Decellularization-Based Quantification of Skeletal Muscle Fatty Infiltration
Summary
The present study describes decellularization-based methodologies for visualizing and quantifying intramuscular adipose tissue (IMAT) deposition through intact muscle volume, as well as quantifying metrics of individual adipocytes that comprise IMAT.
Abstract
Fatty infiltration is the accumulation of adipocytes between myofibers in skeletal muscle and is a prominent feature of many myopathies, metabolic disorders, and dystrophies. Clinically in human populations, fatty infiltration is assessed using noninvasive methods, including computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound (US). Although some studies have used CT or MRI to quantify fatty infiltration in mouse muscle, costs and insufficient spatial resolution remain challenging. Other small animal methods utilize histology to visualize individual adipocytes; however, this methodology suffers from sampling bias in heterogeneous pathology. This protocol describes the methodology to qualitatively view and quantitatively measure fatty infiltration comprehensively throughout intact mouse muscle and at the level of individual adipocytes using decellularization. The protocol is not limited to specific muscles or specific species and can be extended to human biopsy. Additionally, gross qualitative and quantitative assessments can be made with standard laboratory equipment for little cost, making this procedure more accessible across research laboratories.
Introduction
The accumulation of adipocytes between myofibers within the skeletal muscle is a prominent feature of disparate conditions, from type 2 diabetes to sarcopenia to musculoskeletal injury1,2,3,4,5,6,7. Comprehensive assessment of this intramuscular adipose tissue (IMAT) is critical to understanding the pathogenesis of these conditions, as IMAT deposition is strongly correlated with insulin resistance3,8,9,10 and poor skeletal muscle function11,12,13,14,15. Although these associations have been noted for decades, the mechanisms associated with and the origin of IMAT still remain an area of intense investigation. This is partly because most studies assessing skeletal muscle fatty infiltration have been performed in humans, where mechanistic investigations are limited16,17. However, more recently, small animal models, including mice, have been utilized to help pinpoint cellular regulation of IMAT development and signaling18,19,20. This work aims to provide a new tool for use with small animal models for qualitatively visualizing and quantifying skeletal muscle fatty infiltration.
Clinically in human populations, fatty infiltration is assessed using noninvasive methods, including computed tomography (CT)6,21, magnetic resonance imaging (MRI)16,17,22,23, and ultrasound (US)17,24. These imaging techniques typically identify a defined region of interest (ROI) in a muscle and acquire image slices within that region, although comprehensive approaches have also been employed25,26,27. These image slices are subjected to qualitative grading6 and quantified via pixel thresholding28. Similar approaches have been utilized in animals previously29,30; however, they are costly and require access to small animal imaging systems. Spatial resolution via CT and MRI use also presents a major issue, as they are unable to delineate IMAT adipocytes from skeletal muscle fibers within a voxel and instead rely on subjective separation of primarily muscle regions and primarily IMAT regions31,32. As such, the inability to accurately identify fat or muscle tissue also presents inaccurate quantification of representative amounts of these tissues.
Due to these limitations, current techniques to assess skeletal muscle fatty infiltration in small animal models most commonly rely on histology as an inexpensive and accessible alternative33,34. Standard staining procedures, including hematoxylin and eosin (H&E), oil red O (ORO), and immunostaining for adipocyte markers such as perilipin, allow for simple detection and visualization of adipocytes comprising fatty infiltration within the muscle. However, histology approaches are rarely comprehensive, and typically, IMAT qualitative or quantitative assessment is limited to a single section34. Lipid extraction has also been used to quantify total muscle lipids35; however, this technique fails to distinguish between intramyocellular lipid (IMCL) and intramuscular adipose tissue (IMAT) stores36. In summary, current methodologies to visualize and quantify fat in muscle remain limited either by financial costs or the specific detection of IMAT.
Here, we describe a detailed method for assessing skeletal muscle fatty infiltration both by qualitative visualization and multi-scale quantification. This methodology employs a simple decellularization technique that removes myocellular structures, including IMCL, but keeps the larger IMAT adipocyte-derived lipid droplets intact. Validation of the specificity of this technique has been published37, including using lipid extraction to show the depletion of IMCL with decellularization, µCT to show the retention of IMAT patterning with decellularization, and histology to show the similar size distribution of IMAT lipid droplets compared with those identified with decellularization. Once decellularized, muscles can be stained with lipid-soluble dyes for qualitative visualization of the pattern and the extent of fatty infiltration and/or quantitative imaging of individual IMAT lipid droplets. Dyes can subsequently be extracted with isopropanol, and the optical density (OD) of the resulting solution can be used to estimate the IMAT lipid volume. The stringent validation of this technique has been published elsewhere37. This article provides a detailed protocol to use this methodology with mouse muscles and provides troubleshooting tips to support the adoption of this method in other applications, such as muscles from other species or other tissues.
Protocol
The care and sacrifice of mice were performed in accordance with the National Institutes of Health Guide for the Use and Care of Laboratory Animals. All work was approved by the Animal Studies Committee at Washington University in the St. Louis School of Medicine. Male C57BL/6J mice aged 2-3 months (see Table of Materials) were used to generate the example images included in this protocol. All steps described below are performed at room temperature.
1. Muscle decellularization
- Prepare a 1% w/v sodium dodecyl sulfate (SDS; see Table of Materials) solution in phosphate-buffered saline (PBS). Stir the solution until completely mixed.
NOTE: 1% SDS can be prepared in bulk and stored at room temperature for 1 month. - Dissect the muscle(s) of interest as described previously38,39.
