Developing Drosophila melanogaster Models for Imaging and Optogenetic Control of Cardiac Function
Summary
The present protocol describes the generation of Drosophila melanogaster expressing eNpHR2.0 or ReaChR opsins in the heart for OCT imaging and optogenetic heart pacing. Detailed instructions for Drosophila OCT imaging and heart beating modulation, including the simulation of restorable heart arrest, bradycardia, and tachycardia in live animals at different developmental stages, are reported.
Abstract
Using Drosophila melanogaster (fruit fly) as a model organism has ensured significant progress in many areas of biological science, from cellular organization and genomic investigations to behavioral studies. Due to the accumulated scientific knowledge, in recent years, Drosophila was brought to the field of modeling human diseases, including heart disorders. The presented work describes the experimental system for monitoring and manipulating the heart function in the context of a whole live organism using red light (617 nm) and without invasive procedures. Control over the heart was achieved using optogenetic tools. Optogenetics combines the expression of light-sensitive transgenic opsins and their optical activation to regulate the biological tissue of interest. In this work, a custom integrated optical coherence tomography (OCT) imaging and optogenetic stimulation system was used to visualize and modulate the functioning D. melanogaster heart at the 3rd instar larval and early pupal developmental stages. The UAS/GAL4 dual genetic system was employed to express halorhodopsin (eNpHR2.0) and red-shifted channelrhodopsin (ReaChR), specifically in the fly heart. Details on preparing D. melanogaster for live OCT imaging and optogenetic pacing are provided. A lab-developed integration software processed the imaging data to create visual presentations and quantitative characteristics of Drosophila heart function. The results demonstrate the feasibility of initiating cardiac arrest and bradycardia caused by eNpHR2.0 activation and performing heart pacing upon ReaChR activation.
Introduction
At the end of 2010, the Nature Methods journal selected optogenetics as the Method of the Year1. Using genetic tools (transgenic opsins) regulated by light to control biological tissues of interest with unprecedented precision and speed opened a flood gate for new applications. To date, the majority of accomplishments belong to neuroscience. The technology was introduced as a new method of precise control of single neurons2 and has advanced to discoveries in the area of live organism cognitive functions3. From the start, neuroscientists demonstrated the ability to modulate the whole organism's behavior. Expression and light activation of ChR2 opsin in mice dopaminergic neurons caused their activation and were sufficient to drive behavioral conditioning4. Optogenetic inhibition of a subset of neurons containing halorhodopsin NpHR2.0 delivered to the epileptic focus of the rodent brain resulted in attenuation of electroencephalographic seizures5.
Optogenetic applications in cardiology are developing at a steady pace6. ChR2 was successfully expressed in cardiomyocytes cell culture and in mice; heart pacing was conducted by flashes of blue light (performed using an implanted fiber in live animals)7. In zebrafish, ChR2 was expressed and used to identify the pace-making heart region; NpHR activation induced heart arrest8. Optogenetic cardiac pacing has the unique potential for developing new pacing and resynchronization therapies9. Attempts to establish an autogenous arrhythmia termination system have been reported recently as well10.
Extensive research and the development of new therapeutical treatments require the application of various model systems, from cell culture to mammals. A vertebrate's heart is a very complex organ. Cardiomyocytes (CM) comprise one-third of all cardiac cells; other cells include neurons, vascular smooth muscle cells, and non-excitable cells (i.e., endothelial cells, fibroblasts, and immune cells). Researching CM cell culture limits the translation of the obtained results to human medical applications. Mammalian model organisms' genetic manipulations are limited and time-consuming. Smaller invertebrate models have many advantages; their cardiovascular system carries all the essential histological elements. Drosophila melanogaster (fruit fly) is a simple and powerful genetic model system to investigate the role of genes associated with human diseases, including cardiac diseases11,12,13. As short-lived animals, fruit flies represent an excellent opportunity to model age- or disease-dependent cardiac function changes that can be traced throughout life14,15,16,17. The fruit fly's heart tube is located on the dorsal side of its body within 200 µm of the cuticle surface, allowing visible to near-infrared light to reach the heart tube. This anatomical feature enables non-invasive optical pacing of the Drosophila heart using existing optogenetic tools.
To monitor the Drosophila heart, a custom spectral-domain optical coherence tomography (SD-OCT) imaging system with an integrated red light LED excitation module was developed18. Morphological and rhythmic changes in a relatively simple fruit fly heart can be readily analyzed with this non-invasive biomedical imaging technology12,19,20,21. With enhanced optical sectioning performance and micron-scale spatial resolution, OCT has been successfully used to investigate the structure and monitor the function of the Drosophila heart at different developmental stages, including the 3rd instar larva and early pupa18. This system also enables simultaneous monitoring and stimulation of the Drosophila's cardiac condition in the intact animal. A schematic view of the OCT system is shown in Figure 1. The SD-OCT system uses a superluminescent diode (SLD) as the light source (center wavelength: 850 nm ± 10 nm, FWHM: 165 nm, see Table of Materials). Using a 10x objective lens, the OCT imaging system can achieve an axial resolution of ~4.4 µm in air and ~3.3 µm in tissue and a lateral resolution of ~2.8 µm, sufficient to resolve fine details of the fly heart structures18,22. Interference signals of reflected light from the reference arm and the sample arm are detected using a spectrometer with a 2048-pixel line scan camera (max line rate: 80 kHz, see Table of Materials). The measured system sensitivity is ~95.1 dB. Each B-mode OCT scan generates a cross-sectional image in the xz image plane. Repeated B-mode images are acquired at the same location to create M-mode images capturing the beating heart for over ~30 s18,22,23. The frame rate for M-mode imaging is ~125 frames/s, sufficient to capture the fruit fly heart beating dynamics.
