Quantification of Cytokine-Induced Cell Death in Human Colonic Organoids Using Live Fluorescence Microscopy
Summary
This protocol describes a simple and cost-effective method to investigate and quantify cell death in human colonic organoids in response to cytotoxic perturbagens such as cytokines. The approach employs a fluorescent cell death dye (SYTOX Green Nucleic Acid Stain), live fluorescence microscopy, and open-source image analysis software to quantify single-organoid responses to cytotoxic stimuli.
Abstract
Intestinal epithelial cell (IEC) death is increased in patients with inflammatory bowel diseases (IBD) such as ulcerative colitis (UC) and Crohn’s disease (CD). This can contribute to defects in intestinal barrier function, exacerbation of inflammation, and disease immunopathogenesis. Cytokines and death receptor ligands are partially responsible for this increase in IEC death. IBD-relevant cytokines, such as TNF-α and IFN-γ, are cytotoxic to IECs both independently and in combination. This protocol describes a simple and practical assay to quantify cytokine-induced cytotoxicity in CD patient-derived colonic organoids using a fluorescent cell death dye (SYTOX Green Nucleic Acid Stain), live fluorescence microscopy, and open-source image analysis software. We also demonstrate how to use the Bliss independence mathematical model to calculate a coefficient of perturbagen interaction (CPI) based on organoid cytotoxicity. The CPI can be used to determine if interactions between cytokine combinations or other types of perturbagens are antagonistic, additive, or synergistic. This protocol can be implemented to investigate the cytotoxic activity of cytokines and other perturbagens using patient-derived colonic organoids.
Introduction
The intestinal epithelium creates a physical semipermeable barrier between the contents of the gut lumen and the underlying tissues. To maintain this barrier effectively, intestinal epithelial cells (IECs) undergo an extremely high cellular turnover, with a continuous cycle of cell death and regeneration. However, during inflammatory disorders, such as inflammatory bowel disease (IBD), higher levels of aberrant cell death occur1. This may promote a breakdown in barrier function and activation of the immune system, triggering further inflammation. In Crohn's disease (CD), a form of IBD, it has been shown that cytokine signaling contributes to the increased levels of IEC death2. By studying how cytokine signaling induces cell death of IECs, it is hoped that improved treatments can be developed for patients with IBD and other intestinal inflammatory disorders1.
In biology and drug target discovery research, synergy is generally understood to occur when a biological system treated with combinations of individual stimuli shows a response to the combination that is greater than the combined additive effects of the single stimuli alone. Synergistic interactions between cytokines have been well documented in driving innate antiviral responses3. Cytokines have also been known to induce cell death synergistically, including in IECs4. However, the role synergistic cytotoxic cytokine signaling plays in intestinal inflammatory disorders such as IBD is understudied.
Human intestinal organoids are 3D microtissues produced in vitro that are generated from intestinal epithelial stem cells. Intestinal organoids can be grown from gut mucosal biopsies obtained from patients with IBD and retain many characteristics of the disease5,6. Organoids have proven to be an ideal model system for studying cytokine cytotoxicity in the context of intestinal inflammation7,8. Previously, our group has characterized the synergistic killing effects of the IBD-relevant cytokines IFN-γ and TNF-α in CD patient-derived colonic organoids (colonoids)9,10. However, the exact mechanisms involved in mediating this form of synergistic cell death remain elusive. There are also potentially many more uncharacterized cytotoxic cytokine interactions that are relevant to intestinal inflammatory disorders.
Several protocols are available to study cell death in intestinal organoids10,11,12,13; however, they each have drawbacks. Some of these techniques only measure cell viability and do not measure cell death directly, are incapable of evaluating single-organoid responses, or require expensive equipment and complex protocols. Robust and straightforward methodologies are needed to quantify organoid cell death and perturbagen interactions in intestinal organoids. The protocol we present is a simple and inexpensive approach to measure single-organoid responses to cytotoxic cytokines but can be used for any type of stimulus or perturbagen. We also demonstrate how to use the Bliss independence synergy model to calculate a coefficient of perturbagen interaction (CPI) that describes cytotoxic cytokine interactions.