- Expose the muscles of interest by wetting the hair and skin with 70% ethanol solution, then making a transcutaneous incision of approximately 2 cm followed by removing the covering skin with forceps and spring scissors.
NOTE: The muscle(s) of interest here include the tibialis anterior (TA), extensor digitorum longus (EDL), and diaphragm. Some muscles may lie deep in other musculature, requiring dissection of other muscles (e.g., dissecting the EDL requires removal of the TA). - Dissect the muscle(s) of interest using sharp scissors or blades (see Table of Materials) to ensure that the entire extent of the muscle is obtained and the edges are smooth.
NOTE: This is ideally done under a dissecting microscope to improve visualization. - Inspect the muscles to ensure that no bone chips or ragged edges are included, then trim these away with sharp scissors as needed.
- Weigh the muscle using an analytical balance and record the weight.
- Expose the muscles of interest by wetting the hair and skin with 70% ethanol solution, then making a transcutaneous incision of approximately 2 cm followed by removing the covering skin with forceps and spring scissors.
- Place the dissected muscle in at least 0.1 mL of 1% SDS per milligram of weight; 6-, 12- or 24-well plates work well for the diaphragm, TA, and EDL mouse muscles, respectively. Typical volumes are 6 mL, 3 mL, and 1.5 mL for 6-, 12- and 24-well plates, respectively.
NOTE: The SDS solution will sometimes cause the muscles to deform, so ensure that the muscle is flat/extended at initial immersion. - Place the well plate (or other vessels) on a rocking shaker set to 50-80 Hz.
- Visually inspect the SDS solution periodically. When the solution becomes cloudy, remove the solution with a pipette (taking care not to aspirate the muscle) and replace it with an equal volume of fresh 1% SDS. When the solution remains clear for 24 h, decellularization is complete.
NOTE: The frequency required for solution changes varies by muscle size and the initial solution volume. Larger muscles like the TA need new SDS within a few hours, and smaller muscles like the EDL can go overnight in the original solution. - Remove the final SDS solution from the decellularized muscles with a pipette (taking care not to aspirate the muscle) and replace it with an equal volume of PBS.
- Visually inspect the decellularized muscles under a dissecting microscope and use forceps and scissors to remove any hair or debris that is stuck to the muscle.
NOTE: Also, note the clarity of the decellularized muscle in this step. If the decellularized muscle is not fully transparent and the SDS solution does not increase in cloudiness over 24 h, then the decellularization was not efficient. This creates an artifact in qualitative and quantitative assessments, and so decellularization should always be optimized on practice samples before proceeding with experimental muscles. - Carefully remove the PBS, replace it with an equal volume of 3.7% formaldehyde or 4% paraformaldehyde solution, and return the plate to the rocking shaker for 24 h.
NOTE: Ensure that the formaldehyde solution fully covers the decellularized muscle, otherwise staining will be uneven.
2. Visualization of IMAT with oil red O
- Prepare a solution of ORO.
- Dissolve 0.5 g of ORO powder (see Table of Materials) in 100 mL of isopropanol to generate a stock solution.
NOTE: Add gentle heat to the solution to fully dissolve it. This stock can be stored at room temperature for 1 month. - Combine the ORO stock solution and deionized water at a 60:40 ratio to obtain the working solution needed for all muscles using a volume of at least 0.1 mL/ mg muscle weight. Typical volumes are 6 mL, 3 mL, and 1.5 mL for 6-, 12- and 24-well plates, respectively.
NOTE: Inspect the stock solution prior to mixing. If the stock solution contains substantial particulate, make fresh stock. - Cover the working solution for 10 min to allow the particulate to settle.
- Filter the working solution through a 40 µm mesh, followed by a 0.22 µm syringe filter.
NOTE: The 0.22 µm syringe filter clogs rapidly and must be exchanged if the working solution does not push easily through.
- Dissolve 0.5 g of ORO powder (see Table of Materials) in 100 mL of isopropanol to generate a stock solution.
- Remove the formaldehyde or paraformaldehyde solution and wash the decellularized muscles with three solution changes of an equal volume of PBS.
- Replace the PBS with an equal volume of 60% isopropanol solution and incubate on a rocking shaker for 5 min.
- Replace the 60% isopropanol solution with ORO working solution and incubate on the rocking shaker for 10 min.
NOTE: Ensure that the ORO working solution fully covers the decellularized muscle, otherwise staining will be uneven. - Replace the ORO working solution with an equal volume of 1% SDS. Visually inspect the SDS solution periodically. When the solution becomes noticeably pink, remove the solution with a pipette (taking care not to aspirate the muscle) and replace it with fresh 1% SDS.
- When the solution remains clear for 24 h, replace the 1% SDS with PBS and inspect the stained muscle under a dissecting/stereo microscope at 4x magnification. Remove any obvious debris or particulate stuck to the outside of the muscle. If this is extensive, the decellularized muscle can be rolled gently on a cleaning tissue to pull off the debris/particulate.
- If the staining is satisfactory (bright red spheres floating in a transparent matrix), acquire images of the staining as desired using a camera attached to the stereo microscope (see Table of Materials).
NOTE: If the dissecting microscope doesn't have an attached camera, a phone camera can be used to take pictures through the eyepiece.
3. Visualization of IMAT lipid droplets with BODIPY
NOTE: Confocal imaging is most effective with thin muscles like the EDL or diaphragm (~2 mm thickness). Alternatively, comparable thickness strips of muscles like the TA could be used.