For the optogenetic regulation of Drosophila heart function, an illumination module with a 617 nm LED light source is integrated with the sample arm of the SD-OCT system. The stimulation light is focused on a ~2.2 mm diameter spot on the specimen surface, at the same position as the imaging focus spot. A custom-written software is utilized to control the illumination mode (light intensity, pulse width, and duty cycle), adjust the light pulse stimulation frequency, and synchronize the LED module illumination and M-mode OCT imaging acquisition22.
Recent publications described the Drosophila transgenic system consisting of spatiotemporally regulated ChR2, ReaChR, and eNpHR2.0 opsins using the UAS/GAL4 genetic system. The obtained results have demonstrated the ability to initiate heart arrest and bradycardia caused by red light activation of eNpHR2.0 and higher frequency heart pacing caused by blue light activation of ChR2. Similar pacing experiments were performed with another channelrhodopsin, ReaChR, inducible by red-light illumination22,23,24. The opsin expression in all the described experiments was driven by 24B-GAL4, where opsin expression was observed in a broad range of tissues, including cardiomyocytes and surrounding muscle cells. In the current study, 24B-GAL4 was replaced by a Hand-GAL4 driver to achieve heart-specific eNpHR2.0 and ReaChR opsins expression.
Overall, the presented experimental results demonstrate restorable heart arrest and inducible bradycardia and tachycardia heart conditions. A detailed protocol with step-by-step instructions on creating transgenic Drosophila models and conducting simultaneous OCT imaging and optogenetic pacing experiments in live animals is provided.
Protocol
For the present study, eNpHR2.0 transgenic line w[*]; P{y[+t7.7] w[+mC]=UAS-eNpHR-YFP}attP2, ReaChR transgenic line w[*];P{y[+t7.7] w[+mC]=UAS-ReaChR}su(Hw)attP5/CyO, and heart-specific GAL4 driver containing Hand gene regulatory fragment w[1118]; P{y[+t7.7] w[+mC]=GMR88D05-GAL4}attP2/TM3 Sb[1] (this driver stock will be indicated as Hand-GAL4) were used. y[*] w[*]; P{w[+mC]=UAS-2xEGFP}AH3 was used as the GFP reporter line. The mentioned Drosophila stocks were obtained from the Bloomington Drosophila Stock Center (BDSC, see Table of Materials) and maintained at room temperature or at 18 °C on standard cornmeal media. The Drosophila models developed in this study are available upon request for collaborative work.
1. Drosophila genetic crosses and media preparation
- Change the 3rd chromosome balancer TM3 Sb[1] 세스 TM6 Sb Tb,creating w[1118]; P{y[+t7.7] w[+mC]=GMR88D05-GAL4}attP2/TM6 Sb Tb (Hand-GAL4/TM6 Sb Tb). See Supplementary Figure 1 for the crossing scheme. Set crosses in vials with regular cornmeal media.
NOTE: The presence of a Tb marker allows users to distinguish larvae and pupae containing opsin transgene and the GAL4 driver from animals containing opsin but no driver25. - Keep the genetic crosses in a 25 °C, 70% humidity incubator on specially formulated all–trans retinaI (ATR)-containing media (see Table of Materials) in the dark for 5 days for larvae collection and 6 days for pupae collection.
- Combine five Hand-GAL4 /TM6 Sb Tb virgin females and two to three males from UAS-opsin stocks (eNpHR2.0, or ReaChR) per vial. See the cross diagram for eNpHR2.0 and ReaChR opsin in Figure 2A and Figure 2B, respectively.
- On the next day, prepare ATR-containing media vials.
- Prepare Semi-Defined Food according to the instructions of BDSC26. Instead of sucrose and glucose, add only sucrose (5.14 g/100 mL). Cool to ~60 °C with constant stirring.
- Prepare narrow fly vials, and add 50 µL of 100 mM ATR-ethanol solution to each vial.
- Using a serological pipette, dispose of fly food to narrow fly vials, 5 mL per vial. Vortex at maximum speed for 10 s.
- Plug the vials, and wrap them in the dark fabric to protect them from light. Let vials dry for at least 12 h (overnight).
- The next day, transfer the flies steadily laying eggs from Step 1.3. to the vials with ATR-containing food (Step 1.4.4.). Protect the racks with vials from light.
- After 24-48 h (depending on the number of eggs laid), discard the parents to prevent vial over-population.
- Collect non-Tb progeny and use them for heart imaging.
NOTE: The phenotypic differences at the larval and pupal stages are demonstrated in Figure 2C. The summary and the approximate timeline of the specimen preparation steps are shown in Figure 3.
2. Optogenetic control of the Drosophila heart
- Pick UAS-opsin/Hand-GAL4 larva/pupa from the vial (Step 1.7.), put on tissue, and gently wipe off the media from the body surface using a painting brush.
- Prepare the microscope slide and place a small piece of double-sided tape in the middle.
- Using a brush or fine tweezers, gently place the larva/pupa on the tape surface with the dorsal side up and perpendicular to the long side of the slide. Apply gentle pressure to attach the larva/pupa to the tape surface.
- Set up the slide on the imaging stage, larva/pupa facing down.
- Turn on the OCT light source by laser control software (see Table of Materials). Open the custom-written SD-OCT control software, then click on the Preview window.
- Set the scan parameters in the SD-OCT software.
NOTE: The goal is to align the sample for optimal imaging of the beating heart, so selecting the X range and Y range covers the heart's region. At this step, both the number of A-scans and B-scans are 400. The range in the x and y directions is ~490 µm and ~537 µm, showing the heart's two orthogonal cross-sections (xz and yz), respectively. - Use micromanipulators to control the sample stage to bring the fly heart into focus. Adjust the focal position to minimize light reflection from the fly cuticle surface. Consider applying mineral oil on the larva/pupa surface to minimize reflection.
NOTE: Oil may increase the risk of animal movement by compromising the tape adhesive properties. - Ensure that the fly heart can be fully viewed in the image window without any distortions, being blocked out by tissue, and non-negligible shadows and reflections; otherwise, go back to Step 2.7.