Protocol
Colonic mucosal biopsies were collected from patients with CD undergoing routine colonoscopy as part of the standard of care. Ethical approval for the use of patient tissue samples and the generation of colonic organoid lines from these samples was obtained from the Clinical Research Ethics Committee of the Cork Teaching Hospitals (CREC). Written informed consent was obtained from all patients in agreement with the Declaration of Helsinki. All tissue culture work with patient biopsies and colonoids must be performed inside a biosafety cabinet following BSL2 safety protocols. Ensure all plastic wear is sterile before use. See the Table of Materials for details related to all materials, reagents, instruments, and software used in this protocol.
The protocols our group uses for crypt isolation and organoid culture were adapted from established methods14,15,16 and have been published previously9,10,17. For the following protocol colonoids were cultured using Organoid Proliferation Media (Table 1). Colonoids grown using Organoid Proliferation Media are undifferentiated and enriched for colonic stem cells. The primary component of Organoid Proliferation Media is 50% L-WRN conditioned media, which contains the intestinal stem cell niche growth factors Wnt-3A (W), R-spondin 3 (R), and Noggin (N)15. Organoid Proliferation Media is prepared by combining L-WRN conditioned media and Serum-free Media 1:1, followed by supplementation with nicotinamide and chemical inhibitors (Table 1).
1. Colonic crypt isolation and colonoid culture
- Preincubate a 48-well microtiter plate at 37 °C, 5% CO2 for a minimum of 72 h before seeding with crypts.
NOTE: Preincubating the plate accelerates the polymerization of the basement membrane extract (BME) during seeding. - Defrost the BME on ice at 4 °C the evening before the isolation of crypts.
- Collect the colonic biopsies in a sample collection tube containing 15 mL of Biopsy Collection Medium (Table 1) and store at 4 °C until ready for processing.
- Carefully remove as much Biopsy Collection Medium as possible using a pipette. Wash the biopsies by adding 15 mL of ice-cold DPBS supplemented with 2.5 µg/mL amphotericin B and 100 µg/mL gentamicin to the sample tube. Shake the sample tube vigorously to dissociate any mucous or debris from the biopsies. Let the biopsies settle by gravity and carefully remove as much DPBS as possible using a pipette.
- Repeat washing procedure from step 1.4 two times (2x).
- Add 10 mL of enzyme-free cell dissociation reagent supplemented with 2.5 µg/mL amphotericin B and 200 µg/mL gentamicin to the sample tube and incubate for 15 min at room temperature with rocking at 30 rpm.
- After the incubation, shake the sample tube side to side by hand vigorously to release the colonic crypts. Inspect the tube using a low-powered light microscope and look for the released crypts and crypt fragments in suspension. If not visible, shake the tube and check again; repeat until the crypts are seen in suspension.
- Attach a 70 µm cell strainer to a 50 mL tube and filter the crypt suspension through the strainer. Add 10 mL of ice-cold Organoid Wash Media (Table 1) to the empty sample tube, remove the media, and pass it through the cell strainer.
- Transfer the filtered crypts into two 15 mL tubes (10 mL per tube) and centrifuge at 4 °C for 5 min at 150 × g.
- Remove the supernatant carefully from each 15 mL tube, resuspend the crypt pellets in 500 µL of ice-cold Organoid Wash Media, transfer the crypt solution from both 15 mL tubes to a single 1.5 mL microcentrifuge tube, and centrifuge at 4 °C for 3 min at 400 × g.
- Carefully remove the supernatant from the microcentrifuge tube and resuspend the crypt pellet in 70 µL of BME (20 µL per well and 10 µL of extra dead volume).
- Using the preincubated 48-well plate (step 1.1), seed 20 µL of BME/crypt suspension into the center of each well (1 BME dome per well). Smoothly and steadily invert the plate and incubate at 37 °C, 5% CO2 for 20 min.