- Prepare a 1:200 solution of fluorescent BODIPY 493/503 (see Table of Materials) in PBS to obtain a working solution volume of at least 0.1 mL/ mg muscle weight. Typical volumes are 6 mL, 3 mL, and 1.5 mL for 6-, 12- and 24-well plates, respectively.
- Remove the formaldehyde, paraformaldehyde, or 1% SDS and wash the decellularized muscles with three solution changes of an equal volume of PBS.
- Replace the PBS with the BODIPY working solution and incubate on the rocking shaker for 20 min.
- Wash the decellularized muscles with three solution changes of an equal volume of PBS.
- Place the muscle in a clear-bottomed vessel that is compatible with an available confocal microscope. Ideally, the dish will have a recessed bottom to place a coverslip over the muscle without deforming it (see Table of Materials).
- Obtain image stacks with a standard confocal microscope (see Table of Materials) using the 488 laser. Typical stack sizes for an EDL are 0.5-1 mm with a 10 µm slice thickness, yielding 50-100 images per stack.
NOTE: To quantify total lipid volume, the total number of IMAT lipid droplets, and the nearest neighbor index of clustering, image through the entire muscle volume, taking care to leave some overlap for image registration. To quantify the average lipid droplet volume, one stack may be sufficient. - If desired, stain the muscles with ORO, as per section 2, for a complimentary whole-muscle image of IMAT distribution.
NOTE: Because BODIPY is fluorescent, it won't be visible under the light microscope when images of ORO are acquired.
4. Estimation of total lipid volume by lipid extraction
- Following image acquisition, transfer the muscles to 200 µL of isopropanol in individual wells of a 96-well plate, or adapt the well/volume if the muscle is too large to fit.
- Agitate the solution by tapping the plate, pipetting the solution up and down, and/or mechanically crushing the muscle with a pipette tip until no red/fluorescent spheres can be seen in the decellularized muscle under a microscope.
NOTE: Treat all the muscles with the same combination of tapping, pipetting, and crushing for the best reliability. Also, take care to limit the time spent tapping, pipetting, and crushing, as the isopropanol will evaporate rapidly. - Mix the solution in each well by pipetting up and down, then transfer 75 µL to two clean wells of the plate.
- Cover the plate and read the absorbance of the duplicate 75 µL wells using a spectrophotometer or plate reader. If the construct was stained with ORO, read the solution at 500 nm; if it was stained with BODIPY 493/503, read the plate at 493 nm.
NOTE: If the construct was stained with both BODIPY and ORO, it is recommended to read the plate at 500 nm, as 500 nm reads give similar results between constructs stained with ORO only and ORO plus BODIPY. - Divide the absorbance reading by the recorded weight of the muscle if desired to correct for size differences between samples.
5. Quantification of IMAT lipid droplet metrics from confocal images
NOTE: This section require access to ImageJ version 1.47 (see Table of Materials) or later and basic ImageJ skills40.
- Open confocal stacks in ImageJ.
NOTE: Different confocal microscopes may save confocal images in different formats. ImageJ may require an additional plugin or variant, such as Fiji, to open the stacks41. The algorithms used are part of the ImageJ software package. - Run a thresholding algorithm in ImageJ to identify BODIPY positive pixels. This converts the current image to a binary image.
- Open the threshold user interface by selecting Image > Adjust > Threshold. In the user interface, select Intermodes as the thresholding type and ensure Dark background is selected. No other options need to be selected. Then click on Apply.
- A dialog box opens to convert the stack to binary. Ensure that it has the following options selected: Method: Intermodes; Background: Dark. Calculate the threshold for each image and black background (of binary masks). This generates a binary stack, where white indicates BODIPY positive pixels and black indicates BODIPY negative pixels.
NOTE: The Intermodes thresholding algorithm may not be the best choice for all users. Subjective inspection of the built-in thresholding options of ImageJ helps to select the optimal algorithm for separating BODIPY positive and negative pixels.
- Run the Watershed algorithm in ImageJ to separate touching lipid droplets. Select Process > Binary > Watershed. A dialog box opens asking whether all images in the stack should be processed. Select Yes. This adds thin black lines dividing larger areas of solid white.
- Identify ROIs with the Analyze Particles algorithm in ImageJ.
- Select Analyze > Analyze Particles. A dialog box opens to set the selection settings.
NOTE: Selecting the size range is critical to achieving the best estimate of lipid droplet ROIs. The lower bound eliminates regions that are likely too small to be IMAT-derived lipid droplets (background artifact), and the upper bound eliminates regions that are likely too large to be single IMAT-derived lipid droplets (touching lipid droplets that weren't separated by the Watershed algorithm). - To set these values, open the original image and outline the smallest and largest lipid droplet in view using the Oval tool. Then, add these shapes to the ROI manager by typing "t". Select Analyze, then Set Measurements. A dialog box opens for selection of the settings.
- Check Area only and click OK. Then, select Measure from the ROI Manger. Use the two area measures from this Results window as the size range in Analyze Particles.
- Ensure Add to Manager is checked. No other options are needed.
- Select Analyze > Analyze Particles. A dialog box opens to set the selection settings.
- Overlay the ROIs on the confocal stack by checking Show All in the ROI Manager. Use the slider bar to inspect each stack slice individually and add missing regions by hand using the Oval tool (located in the ImageJ toolbar).
- Output the ROI measurements. Select Analyze > Set Measurements and select Area, Centroid, Fit ellipse and Stack position. Click on OK. Then, select Measure from the ROI Manger. The data in the Results table can be selected and copied into Matlab or Excel for further analysis.
- Refine the ROIs in each stack to a single ROI using the Refine.m Matlab code37 or a similar algorithm.