- Set the scan parameters for M-mode OCT image acquisition.
NOTE: The number of A-scans is reduced compared to Step 2.7. for the faster frame rate to capture the beating dynamics of the fly heart (several Hz). The number of B-scans denotes the number of repeated frames for M-mode imaging, which can be adjusted based on the recording time and available system memory. In this experiment, 128 A-scans can allow a speed of ~125 frames/s, and 4,000 repeated B-scans are recorded, providing a continuous recording of ~32 s. - Acquire five sets of control data without red light stimulation pulses to calculate the resting heart rate (RHR).
- Design the light pulse for the pacing stimulation in the custom OCT control software. In "Settings", add the designed light pulse sequences to control pulse frequency, pulse width, stimulation duration, and waiting time according to different stimulation protocols.
NOTE: The RHR is measured from the control experiment without light illumination and used to calculate the frequency at which light should be pulsed for tachypacing and bradypacing experiments22. - Open the light controller software (see Table of Materials) to generate red light pulses. Choose the Pulse Mode in "Mode Selection". Double-click the figure for the "Pulse Profile" settings and choose Follower Mode. Keep the OFF intensity as 0, and set the ON intensity percentage upon calculating the actual power density.
NOTE: Stimulation light pulses are triggered by a signal from the OCT control software according to the settings in Step 2.12. - Acquire M-mode videos of the beating Drosophila heart with light stimulation by clicking on Acquire in the OCT control software. Repeat the measurements 5x.
3. Image analysis
- Open the custom-developed fly heart segmentation software.
- Click on Select File and then select the file to be analyzed in the GUI that appears.
- Enter both the vertical and horizontal boundaries of the heart region in pixels in the top text boxes. Click on Resize. Using the slider on the bottom, ensure that the entire heart region is visible and that it fills up the whole box for the entire collection. If it is not, repeat this process and adjust the boundaries.
- Click on Predict to predict the heart region. The program will now go through every slice in the collection and select the heart region, taking approximately 3 min.
- Click on HR Plot once the prediction is completed. This will open a new window displaying a plot of the heart area over time. Ensure the correct peak or valley areas are selected. Choose Pulse and then HR to generate a final figure, and the functional parameters will be saved in the .csv files simultaneously.
Representative Results
D. melanogaster animals expressing red light-sensitive opsins eNpHR2.0 or ReaChR in the heart tube were generated by obtaining progeny from the cross between each UAS-opsin transgenic line and Hand-GAL4 driver. The tissue specificity of the GAL4 driver was verified by imaging GFP expression (Figure 4). Drosophila 3rd instar larva and early pupa developmental stages were used to demonstrate the effects of eNpHR2.0 and ReaChR activation by red light. Designed ~617 nm red-light pulses, delivered by LED, illuminated the larva/pupa and activated the eNpHR2.0 and ReaChR in the heart. Although the reported maximum response wavelength of NpHR is ~580 nm and of ReaChR is ~600 nm, 617 nm light illumination can penetrate deeper with enhanced light energy delivery toward the opsin-expressing heart tissue22.
Mounted on the microscope slide with the dorsal side down in the inverted microscope setup, the larva/pupa was illuminated by an LED light beam directed to the A7 body segment. Examples of the body cross-section images are shown in Figure 5A and Figure 6A. The heart appears as a contracting and dilating circular shape in the video recordings consisting of 4,000 frames (Supplementary videos 1–6). To mimic different heart conditions, four types of light pulses were designed. A single pulse lasting 10 s after 5 s waiting time generated restorable heart arrest induced by eNpHR2.0, as shown in Figure 5B. For the heart pacing at frequencies slower than the resting heart rate (RHR), mediated by eNpHR2.0, two light-pulse sequences with pacing frequencies equal to RHR/2 and RHR/4 lasting 8 s with a waiting time of 6 s in between were used (Figure 5C). The duty cycle of each light pulse sequence was 90%. This light stimulation regimen caused a heart condition reminiscent of bradycardia. The stimulation pattern to increase the heart rate due to ReaChR activation consisted of three sequences of light pulses at frequencies of RHR + 0.5 Hz, RHR + 1 Hz, and RHR + 1.5 Hz, respectively, with a pulse width of 20 ms (Figure 5D). This pulse regimen was aimed at causing a tachycardic heart condition. The light power density was 7.49 mW/mm2 during all the experiments. For control experiments, no light illumination was set.
Each experimental variant was recorded five times. M-mode videos of the fly heart were processed into 2D masks using FlyNet 2.027. This software automatically segments the heart region to produce the cardiac function data sets. The program provides a mask of the heart in each frame, which can be further corrected manually, if needed, to generate accurate quantification of the functional parameters of the beating heart, such as heart rate (HR), end-diastolic dimension (EDD), and end-systolic dimension (ESD), fractional shortening (FR), end-diastolic area (EDA), end-systolic area (ESA), etc. The heart rate is measured by analyzing the heart area over time. The control video with no light pulses is used to establish a baseline heart rate (e.g., RHR) for each animal.
Figure 5B and Figure 6B show 10 s long heart arrest caused by Hand>eNpHR2.0 activation using red light (617 nm) in larva and pupa, respectively. When the red light was turned on, the Drosophila's heart stopped beating and remained in this state until the end of the light illumination. The heart function was restored after the red light was turned off. Animals that did not have opsin expressed ("no opsin" control) did not respond to the red-light illumination (Supplementary Figure 2A and Supplementary Figure 3A). The control experiments with Hand>eNpHR2.0 animals where the 10 s red light illumination was not turned on ("no light" control) showed the heart beating normally (Supplementary Figure 4A and Supplementary Figure 4C).
Using Hand>eNpHR2.0 animals, red-light pulses at frequencies lower than the RHR were applied. The heart contraction frequency was reduced following the light signals; this slower heart rate mimics one type of heart arrhythmia, bradycardia (Figure 5C and Figure 6C for larva and pupa, respectively). The slower heart pacing was not observed in "no opsin" (Supplementary Figure 2B and Supplementary Figure 3B) and in "no light" (Supplementary Figure 4A and Supplementary Figure 4C) control experiments.