NOTE: Inverting the plate prevents cells from adhering to the plastic surface of the well and ensures their distribution within the BME. - Remove the plate from the incubator. Ensure the BME has fully polymerized and then overlay the domes with 350 µL of prewarmed Organoid Proliferation Media supplemented with 100 µg/mL of a broad-range antimicrobial reagent. Incubate the plate at 37 °C, 5% CO2.
NOTE: The broad-range antimicrobial reagent is to prevent contamination from mucosal microbes associated with the colonic biopsies. It is only required for the first week of culture following crypt isolation. - Change the media 2-3x in a week using prewarmed Organoid Proliferation Media, supplementing with the broad range antimicrobial reagent for the first week of culture; following this period, remove the reagent.
- Once the colonoid culture is fully established (1-2 weeks post isolation), dissociate the colonoids using the enzymatic dissociation reagent supplemented with 10 µM of the ROCK-I/II inhibitor Y-27632 (see section 2 for full details on colonoid dissociation).
NOTE: For the first two passages (P0-1, P1-2), colonoids should be expanded using a 1:1/2 ratio; after P2, colonoids can be passaged using a 1:3/4 ratio. - Seed and maintain the colonoids following steps 1.11-1.13 (do not supplement Organoid Proliferation Media with the broad range antimicrobial reagent).
2. Preparation of colonoids for cell death assay
NOTE: The cell death assay protocol takes 4 days to complete (Figure 1A).
- Expand the colonoids using a 48-well plate format, seeding 20 µL of BME/crypt suspension per well, incubating at 37 °C, 5% CO2, and changing the medium 2-3x a week (350 µL per well).
- Passage the colonoids approximately 1 week prior to harvesting for the cell death assay. Before harvesting the colonoids, ensure they are propagated to a high density (Figure 1Bi), that they are approximately 25-50 µm in diameter, and are actively proliferating.
NOTE: We have used colonoids for this assay from passage 3 to passage 14 with consistent results. However, it has been demonstrated that the transcriptional response of colonoids to cytokines can change depending on culture duration18. On this basis, we recommend not to use colonoids for this assay after passage 15. - Preincubate 96-well microtiter plates at 37 °C, 5% CO2 for a minimum of 72 h before seeding with colonoid cells.
- Defrost the BME on ice at 4 °C the evening before starting the experiment.
- Prepare a sufficient volume of the enzymatic dissociation reagent (500 µL per well of colonoids) by supplementing with 10 µM of the ROCK-I/II inhibitor Y-27632. Gently remove the medium from the wells containing colonoids; pipette from the edge of the well to avoid damaging the colonoid dome. Add 300 µL of enzymatic dissociation reagent to each well.
- For each well, break apart the colonoid dome by scraping the surface of the well with the tip of a P1000 pipette and pipetting the cell suspension up and down; try to avoid generating air bubbles. Collect the cell suspension in a 15 mL tube.
NOTE: This 15 mL tube will be used to collect colonoids from 10 wells of the 48-well plate. - Wash the same well with another 200 µL of the enzymatic dissociation reagent to ensure that all colonoid material is collected and transferred to the same 15 mL tube. Repeat this process for each well of expanded colonoids being harvested and collect them in the same single 15 mL tube.
NOTE: For efficient dissociation, a maximum of 10 wells of colonoids should be collected per tube. If collecting >10 wells, split the collected colonoids equally across multiple 15 mL tubes.
- For each well, break apart the colonoid dome by scraping the surface of the well with the tip of a P1000 pipette and pipetting the cell suspension up and down; try to avoid generating air bubbles. Collect the cell suspension in a 15 mL tube.
- Incubate the 15 mL tube with collected colonoids from step 2.5.2 in a water bath at 37 °C for 5 min. Following the incubation, centrifuge the tube for 3 min at 400 × g. Gently remove the supernatant, leaving approximately 1.2 mL behind in the tube.