NOTE: This step is required as steps 5.2-5.4 identify the same lipid droplet in adjacent slices as distinct ROIs. However, ROIs can alternatively be identified by hand only, or duplicate ROIs can be eliminated by hand in ImageJ to avoid the need for Matlab. - Obtain the summary statistics in the ROI using Matlab or Excel.
- Estimate the total number of lipid droplets as the total number of ROIs.
- Estimate lipid droplet volume as the volume of the ellipse fitted to each 2D ROI, assuming the depth of the shape is the average of the major and minor axes of the ellipse.
- Estimate the total lipid volume, which is the sum of individual estimated lipid droplet volumes.
Representative Results
Qualitative visualization of skeletal muscle fatty infiltration
Properly decellularized muscles are white and semi-transparent (section 1; Figure 1). When decellularized muscles are stained with ORO to visualize IMAT (section 2), IMAT lipid droplets are apparent within the clear muscle structures as red spheres (Figure 1). Healthy mouse hindlimb muscles has little natural IMAT, evidenced by little to no red, ORO positive lipid (Figure 1A). By comparison, hindlimb muscles injected with cardiotoxin (CTX; Figure 1B) or glycerol (GLY; Figure 1C) 14 days prior to decellularization have an increased accumulation of IMAT, with a larger concentration of IMAT following CTX compared with GLY as previously noted37.
Incomplete decellularization can be identified immediately following initial SDS treatment or after washout of the ORO staining as semi-opaque, light pink fibers (Figure 2B compared with Figure 2A). Incomplete clearance of ORO can be identified following ORO washout as a pink or red uniform background, rather than distinct fiber lines (Figure 2C). Figure 2A,B also contains epimuscular adipose (asterisks), a clump of lipid droplets outside the decellularized muscle. Figure 2C also demonstrates the muscle folding that can occur if muscles are not spread out during decellularization. Incomplete decellularization, incomplete ORO clearance, and residual epimuscular adipose all artifactually increase the (OD) of extracted lipid, but do not necessarily impede qualitative assessment of fatty infiltration if they are recognized as an artifact.
Quantitative imaging of skeletal muscle fatty infiltration
BODIPY fluorescently labeled lipid droplets can be imaged via confocal microscopy for a more detailed assessment of individual lipid droplet metrics and distribution (Figure 3). This process is semi-automated, as previously described, including Matlab code37. Good BODIPY staining results in bright elliptical shapes delineated from neighboring shapes when imaged in plane (Figure 3A). Thresholding and shape segmentation provides a good first pass for generating ROIs for each lipid droplet (Figure 3B), but manual edits are needed to correct errors. The most pervasive error is lipid droplets that are deep in the tissue and thus are not bright on any slices (Figure 3B; pink double arrows). This can be remedied by adding ROIs by hand using the Oval tool in ImageJ (Figure 3C). The second is identifying a group of lipid droplets as a single ROI (Figure 3B; red asterisk). This can be corrected by deleting and replacing the original ROI with multiple new ROIs (Figure 3D). Finally, a single lipid droplet is identified as a unique ROI in multiple slices, so the duplicate ROIs must be consolidated into a single ROI (Figure 3E; blue arrow). This is most easily done using a data processing tool such as Matlab, but could also be done by hand by identifying the largest ROI and deleting the rest. Average values for each metric in the C57BL6/J and 129Sv mouse can be found in Biltz et al.37.
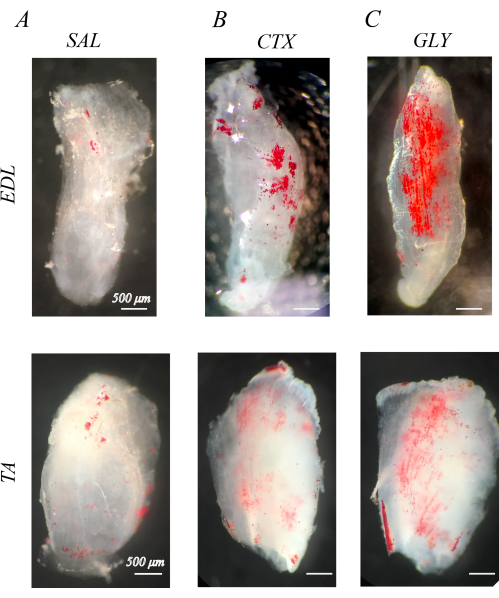
Figure 1: Example oil red O (ORO) staining of decellularized muscles. ORO-stained muscles 14 days post-injection with saline (SAL), cardiotoxin (CTX), or glycerol (GLY). Mouse extensor digitorum longus (EDL) and tibialis anterior (TA) muscles have little IMAT (red spheres) with SAL treatment (A), but accumulate IMAT in response to CTX (B) and GLY (C) treatment. There is a complete decellularization and ORO washout, evidenced by distinct ORO positive lipid droplets in a transparent white muscle background. Scale bars = 500 µm. Please click here to view a larger version of this figure.