Increasing the heart rates can be achieved by activating Hand>ReaChR opsin with red-light pulse trains at a frequency higher than the RHR of the given animal. A series of three light pulse trains at different stimulation frequencies (e.g., RHR + 0.5 Hz, RHR + 1 Hz, RHR + 1.5 Hz) were applied on Hand>ReaChR larvae and pupae hearts. The obtained data clearly shows increased heart rate following the light pulses (Figure 5D and Figure 6D for larva and pupa, respectively). The heart condition demonstrated in these experiments mimics tachycardia. Negative control experiments are shown in Supplementary Figure 2C, Supplementary Figure 3C, and Supplementary Figure 4B,D.
Overall, the results demonstrate the feasibility of non-invasive and specific optogenetic control of the heart rhythm at various developmental stages in transgenic animal models of D. melanogaster.
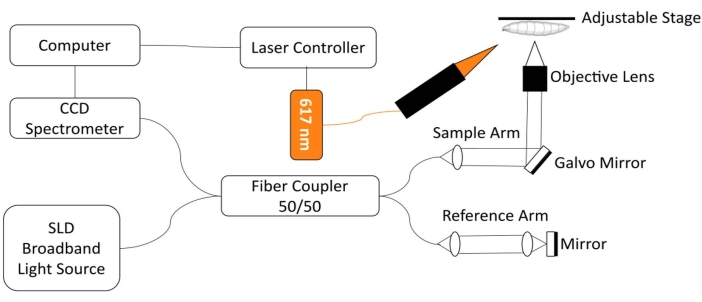
Figure 1: OCT imaging system integrated with 617 nm LED module for optogenetic control of Drosophila heart function. Please click here to view a larger version of this figure.
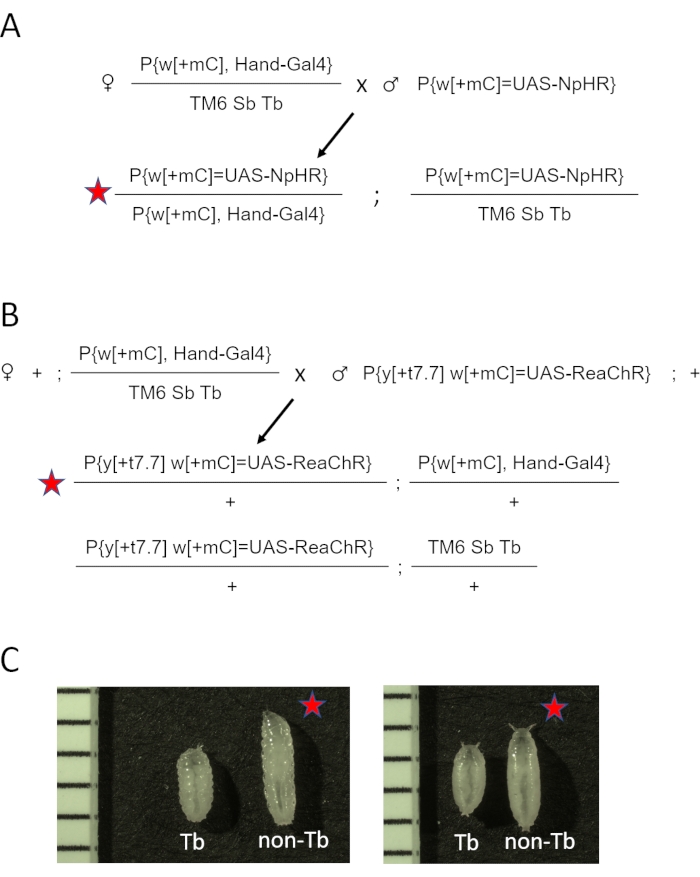
Figure 2: Generating D. melanogaster animals expressing opsin in the heart. (A) Genetic cross diagram. Females Hand-GAL4/TM6 SbTb were crossed to males carrying eNpHR2.0. The resulting Hand-GAL4/eNpHR2.0 progeny (marked by the red star) were collected for OCT imaging, and Hand-GAL4/TM6 Sb Tb were discarded based on their phenotypic appearance. (B) Genetic cross diagram. Females Hand-GAL4/TM6 SbTb were crossed to males carrying ReaChR. The resulting Hand-GAL4/ReaChR progeny (marked by the red star) were collected for OCT imaging, and Hand-GAL4/TM6 Sb Tb were discarded based on their phenotypic appearance. (C) Phenotypic differences between Hand-GAL4/opsin (red star) and Hand-GAL4/TM6 Tb progeny. Animals carrying the Tb gene mutation on the TM6 chromosome have a "tubby" body shape compared to normal, non-Tb larva or pupa. The left panel shows larvae; the right panel shows early pupae. Images also include a ruler with 1 mm marks. Please click here to view a larger version of this figure.
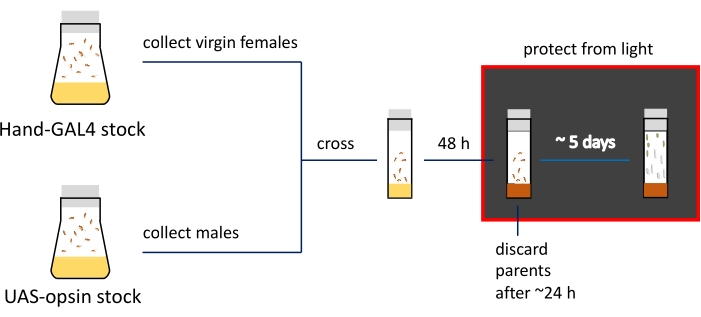
Figure 3: Schematic presentation and timeline of the imaging preparation procedures. Parental stocks are kept in fly bottles; virgin females and males are crossed in narrow vials filled with regular food (indicated by yellow color). Actively egg-laying flies are transferred to ATR-containing media (shown in brown) vials. Vials with developing progeny need to be kept in the dark from this step. 3rd instar larvae and early pupa are collected from the vial walls for imaging. Please click here to view a larger version of this figure.