NOTE: The next step requires the physical dissociation of colonoids using rapid pipetting. For this to be effective, you must have a small volume of cell suspension in the tube – the 1.2 mL left in the 15 mL tube is sufficient for this purpose. - Resuspend the colonoid pellet in the remaining 1.2 mL of the enzymatic dissociation reagent. Using a P1000 pipette set to 1,000 µL, place the tip of the pipette into the suspension, holding it just above the bottom of the 15 mL tube, then rapidly pipette the suspension in and out of the tip. To rapidly pipette, quickly depress the plunger button to the first stop, release the button until it is approximately halfway to the top position, and repeat. Rapidly pipette for approximately 10 s (30-40 depressions), then resuspend the suspension fully and repeat the pipetting. Perform 2-3 rounds of rapid pipetting.
- Check the sample under the microscope to confirm that there are no whole colonoids left and that the majority of colonoid fragments are approximately 30-40 µm in size (Figure 1Bii).
- If the sample requires further dissociation, incubate in the water bath at 37 °C for another 3 min, repeat the pipetting technique in step 2.7, and check the sample under the microscope. Repeat this process until the majority of the colonoid fragments are of the optimal size.
NOTE: Be careful not to dissociate the colonoids too much as this will result in excessive cell death, low plating efficiency, and undersized colonoids. - Add 10 mL of ice-cold Organoid Wash Media (Table 1) to the 15 mL tube. Centrifuge the tube for 3 min at 400 × g, remove the supernatant, resuspend in 1 mL of ice-cold Organoid Wash Media, and transfer to a 1.5 mL microcentrifuge tube (referred to as the colonoid fragment tube).
- Mix the contents of the colonoid fragment tube by pipetting and take a 50 µL sample; transfer this 50 µL sample to a new 1.5 mL microcentrifuge tube (referred to as the cell count tube). Store the colonoid fragment tube on ice from this point until the completion of step 2.15.
- Centrifuge the cell count tube for 3 min at 400 × g, remove the supernatant, and resuspend in 500 µL of enzymatic dissociation reagent supplemented with 10 µM of the ROCK-I/II inhibitor Y-27632.
- Incubate the single cell count tube in the water bath at 37 °C for 5 min. Using a P1000 pipette set to 400 µL, pipette the sample as described in step 2.7 and check the sample under the microscope to ensure there is a single-cell suspension. If not, repeat this process until the colonoids are fully dissociated into a single-cell suspension.
- Add 1 mL of Organoid Wash Media to the single cell count tube, centrifuge for 3 min at 400 × g and carefully remove the supernatant. Resuspend the cells in 50 µL of Organoid Wash Media and then add 50 µL of trypan blue.
- Count the cells using a hemocytometer. Calculate the number of cells in the 50 µL sample and use this to calculate the concentration of cells in the colonoid fragment tube.
- Calculate the volume of cell suspension required for the experiment where 0.5 × 104 colonoid cells will be seeded per well of a 96-well microtiter plate. Add approximately 15% extra to this calculated volume to account for dead volume. Transfer this total volume from the colonoid fragment tube (step 2.11) to a new 1.5 mL microcentrifuge tube.
- Centrifuge the new 1.5 mL microcentrifuge tube containing colonoid fragments for 3 min at 400 × g, remove the supernatant, and resuspend in BME (use 10 µL of BME per 0.5 × 104 cells).
NOTE: Due to the physical characteristics of the BME (high viscosity, temperature-dependent polymerization), a significant quantity of material can be lost through pipetting (dead volume).
- Centrifuge the new 1.5 mL microcentrifuge tube containing colonoid fragments for 3 min at 400 × g, remove the supernatant, and resuspend in BME (use 10 µL of BME per 0.5 × 104 cells).
- Set up the tube or reservoir containing the colonoid/BME suspension on ice in a sterile container inside the biosafety cabinet.
NOTE: Keeping the cells on ice while seeding prevents the cell substrate from polymerizing prematurely. - Using a preincubated 96-well microtiter plate (from step 2.3), reverse pipette 10 µL of the colonoid/BME solution per well. Be sure to position the tip just above the surface of the well and pipette into the center to avoid hitting the wall of the well. Mix the colonoid/BME suspension regularly to prevent uneven seeding.
NOTE: Do not seed colonoids in the outer edge wells of the microtiter plate.- To reverse pipette:
- Set the pipette and then press the plunger button past the first stop to the second stop.