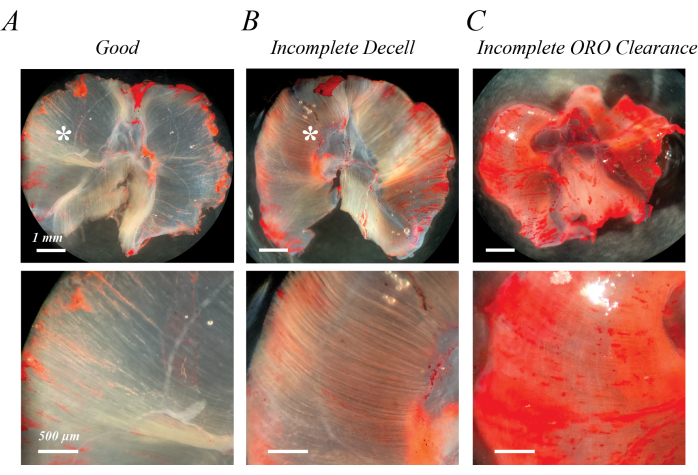
Figure 2: Examples of poor ORO staining results. Incomplete decellularization or incomplete ORO clearance leads to a semi-opaque pink/red background. Compared with the transparent white background of fully decellularized mouse diaphragm muscle (A), incomplete decellularization is characterized by light pink/red fiber tracks (B), and incomplete ORO clearance is characterized by a diffuse pink/red background (C). Scale bars: upper panels = 1 mm; lower panels = 500 µm. Please click here to view a larger version of this figure.
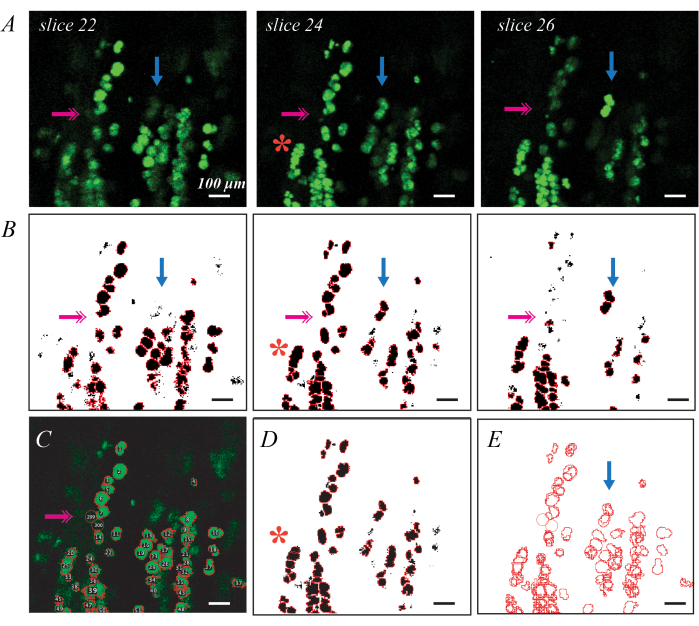
Figure 3: Examples of individual lipid droplet identification with fluorescent BODIPY staining and confocal microscopy Individual BODIPY stained lipid droplets can be identified and quantified in decellularized muscles using confocal microscopy. Individual slices through a confocal stack show lipid droplets in plane as bright green ellipses, and lipid droplets out of plane as fainter shapes (A; blue arrows). Thresholding combined with watershed object segmentation and ROI identification can map BODIPY-stained ROIs (B). Thresholding may miss some fainter lipid droplets (B; pink double arrow), requiring identification by hand (C). Watershed segmentation may group several lipid droplets together (B; red asterisk), requiring deletion of the ROI and re-estimation by hand (D). The same lipid droplet is identified in multiple slices requiring image registration (E) to delete the duplicate ROIs. Scale bars = 100 µm. Please click here to view a larger version of this figure.
Discussion
This manuscript describes methods to qualitatively visualize and quantify skeletal muscle fatty infiltration in small animal models that can be applied to further understanding the pathogenesis of intramuscular adipose tissue (IMAT) development and pathological expansion. The use of whole-muscle decellularization and lipid-soluble staining allows for a cost-effective, reproducible, and simple methodology to comprehensively assess the presence of IMAT in whole muscles.
The basis for this protocol is that decellularization of muscle with SDS removes the cellular components of myofibers, including the small lipid droplets of IMCL, but spares the large lipid droplets in intramyocellular adipocytes. SDS has been used extensively42 in tissue engineering to decellularize matrices. Tissues such as adipose and skeletal muscle typically require additional mechanical dissociation and/or alcohol extraction to remove the residual adipocyte lipid42,43. We have previously shown that this is because while decellularization with SDS eliminates IMCL, it spares the large lipid droplet in adipocytes37. Imaging of osmium tetroxide-stained intact muscle pre- and post-decellulariztion with µCT verified that the spatial pattern of IMAT was not disrupted by decellularization. Further, intramuscular triglyceride quantification in a decellularized muscle with negligible IMAT was ~5% of the intact muscle values, verifying the removal of IMCL. Therefore, this methodology retains IMAT lipid droplets in their original anatomical distribution through a semi-transparent muscle matrix.
Proper decellularization is the most critical step in this protocol. If the decellularization is incomplete, IMAT lipid droplets will be difficult to visualize and residual IMCL will cause high background staining with either ORO or BODIPY (Figure 2). Common errors by inexperienced users are inadequate SDS coverage per muscle (within each well), such that each muscle is not completely covered in SDS solution, not using a rocker to agitate the solution during decellularization, and not performing solution changes frequently enough. In this manuscript, we have recommended the amount of SDS needed per unit muscle mass, but the user will still need to ensure that muscles are completely covered by solutions, as each muscle has a unique geometry. Users are also recommended to change the solutions liberally (as much as twice per day) to ensure that decellularization is complete. Good quality staining of IMAT lipid droplets has been achieved after as many as 4 days of SDS treatment. For high-quality ORO staining results, adequate fixation and ORO solution prep are also important. Similar to the SDS treatment described above, adequate coverage of 3.7% formaldehyde solution for each muscle sample is needed. If the muscle is removed from the fixative too early, lipid droplets will only weakly stain with ORO. A total of 1-2 h should be sufficient, but overnight fixation is recommended to ensure the fixative penetrates to the center of the muscle and fully fixes all lipid droplets. An additional challenge with ORO staining is that when the alcohol concentration is reduced to 60%, a particulate begins to form. This particulate can settle on the surface and become stuck on the border of muscle. The best way to avoid this is to make a fresh working solution for each staining and use both 40 mesh µm and 0.22 µm filters. Then, maintaining agitation with the rocker and limiting the staining time to 10 min will help keep any particulate that forms from settling. If the problem persists, making a fresh ORO stock solution may help. If some artifact remains stuck to the decellularized muscle surface, a stereo microscope, forceps, and surgical scissors can be used to remove this artifact. Failing to eliminate artifacts will impact the image quality of muscles and overestimate IMAT content during the lipid extraction portion in preparation for OD reading.