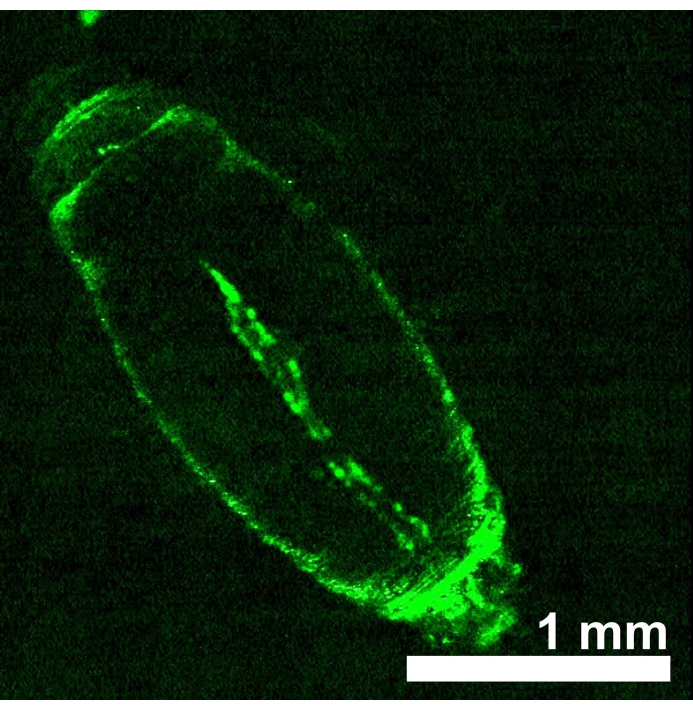
Figure 4: D. melanogaster early pupa expressing UAS-GFP (BDSC 6658) driven by Hand-GAL4 (BDSC 48396). The fluorescence pattern confirms the heart specificity of the Hand-GAL4 driver. Please click here to view a larger version of this figure.
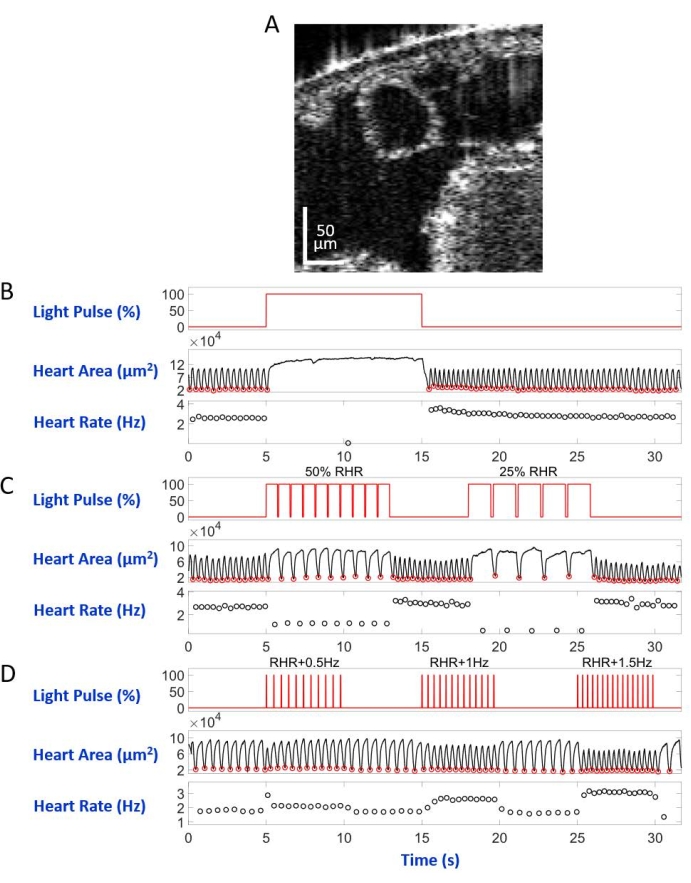
Figure 5: Simulation of heart arrest, bradycardia, and tachycardia in D. melanogaster larva. (A) OCT image of a larval body cross-section. The heart appears as a circle below the body's surface. (B) Graphic presentation of the restorable heart arrest. The upper panel shows the timing (X-axis) of the red-light illumination (Y-axis, light source power level percentage). The middle panel indicates the change in heart area (Y-axis, square micrometers) over time (X-axis). The lower panel shows the heart rate change (Y-axis, hertz) over time (X-axis). (C) Graphic presentation of eNpHR2.0-mediated restorable bradycardia. The upper panel shows pulses of the red-light illumination, inducing two periods of bradycardia: 50% of the RHR and 25% of the RHR. Heart area and heart rate changes are shown on the middle and lower panels, respectively. (D) Graphic presentation of the heart pacing by activated ReaChR. The upper panel shows a series of 20 ms red light pulses occurring at RHR + 0.5 Hz, RHR + 1 Hz, and RHR + 1.5 Hz frequencies. The heart contractions follow the light pulse frequencies, as shown on the middle and lower panels. Please click here to view a larger version of this figure.
