vivo colonization of intestinal tissue was required. As shown in (Figure 3a), typical SFB filaments were detected in close association with the SI villi using fluorescence in situ hybridization (FISH), 2 h after SFB introduction. Additionally, a transmission electron microscopy presented SFB within a few microns of the SI epithelium brush border (Figure 3b). At that point, Yissachar et al. examined whether the primary association of SFB with the small intestine epithelium activated a transcriptional response in the tissue. Gene expression profiles of whole-tissue samples were produced, in triplicate, 2 h after infusion with SFB. Control cultures were infused with fecal suspensions of GF or B.fragilis-monocolonized mice (as a non-Th17-inducing control). Figure 3c shows that the changes persuaded by SFB were mainly of small amplitude, compared to the GF control.
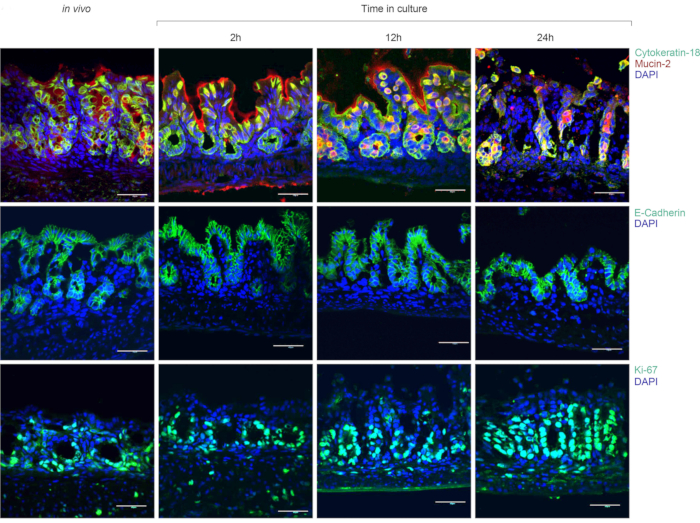
Figure 2: The gut organ culture system maintains tissue viability ex vivo. Immunofluorescence staining of Mucin-2 and Cytokeratin-18 (top), E-cadherin (middle) and Ki-67 (bottom). Confocal imaging of freshly dissected colon tissues and tissues cultured for 2/12/24 h in the gut organ culture device. Scale bar, 40 mm. This figure has been modified from Yissachar et al. 2017. Please click here to view a larger version of this figure.
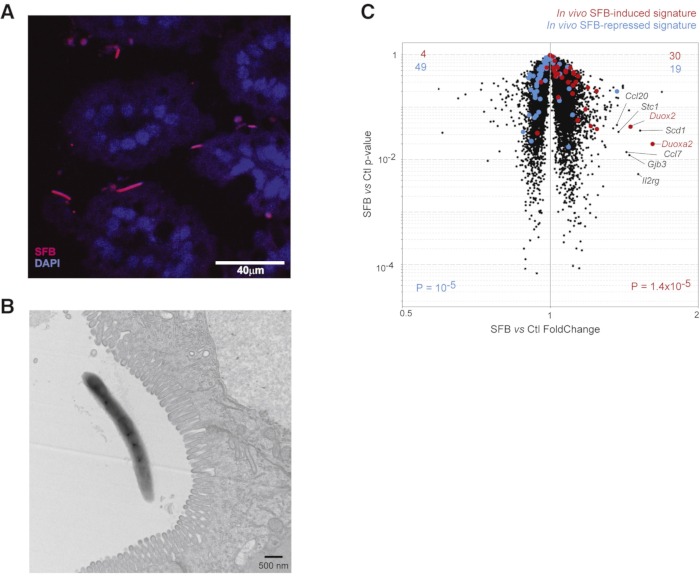
Figure 3: Mucosal association and rapid transcriptional triggering of a typical signature by SFB. (A, B) SFB associates with the intestinal epithelium after 2 h in cultured small intestine segment visualized by (A) FISH with an SFB-specific probe (red) or (B) electron microscopy. (C) Induction of intestinal gene expression by SFB. SI organ cultures were infused with microbes containing supernatant of feces from SFB monocolonized mice or GF controls and cultured for 2 h before microarray profiling of gene expression in the entire tissue. Modifications in gene expression on a ''volcano plot'' comparing SFB or GF infused cultures. Transcripts up or down-regulated by SFB in whole SI in vivo are highlighted (red and blue, respectively). This figure has been modified from Yissachar et al. 2017. Please click here to view a larger version of this figure.
Discussion
This article describe an optimized protocol for ex vivo gut organ cultures that Yissachar et al. have recently developed (published9 and unpublished data). The gut organ culture system supports multiplexed culture of intact intestinal fragments while maintaining luminal flow. It provides full control over the intra- and extra-luminal environment (including stimulation dose, exposure time and flow rate) and preserves the naïve intestinal tissue structure and its cellular complexity9.
Critical steps in the protocol include the following. (1) Tissue dissection should be performed in minimal time, and very gently, to minimize tissue damage (which may lead to cell death and leaking of luminal content through damaged sites). (2) Maintain tissue orientation in the device by connecting the input port to the proximal colon, and the output port to the distal end of the colon. (3) After connecting the gut to the device, make sure that the tissue is intact and that the connection points to the intra-luminal ports are not leaking. That will keep the bacterial stimulation inside the lumen and will not contaminate the external environment, which should be maintained as sterile as possible. (4) Tissues should be dissected from mice aged 12-14 days. In younger mice, the colon is smaller and gentler and requires different handling and culture conditions (data not shown). Tissues from older mice are significantly larger, which affects tissue viability and culture duration. (5) To reduce variability in gut responses, use tissues dissected from littermate mice in each device (reduce variability in endogenous microbiome composition, similar to in vivo experimental design).