Overall, this technique is straightforward and offers several advantages over gold standard methods for visualizing and quantifying skeletal muscle fatty infiltration. Noninvasive techniques, such as CT, MRI and US, which are used extensively in humans and sometimes in animal models, have limited spatial resolution and are unable to distinguish lipid droplets from muscle fibers. Thus, a pixel or voxel of intermediate signal intensity is assigned as “muscle” or “fat”, while in actuality it is likely a mix of myofibers and adipocytes. More commonly, fatty infiltration in animal muscle is assessed by histology, most frequently by ORO in muscle cryosections. However, this is typically only performed in a single representative section and is difficult to quantify due to lipid scatter over the section. By contrast, ORO staining of an entire decellularized muscle provides a comprehensive assessment of IMAT with similar costs and effort as intact morphology. Furthermore, in addition to enhancing visualization, ORO staining of decellularization enables the quantification of fatty infiltration by lipid extraction. For a deeper dive into the features of fatty infiltration, a fluorescent stain, BODIPY, can be used in conjunction with confocal microscopy. This enables the reconstruction of individual IMAT lipid droplets to map the 3D landscape, which is not possible with histology unless sections are analyzed over the length of the muscle. While a confocal microscope is not standard lab equipment, it is more likely to be accessible in a university or industry setting than small animal MRI or CT. Furthermore, much of this process can be automated, reducing the time cost compared with sequential histology. Optimizing the settings on the confocal microscope is an additional consideration for BODIPY staining. These are unique to each microscope. The critical value is laser intensity, which must be high enough to detect the lipid droplets on the distant surface of the muscle while also not saturating the signal from the lipid droplets on the near side. Because of this, it is suggested that using BODIPY staining with confocal microscopy is best suited on thinner muscles, including the EDL or diaphragm.
Several limitations of this approach warrant discussion. First, while it is anticipated that this technique has broad applicability beyond injury models (cardiotoxin and glycerol) in mice presented here, new applications (e.g., the mdx model) may require optimization, as the size and composition of the muscle (e.g., fibrosis) could affect decellularization, requiring increased SDS concentration or incubation times. Other disease models with altered muscle mass would also require analysis of both absolute and normalized (to muscle mass) metrics of fatty infiltration to determine the absolute amount of lipid or percentage of lipid relative to the muscle volume to provide a more meaningful outcome measure. Furthermore, this technique is anticipated to be broadly applicable to larger animal models and human biopsies, but this may require optimization for each new application. Second, in this strategy, the entire muscle must be dedicated to this assay and cannot be used to assess another pathological feature. Studies that aim to assess longitudinal changes in IMAT are better served with noninvasive imaging techniques and studies whose primary aim requires the muscle for other purposes (histology, quantitative polymerase chain reaction, western blotting) are better served by histological assessment, as the remainder of the frozen muscle can be allocated to other assays. However, this assay is well suited to pair with in vivo testing, such as treadmill running, or ex vivo contractile testing ,since these measures can be made before decellularization44. Third, although the use of BODIPY stain with confocal microscopy provides high-resolution visualization and quantification of lipid droplets, it cannot conclusively identify lipid droplets as individual adipocytes, as the cell membrane is removed and endogenous adipocyte proteins are lost. Multilocular adipocytes, representing immature adipocytes or a “brown/beige” phenotype, may be identified as multiple lipid droplets. Finally, the protocol does not work well on previously frozen muscle. These limitations are probably most profound for human biopsies, as while the entire biopsy can be decellularized, the spatial distribution of IMAT in the biopsy is not likely to be more representative of the whole muscle than a histological slice. However, since this technique is relatively insensitive to unfrozen biopsy handling conditions (e.g., hours on ice in PBS), the biopsy could be divided later for various assays, including a portion for decellularization, which would provide a better resolution of individual lipid droplets.
In conclusion, a novel method for qualitative and quantitative analysis of skeletal muscle fatty infiltration has been developed by staining and imaging the retained lipid of decellularized constructs. This methodology offers improvements over gold-standard approaches, in that it enables comprehensive imaging of three-dimensional fatty infiltration within muscle and quick, cheap quantification with ORO staining. For more detailed measures, a second lipid-soluble BODIPY stain provides a more detailed quantification of lipid droplet number, volume, and distribution pattern, as imaged by confocal microscopy. Together, these measures provide researchers with a way to precisely measure skeletal muscle fatty infiltration at the level of the individual lipid droplets without sampling or expensive noninvasive imaging.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by R01AR075773 to GAM.