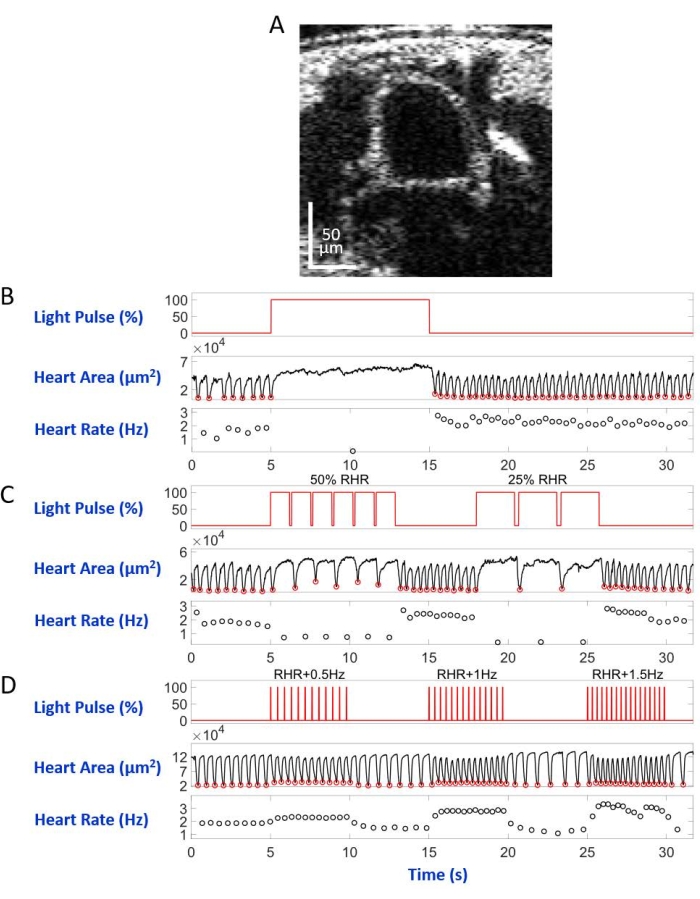
Figure 6: Simulation of heart arrest, bradycardia, and tachycardia in D. melanogaster pupa. (A) OCT image of pupal body cross-section. The heart appears as a circle below the body's surface. (B) Graphic presentation of the restorable heart arrest. The upper panel shows the timing (X-axis) of the red-light illumination (Y-axis, light source power level percentage). The middle panel indicates the change in heart area (Y-axis, square micrometers) over time (X-axis). The lower panel shows the heart rate change (Y-axis, hertz) over time (X-axis). (C) Graphic presentation of eNpHR2.0-mediated restorable bradycardia. The upper panel shows pulses of the red-light illumination, inducing two periods of bradycardia: 50% of the RHR and 25% of the RHR. The middle and lower panels show heart area and heart rate changes, respectively. (D) Graphic presentation of the heart pacing by activated ReaChR. The upper panel shows a series of 20 ms red light pulses at RHR + 0.5 Hz, RHR + 1 Hz, and RHR + 1.5 Hz frequencies. The heart contractions follow the frequencies of the light pulse, as shown on the middle and lower panels. Please click here to view a larger version of this figure.
Supplementary Figure 1: Genetic crosses to replace TM3 Sb balancer chromosome with TM6 Sb Tb. Virgin females Hand-GAL4 w+/ TM3 Sb were crossed with nub-GAL4NP3537 tub-GAL80ts w+/ TM6 Sb Tb males. Hand-GAL4 w+/ TM6 Sb Tb progeny, including virgin females and males, were selected (screening for pigmented eyes combined with tubby body shape). Selected flies were self-crossed to establish a stable stock. Please click here to download this File.
Supplementary Figure 2: In control experiments, the wild-type (wt) larva's heart rhythm does not change upon red light illumination. (A) No heart arrest was observed during the red-light illumination in wt larva. The upper panel shows the M-mode heart images. The red line indicates the illumination timing. The middle and lower panels show the heart area and the heart rates during the 32 s imaging time. (B,C) Red light pulses do not change the heart rates in wt larva. The upper panels show the M-mode heart images. The red line indicates the illumination timing. The middle and lower panels show the heart area and the heart rates during the 32 s imaging time. Please click here to download this File.
Supplementary Figure 3: In control experiments, the wild-type (wt) pupa's heart rhythm does not change upon red light illumination. (A) No heart arrest was observed during the red-light illumination in wt larva. The upper panel shows the M-mode heart images. The red line indicates the illumination timing. The middle and lower panels show the heart area and the heart rates during the 32 s imaging time. (B,C) Red light pulses do not change the heart rates in the wt pupa. The upper panels show the M-mode heart images. The red line indicates the illumination timing. The middle and lower panels show the heart area and heart rates during the 32 s imaging time. Please click here to download this File.
Supplementary Figure 4: D. melanogaster larvae and pupae expressing Hand>eNpHR2.0 or Hand>ReaChR do not show significant HR changes during OCT imaging without red light illumination. (A) The heart rates of Hand>eNpHR2.0 larva. (B) The heart rates of Hand>ReaChR larva. (C) The heart rates of Hand>eNpHR2.0 pupa. (D) The heart rates of Hand>ReaChR pupa. Please click here to download this File.
Supplementary Video 1: Activated eNpHR2.0 causes heart arrest in D. melanogaster larva. Please click here to download this Video.
Supplementary Video 2: Activated eNpHR2.0 causes heart arrest in D. melanogaster pupa. Please click here to download this Video.
Supplementary Video 3: eNpHR2.0-mediated restorable bradycardia in D. melanogaster larva. Please click here to download this Video.
Supplementary Video 4: eNpHR2.0-mediated restorable bradycardia in D. melanogaster pupa. Please click here to download this Video.
Supplementary Video 5: Heart pacing by activated ReaChR in D. melanogaster larva. Please click here to download this Video.
Supplementary Video 6: Heart pacing by activated ReaChR in D. melanogaster pupa. Please click here to download this Video.
Discussion
Compared to our previous reports where the expression of opsins was driven not just in the heart but also in the surrounding muscle tissues, the present work reports using a heart-specific driver, Hand-GAL4. This new Hand> opsin genetic configuration used for optogenetic heart regulation further confirms previously reported results and establishes a better Drosophila cardiovascular research model.