Current limitations of the gut culture system include the following. (1) Each device contains six channels, which limit the number of different conditions that can be tested in one experiment. (2) Tissues should be dissected from mice aged 12-14 days (see above); thus, the enteric immune system and the epithelial barrier have not reached final maturation. (3) Tissue viability in culture is impaired if culture duration exceeds 24 h (for colon tissues, less for small intestine). (4) This protocol requires the purchase of special equipment (i.e., syringe pumps, custom-made incubator, and others).
In summary, the gut organ culture system serves as an intermediate experimental step between simple in vitro assays to complex in vivo experiments. It offers a combination of high controllability (as in simple in vitro assays) with preservation of intestinal physiology (closely resembles in vivo models) – a significant and unique advantage of this system. This advantage facilitates experimentations that cannot be readily performed in mice (i.e., tracking early responses of gut-residing cells in high temporal resolution). Recently, it has enabled us to discover some surprising connections between the gut microbiome (specific microbes and whole human-derived microbiota) and the intestinal immune and nervous systems9,10. Overall, this powerful and unique tool can be combined with a wide range of readout techniques (including next-generation sequencing, imaging, cell sorting, and more) and provides some novel insights into host-microbiome interactions in health and disease.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank past and present members of the Yissachar lab for their valuable contributions in optimizing the gut organ culture system protocol. We thank Yael Laure for critical editing of the manuscript. This work was supported by the Israel Science Foundation (grant No. 3114831), the Israel Science Foundation – Broad Institute Joint Program (grant No. 8165162), and the Gassner Fund for Medical Research, Israel.
Materials
| Device | |||
| 18 Gauge Blunt Needle | Mcmaster | 75165a754 | |
| 22 Gauge Blunt Needle | Mcmaster | 75165a758 | |
| All Purpose Adhesive Selant 100% Silicone | DAP | 688 | |
| Cubic Vacuum Desiccator VDC-21+ 2 Shelves | AAAD4021 | ||
| Glass Slide 1 mm Thick | Corning | 2947-75X50 | |
| Mini Incubator im-10 | AAH24315K | ||
| MPC 301E Vacuum PUMP | VI-412711 | ||
| Plastic Quick Turn Tube Coupling Plugs | Mcmaster | 51525k121 | |
| plastic Quick Turn Tube Coupling Sockets | Mcmaster | 52525k211 | |
| Sylgard 184 Silicone Elastomer | Dow | Polydimethylsiloxane, PDMS | |
| Tubing | Mcmaster | 6516t11 | |
| Zortrax M200 | Zortrax | Zortrax Z-SUITE, Autodesk Fusion 360 | |
| Zortrax M200 Materials: z-ultrat | Zortrax | ||
| Medium | |||
| B27 Supplement (50x), Serum Free | Thermo Fisher Scientific | 17504044 | |
| HEPES Buffer (1M) | Thermo Fisher Scientific | 15630056 | |
| Iscove's Mod Dulbecco's Medium With Phenol Red (1x) | Thermo Fisher Scientific | 12440061 | |
| Knock-Out Serum | Thermo Fisher Scientific | 10828028 | |
| N2 Supplement (100x) | Thermo Fisher Scientific | A1370701 | |
| Non Essential Amino Acid (100x) | Thermo Fisher Scientific | 11140035 | |
| Surgical Tools | |||
| Large Scissors | Aseltech | 11-00-10 | |
| Sharp Forceps | F.S.T | 11297-10 | |
| Silk Braided Surgical Thread | SMI | 8010G | |
| Straight Scissors | F.S.T | 14091-09 | |
| Thin Forceps | F.S.T | 11051-10 | |
| Organ System | |||
| 0.1 µm Filter | Life Gene | ||
| 0.22 µm Filter | Life Gene | ||
| 5 mL Luer Lock Syringe | B-D | 309649 | |
| Greenough Stereo Microscope | ZEISS | Stemi 305 | |
| Recirculating Precision Air Heater "CUBE" | CUBE-2-LIS | ||
| Syringe Pump | new era pump systems inc | nep-ne-1600-em |
References
- Mowat, A. M., Agace, W. W. Regional specialization within the intestinal immune system. Nature Reviews Immunology. 14 (10), 667-685 (2014).
- Pearce, S. C., et al. Intestinal in vitro and ex vivo Models to Study Host-Microbiome Interactions and Acute Stressors. Frontiers in Physiology. 9 (1584), (2018).
- Hooper, L. V., et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 291 (5505), 881-884 (2001).
- Haller, D., et al. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 47 (1), 79-87 (2000).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Tsilingiri, K., et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut. 61 (7), 1007-1015 (2012).
- Gazzaniga, F. S., et al. Harnessing Colon Chip Technology to Identify Commensal Bacteria That Promote Host Tolerance to Infection. Frontiers in Cellular and Infection Microbiology. 11, 638014 (2021).
- Yissachar, N., et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell. 168 (6), 1135-1148 (2017).
- Duscha, A., et al. Propionic Acid Shapes the Multiple Sclerosis Disease Course by an Immunomodulatory Mechanism. Cell. 180 (6), 1067-1080 (2020).
- Grosheva, I., et al. High-Throughput Screen Identifies Host and Microbiota Regulators of Intestinal Barrier Function. Gastroenterology. 159 (5), 1807-1823 (2020).
- Blaize, J. F., Corbo, C. P. Serial Dilutions and Plating: Microbial Enumeration. Journal of Visualized Experiments. , (2021).
- Ivanov, I. I., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139 (3), 485-498 (2009).
- Schnupf, P., et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 520 (7545), 99-103 (2015).
- Chung, H., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149 (7), 1578-1593 (2012).
- Atarashi, K., et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 163 (2), 367-380 (2015).