Materials
| 0.22 µm Syringe Filter | Fisher Scientific | SLGP033RS | |
| 1 mL LuerLock Syringes | Fisher Scientific | 14823434 | |
| 12 mm Coverslips | Fisher Scientific | 12545F | |
| 12 well plates | Fisher Scientific | 08-772-29 | |
| 24 well plates | Fisher Scientific | 08-772-1H | |
| 2-Propanol (Isopropanol) | Sigma Aldrich | I9516 | 0.5 mg/mL stock solution can be stored at room temperature for 1 month. Working solution must be made fresh. |
| 37% Formaldehyde Solution | Sigma Aldrich | 8187081000 | |
| 40 µm Mesh Filter | Fisher Scientific | 87711 | |
| 6 well plates | Fisher Scientific | 08-772-1B | |
| 96 well plates | Fisher Scientific | 08-772-2C | |
| BODIPY 493/503 | Fisher Scientific | D-3922 | |
| C57BL/6J Mice | Jackson Laboratory | 000664 | |
| Confocal Imaging Dish | VWR | 734-2905 | |
| Confocal Microscope | Leica | TCS SPEII | |
| Dissecting/stereo Microscope | Zeiss | 4107009123001000 | |
| Dissection scissors | Fine Science Tools | 14060-09 | |
| Dumont #5 forceps | Fine Science Tools | 11254-20 | |
| Ethanol | Fisher Scientific | 033361.K2 | |
| ImageJ | NIH | ||
| Matlab | Mathworks | ||
| Oil Red O Powder | Sigma Aldrich | O0625 | |
| Plate reader | Bio-tek | Synergy II | |
| Rocker/Shaker | Reliable Scientific | 55D | |
| Sodium Dodecyl Sulfate (SDS) | Sigma Aldrich | L3771 | 1% Solution can be stored at room temperature for 1 month |
| Transfer pipettes | Fisher Scientific | 137119D | |
| Vannas spring scissors | Fine Science Tools | 15000-00 |
References
- Delmonico, M. J., et al. Longitudinal study of muscle strength, quality, and adipose tissue infiltration. The American Journal of Clinical Nutrition. 90 (6), 1579-1585 (2009).
- Goodpaster, B. H., et al. Obesity, regional body fat distribution, and the metabolic syndrome in older men and women. Archives of Internal Medicine. 165 (7), 777-783 (2005).
- Freda, P. U., et al. Lower visceral and subcutaneous but higher intermuscular adipose tissue depots in patients with growth hormone and insulin-like growth factor I excess due to acromegaly. The Journal of Clinical Endocrinology and Metabolism. 93 (6), 2334-2343 (2008).
- Garg, A., Peshock, R. M., Fleckenstein, J. L. Adipose tissue distribution pattern in patients with familial partial lipodystrophy (Dunnigan variety). The Journal of Clinical Endocrinology and Metabolism. 84 (1), 170-174 (1999).
- Gorgey, A. S., Dudley, G. A. Skeletal muscle atrophy and increased intramuscular fat after incomplete spinal cord injury. Spinal Cord. 45 (4), 304-309 (2007).
- Goutallier, D., Postel, J. M., Bernageau, J., Lavau, L., Voisin, M. C. Fatty muscle degeneration in cuff ruptures. Pre- and postoperative evaluation by CT scan. Clinical Orthopaedics and Related Research. (304), 78-83 (1994).
- Liu, W., Liu, Y., Lai, X., Kuang, S. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. 발생학. 361 (1), 27-38 (2012).
- Goodpaster, B. H., Thaete, F. L., Kelley, D. E. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. The American Journal of Clinical Nutrition. 71 (4), 885-892 (2000).
- Elder, C. P., Apple, D. F., Bickel, C. S., Meyer, R. A., Dudley, G. A. Intramuscular fat and glucose tolerance after spinal cord injury-a cross-sectional study. Spinal Cord. 42 (12), 711-716 (2004).
- Albu, J. B., et al. Independent association of insulin resistance with larger amounts of intermuscular adipose tissue and a greater acute insulin response to glucose in African American than in white nondiabetic women. The American Journal of Clinical Nutrition. 82 (6), 1210-1217 (2005).
- Tuttle, L. J., Sinacore, D. R., Mueller, M. J. Intermuscular adipose tissue is muscle specific and associated with poor functional performance. Journal of Aging Research. 2012, 172957 (2012).
- Gerber, C., Schneeberger, A. G., Hoppeler, H., Meyer, D. C. Correlation of atrophy and fatty infiltration on strength and integrity of rotator cuff repairs: a study in thirteen patients. Journal of Shoulder and Elbow Surgery. 16 (6), 691-696 (2007).
- Hilton, T. N., Tuttle, L. J., Bohnert, K. L., Mueller, M. J., Sinacore, D. R. Excessive adipose tissue infiltration in skeletal muscle in individuals with obesity, diabetes mellitus, and peripheral neuropathy: association with performance and function. Physical Therapy. 88 (11), 1336-1344 (2008).
- Gaeta, M., et al. Muscle fat-fraction and mapping in Duchenne muscular dystrophy: evaluation of disease distribution and correlation with clinical assessments. Preliminary experience. Skeletal Radiology. 41 (8), 955-961 (2012).
- Buford, T. W., et al. Age-related differences in lower extremity tissue compartments and associations with physical function in older adults. Experimental Gerontology. 47 (1), 38-44 (2012).
- Addona, J., et al. Estimating 3D supraspinatus intramuscular fatty infiltration in older adults: a pilot study. Acta Radiologica. , 2841851221139597 (2022).
- Crook, J., et al. Comparison of multifidus muscle intramuscular fat by ultrasound echo intensity and fat-water based MR images in individuals with chronic low back pain. Musculoskeletal Science & Practice. 63, 102717 (2023).