Media preparation is essential for the success of the experiments. Opsin proteins require a ligand, all-trans retinal (ATR), to function28. Flies do not produce enough ATR, so ATR has to be supplemented to the fly media. In this study, the previously reported instant food was replaced with Semi-Defined media29. The new recipe of ATR-containing media was introduced to ensure a uniform distribution of ATR. ATR is not soluble in water; when ethanol-based 100 mM ATR stock is added to water-based media, it is dispersed by vortexing the vials containing warm Semi-Defined media. Also, the previously reported ATR concentration was reduced from 10 mM for eNpHR2.0 and 3 mM for ReaChR22 to a 1 mM final concentration for both. This concentration is sufficient to ensure proper eNpHR2.0 and ReaChR function.
A vital component of the experimental success is the improved data processing with FlyNet 2.027. The lab has continued to develop this software to improve both the computational efficiency and accuracy of the automated fly heart segmentation algorithm. The cross-sectional masks produced by this software are used to derive Drosophila physiological data such as fractional shortening and heart wall velocity. This approach has enabled efficient data analysis with minimal human supervision, making it quicker and more reliable to characterize heart function for large fly heart imaging datasets.
Myocardial infarction remains the leading cause of death, and myocardial ischemia contributes to two-thirds of all cases of heart failure, which is rapidly emerging among the leading causes of mortality and morbidity in the United States30. The development of new therapeutics and medical devices requires deep knowledge of the mechanisms of heart disorders on physiological and biochemical levels. These goals can be accomplished with the help of model organisms. D. melanogaster has established itself as one of the most reliable and efficient models31,32,33,34,35. This work has generated the simulated Drosophila heart disorders models induced by a non-invasive optogenetic approach. The development of non-invasive optical heart pacing technologies provides a basis for developing an alternative to traditional electric heart pacing devices. Using OCT to observe the heart function in real-time allows studies to accurately characterize relevant heart physiology in Drosophila models for advanced investigations, including drug candidate screening. OCT imaging has a penetration depth of ~1 mm, which works well for Drosophila heart studies but limits its use to characterize heart function in larger animal models. Furthermore, directly translating Drosophila research to mammalian models represents a challenge. New optogenetic tools need to be developed to improve the opsins' sensitivity and translate them to various model systems, including zebrafish, mice, rats, and human heart organoids, for cardiovascular research.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Andrey Komarov, Yuxuan Wang, and Jiantao Zhu for their assistance in data analysis and thank the Zhou lab members for their valuable discussions. Work in Dr. Zhou's laboratory was supported by a start-up fund from the Washington University in St. Louis, the National Institutes of Health (NIH) grants R01-EB025209 and R01-HL156265, and the Clayco Foundation Innovative Research Award.
Materials
| All-trans retinal | Cayman Chemicals | 18449 | |
| Bacto Peptone | Gibco | 02-10-2025 | |
| BioLED Light Source Control Module, 4-channel | Migtex Systems | BLS-SA04-US | Part of the optogenetic stimulation module |
| Broadband Light Source Module | Superlum | cBLMD-T-850-HP | Part of the SD-OCT imaging system |
| Cobra-S 800 OCT Spectrometers | Wasatch Photonics | CS800-840/180-80-OC2K-U3 | Part of the SD-OCT imaging system |
| Delicate Task Wipers | Kimberly-Clark Professtional | 34155 | tissues |
| Drosophila agar | Genesee Scientific | 66-103 | |
| Drosophila culture bottles | Genesee Scientific | 32-131 | |
| FlyNet 2.0 Software | Z-Lab | Custom software for fly heart segmentation and heart function analysis developed in the Zhou lab | |
| High-Power LED Collimator Sources | Migtex Systems | BLS-LCS-0617-03-22 | Part of the optogenetic stimulation module |
| Inactive dry yeast | Genesee Scientific | 62-106 | |
| Microscope slides | AmScope | BS-72P | |
| Narrow plugs for Drosophila culture | Genesee Scientific | 59-200 | |
| Narrow vials for Drosophila culture | Genesee Scientific | 32-116SB | |
| Permanent double-sided tape | Scotch | ||
| Plugs for Drosophila bottles | Genesee Scientific | 59-194 | |
| Propionic Acid | Sigma | P1386-1L | |
| SD-OCT control software | Z-Lab | Custom software for image acquisition and pacing control developed in the Zhou lab | |
| SD-OCT imaging and optogenetic pacing system | Z-Lab | Imaging and optogenetic pacing system developed in the Zhou lab (~$50k BOM) | |
| Sucrose | Carolina | 89-2871 | |
| w[*]; P{y[+t7.7] w[+mC]=UAS-eNpHR-YFP}attP2 | Bloomington Drosophila Stock Center (BDSC) | stock # 41752 | eNpHR2.0 transgenic line |
| w[*]; P{y[+t7.7] w[+mC]=UAS-ReaChR}su(Hw)attP5/CyO | Bloomington Drosophila Stock Center (BDSC) | stock # 53748 | ReaChR transgenic line |
| w[1118]; P{y[+t7.7] w[+mC]=GMR88D05-GAL4}attP2/TM3 Sb[1] | Bloomington Drosophila Stock Center (BDSC) | stock # 48396 | Heart specific GAL4 driver containing Hand gene regulatory fragment |
| y[*] w[*]; P{w[+mC]=UAS-2xEGFP}AH3 | Bloomington Drosophila Stock Center (BDSC) | stock #6658 | GFP reporter line |
| Yeast extract | Lab Scientific bioKEMIX | 978-907-4243 |
References
- Nature Methods. Method of the Year 2010. Nature Methods. 8, 1 (2011).
- Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience. 8 (9), 1263-1268 (2005).
- Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nature Neuroscience. 18 (9), 1213-1225 (2015).
- Tsai, H. -. C. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 324 (5930), 1080-1084 (2009).
- Wykes, R. C., et al. Optogenetic and potassium channel gene therapy in a rodent model of focal neocortical epilepsy. Science Translational Medicine. 4 (161), (2012).