- Lee, C., et al. Beige FAPs transplantation improves muscle quality and shoulder function after massive rotator cuff tears. Journal of Orthopaedic Research. 38 (5), 1159-1166 (2020).
- Lee, C., et al. Beige fibro-adipogenic progenitor transplantation reduces muscle degeneration and improves function in a mouse model of delayed repair of rotator cuff tears. Journal of Shoulder and Elbow Surgery. 29 (4), 719-727 (2020).
- Kopinke, D., Roberson, E. C., Reiter, J. F. Ciliary hedgehog signaling restricts injury-induced adipogenesis. Cell. 170 (2), 340-351 (2017).
- Overend, T. J., Cunningham, D. A., Paterson, D. H., Lefcoe, M. S. Thigh composition in young and elderly men determined by computed tomography. Clinical Physiology. 12 (6), 629-640 (1992).
- Li, W., et al. Progression and variation of fatty infiltration of the thigh muscles in Duchenne muscular dystrophy, a muscle magnetic resonance imaging study. Neuromuscular Disorders. 25 (5), 375-380 (2015).
- Davis, D. L., et al. Supraspinatus fatty infiltration on MRI among older adults receiving physical therapy as initial management for clinically suspected rotator cuff tear: A pilot study. Journal of Clinical Imaging Science. 12, 66 (2022).
- Salaffi, F., et al. Ultrasound and magnetic resonance imaging as diagnostic tools for sarcopenia in immune-mediated rheumatic diseases (IMRDs). La Radiologia Medica. 127 (11), 1277-1291 (2022).
- Gallagher, D., et al. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. The American Journal of Clinical Nutrition. 81 (4), 903-910 (2005).
- Tuttle, L. J., Sinacore, D. R., Cade, W. T., Mueller, M. J. Lower physical activity is associated with higher intermuscular adipose tissue in people with type 2 diabetes and peripheral neuropathy. Physical Therapy. 91 (6), 923-930 (2011).
- Matsumura, N., et al. Quantitative assessment of fatty infiltration and muscle volume of the rotator cuff muscles using 3-dimensional 2-point Dixon magnetic resonance imaging. Journal of Shoulder and Elbow Surgery. 26 (10), 309-318 (2017).
- Cheuy, V. A., Hastings, M. K., Commean, P. K., Ward, S. R., Mueller, M. J. Intrinsic foot muscle deterioration is associated with metatarsophalangeal joint angle in people with diabetes and neuropathy. Clinical Biomechanics. 28 (9-10), 1055-1060 (2013).
- Samagh, S. P., et al. MRI quantification of fatty infiltration and muscle atrophy in a mouse model of rotator cuff tears. Journal of Orthopaedic Research. 31 (3), 421-426 (2013).
- Gerber, C., Meyer, D. C., Schneeberger, A. G., Hoppeler, H., von Rechenberg, B. Effect of tendon release and delayed repair on the structure of the muscles of the rotator cuff: an experimental study in sheep. The Journal of Bone and Joint Surgery. American Volume. 86 (9), 1973-1982 (2004).
- Goodpaster, B. H., Kelley, D. E., Thaete, F. L., He, J., Ross, R. Skeletal muscle attenuation determined by computed tomography is associated with skeletal muscle lipid content. Journal of Applied Physiology. 89 (1), 104-110 (2000).
- Torriani, M., et al. Lower leg muscle involvement in Duchenne muscular dystrophy: an MR imaging and spectroscopy study. Skeletal Radiology. 41 (4), 437-445 (2012).
- Kim, H. M., Galatz, L. M., Lim, C., Havlioglu, N., Thomopoulos, S. The effect of tear size and nerve injury on rotator cuff muscle fatty degeneration in a rodent animal model. Journal of Shoulder and Elbow Surgery. 21 (7), 847-858 (2012).
- Rowshan, K., et al. Development of fatty atrophy after neurologic and rotator cuff injuries in an animal model of rotator cuff pathology. The Journal of Bone and Joint Surgery. American Volume. 92 (13), 2270-2278 (2010).
- Li, B., et al. Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nature Medicine. 6 (10), 1115-1120 (2000).
- Goodpaster, B. H., Theriault, R., Watkins, S. C., Kelley, D. E. Intramuscular lipid content is increased in obesity and decreased by weight loss. Metabolism. 49 (4), 467-472 (2000).
- Biltz, N. K., Meyer, G. A. A novel method for the quantification of fatty infiltration in skeletal muscle. Skeletal Muscle. 7 (1), 1 (2017).
- Hakim, C. H., Wasala, N. B., Duan, D. Evaluation of muscle function of the extensor digitorum longus muscle ex vivo and tibialis anterior muscle in situ in mice. Journal of Visualized Experiments. (72), 50183 (2013).
- Moorwood, C., Liu, M., Tian, Z., Barton, E. R. Isometric and eccentric force generation assessment of skeletal muscles isolated from murine models of muscular dystrophies. Journal of Visualized Experiments. (71), e50036 (2013).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Crapo, P. M., Gilbert, T. W., Badylak, S. F. An overview of tissue and whole organ decellularization processes. Biomaterials. 32 (12), 3233-3243 (2011).
- Ungerleider, J. L., Johnson, T. D., Rao, N., Christman, K. L. Fabrication and characterization of injectable hydrogels derived from decellularized skeletal and cardiac muscle. Methods. 84, 53-59 (2015).
- Biltz, N. K., et al. Infiltration of intramuscular adipose tissue impairs skeletal muscle contraction. The Journal of Physiology. 598 (13), 2669-2683 (2020).