- Entcheva, E., Kay, M. W. Cardiac optogenetics: a decade of enlightenment. Nature Reviews Cardiology. 18 (5), 349-367 (2021).
- Bruegmann, T., et al. Optogenetic control of heart muscle in vitro and in vivo. Nature Methods. 7 (11), 897-900 (2010).
- Arrenberg, A. B., Stainier, D. Y. R., Baier, H., Huisken, J. Optogenetic control of cardiac function. Science. 330 (6006), 971-974 (2010).
- Nussinovitch, U., Gepstein, L. Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nature Biotechnology. 33 (7), 750-754 (2015).
- Nyns, E. C. A., et al. An automated hybrid bioelectronic system for autogenous restoration of sinus rhythm in atrial fibrillation. Science Translational Medicine. 11 (481), (2019).
- Bier, E., Bodmer, R. Drosophila, an emerging model for cardiac disease. Gene. 342 (1), 1-11 (2004).
- Wolf, M. J., Amrein, H., Izatt, J. A., Choma, M. A., Reedy, M. C., Rockman, H. A. Drosophila as a model for the identification of genes causing adult human heart disease. Proceedings of the National Academy of Sciences of the United States of America. 103 (5), 1394-1399 (2006).
- Yu, L., Lee, T., Lin, N., Wolf, M. J. Affecting rhomboid-3 function causes a dilated heart in adult Drosophila. PLOS Genetics. 6 (5), 1000969 (2010).
- Cooper, A. S., Rymond, K. E., Ward, M. A., Bocook, E. L., Cooper, R. L. Monitoring heart function in larval Drosophila melanogaster for physiological studies. Journal of Visualized Experiments. (33), e1596 (2009).
- Zhu, Y. C., Yocom, E., Sifers, J., Uradu, H., Cooper, R. L. Modulatory effects on Drosophila larva hearts: Room temperature, acute and chronic cold stress. Journal of Comparative Physiology. B, Biochemical, Systemic, and Environmental Physiology. 186 (7), 829-841 (2016).
- Zhu, Y. C., Uradu, H., Majeed, Z. R., Cooper, R. L. Optogenetic stimulation of Drosophila heart rate at different temperatures and Ca2+ concentrations. Physiological Reports. 4 (3), 12695 (2016).
- Malloy, C., et al. Using optogenetics to assess neuroendocrine modulation of heart rate in Drosophila melanogaster larvae. Journal of Comparative Physiology. A, Neuroethology, Sensory, Neural, and Behavioral Physiology. 203 (10), 791-806 (2017).
- Men, J., et al. Drosophila preparation and longitudinal imaging of heart function in vivo using optical coherence microscopy (OCM). Journal of Visualized Experiments. (118), e55002 (2016).
- Choma, M. A., Izatt, S. D., Wessells, R. J., Bodmer, R., Izatt, J. A. In vivo imaging of the adult Drosophila melanogaster heart with real-time optical coherence tomography. Circulation. 114 (2), 35-36 (2006).
- Li, A., et al. Changes in the expression of the Alzheimer’s disease-associated presenilin gene in drosophila heart leads to cardiac dysfunction. Current Alzheimer Research. 8 (3), 313-322 (2011).
- Li, A., et al. Silencing of the Drosophila ortholog of SOX5 in heart leads to cardiac dysfunction as detected by optical coherence tomography. Human Molecular Genetics. 22 (18), 3798-3806 (2013).
- Men, J., Li, A., Jerwick, J., Li, Z., Tanzi, R. E., Zhou, C. Non-invasive red-light optogenetic control of Drosophila cardiac function. Communications Biology. 3 (1), 1-10 (2020).
- Alex, A., Li, A., Tanzi, R. E., Zhou, C. Optogenetic pacing in Drosophila melanogaster. Science Advances. 1 (9), 1500639 (2015).
- Stanley, C. E., Mauss, A. S., Borst, A., Cooper, R. L. The effects of chloride flux on Drosophila heart rate. Methods and Protocols. 2 (3), 73 (2019).
- Lindsley, D. L., Zimm, G. G. . The Genome of Drosophila melanogaster. , (1992).
- . Bloomington Drosophila Stock Center Available from: https://bdsc.indiana.edu/information/recipes/germanfood.html (2022)
- Dong, Z., et al. FlyNet 2.0: Drosophila heart 3D (2D + time) segmentation in optical coherence microscopy images using a convolutional long short-term memory neural network. Biomedical Optics Express. 11 (3), 1568-1579 (2020).
- Deisseroth, K. Optogenetics. Nature Methods. 8 (1), 26-29 (2011).
- Backhaus, B., Sulkowski, E., Schlote, F. W. A semi-synthetic, general-purpose medium for Drosophila melanogaster. Drosophila Information Service. 60, 210-212 (1984).
- Benjamin, E. J., et al. Heart disease and stroke statistics-2019 update: A report from the American Heart Association. Circulation. 139 (10), 56 (2019).
- Wolf, M. J., Rockman, H. A. Drosophila, genetic screens, and cardiac function. Circulation Research. 109 (7), 794-806 (2011).
- Choma, M. A., Suter, M. J., Vakoc, B. J., Bouma, B. E., Tearney, G. J. Physiological homology between Drosophila melanogaster and vertebrate cardiovascular systems. Disease Models & Mechanisms. 4 (3), 411-420 (2011).
- Ocorr, K., Vogler, G., Bodmer, R. Methods to assess Drosophila heart development, function and aging. Methods [Supplement to Methods in Enzymology]. 68 (1), 265-272 (2014).
- Ugur, B., Chen, K., Bellen, H. J. Drosophila tools and assays for the study of human diseases. Disease Models & Mechanisms. 9 (3), 235-244 (2016).
- Rotstein, B., Paululat, A. On the morphology of the Drosophila heart. Journal of Cardiovascular Development and Disease. 3 (2), 15 (2016).