- Holding this position, immerse the tip in the colonoid/BME suspension and slowly release the plunger to the top.
- Dispense the suspension by pressing the plunger button gently and steadily to the first stop. If seeding more wells, hold this position and repeat steps 2.18.1.2-2.18.1.3.
- Once finished, expel the small amount of remaining suspension by pressing the plunger button to the second stop.
NOTE: We recommend using the reverse pipetting technique due to the high viscosity of the BME.
- To reverse pipette:
- Smoothly and steadily invert the plate and incubate at 37 °C, 5% CO2 for 20 min.
- Remove the plate from the incubator. Ensure the BME has fully polymerized and then, overlay the domes with 200 µL of prewarmed Organoid Proliferation Media. If using a 96-well microtiter plate with a surrounding moat (that reduces evaporation), fill each reservoir with 2 mL of Organoid Wash Media.
- Incubate the colonoids at 37 °C, 5% CO2 for 3 days and inspect microscopically once a day to ensure the colonoids have recovered and are proliferating.
3. Colonoid treatments for cell death assay
- Prepare a 2.5 µM solution of fluorescent cell death dye (SYTOX Green Nucleic Acid Stain) and DMSO solution by adding fluorescent cell death dye or DMSO to prewarmed Organoid Proliferation Media (Table 2).
NOTE: Protect the fluorescent cell death dye stock and diluted solution from light. SYTOX Green Nucleic Acid Stain is solubilized in DMSO; the DMSO solution is used to prepare the No Dye condition to control for solvent effects. - Use the 2.5 µM solution of fluorescent cell death dye and DMSO solution from step 3.1 to prepare treatments as shown in Table 2.
- Gently remove the medium from the 96-well microtiter plate seeded with colonoids by tilting the plate and pipetting from the edge of the wells; then, add 200 µL of treatment medium per well. Be sure to have extra PBS/BSA control wells for the Max Toxicity condition(s) (see Table 2 for the plate map).
- Incubate the colonoids at 37 °C, 5% CO2 for the required treatment time points until ready for imaging.
- At least 2 h before imaging, prepare a 10% v/v solution of Triton-X 100 in sterile cell-culture grade water and add 22 µL of the 10% Triton-X 100 directly to the medium of the control wells for the Max Toxicity condition(s) to a final concentration of 1% v/v 1 h before imaging.
NOTE: Using a 10% solution reduces pipetting errors that can occur due to the high viscosity of the surfactant.
4. Image acquisition
- Remove the 96-well microtiter plate seeded with colonoids from the incubator and transfer to the stage of a digital inverted epifluorescence microscope. Allow the plate to come to room temperature.
- Confirm that the Max Toxicity condition colonoids are fully lysed by examining them under the microscope (Figure 1Biii).
- Select a suitable objective, such as a long working distance 40x fluorescence objective. Optimize the imaging settings of the microscope before beginning.
- Using the transmission channel, focus on a colonoid with SYTOX-positive cells, switch to the Green Fluorescent Protein (GFP)
channel (488 nm), and adjust the light intensity and exposure time to maximize the fluorescent signal while minimizing the background. First, try a low light intensity, then gradually increase the exposure time; if the length of exposure is impractical, then increase the light intensity slightly.
NOTE: Increasing the exposure time instead of the light intensity will reduce phototoxicity and photobleaching of the samples19. Only expose the samples to fluorescent light when necessary.
- Using the transmission channel, focus on a colonoid with SYTOX-positive cells, switch to the Green Fluorescent Protein (GFP)
- Using the optimized imaging settings for the GFP channel, observe the No Dye and Max Toxicity conditions to ensure the samples are not over- or underexposed. Once finalized, keep the imaging settings consistent between conditions.
- Acquire images using a random sampling approach: do this by selecting fields of view (FOVs) that follow a fixed grid pattern that covers the colonoid dome. Acquire images of colonoids from a minimum of 10 FOVs. Ensure the central plane of the colonoid is in focus and acquire images in both transmission and GFP channels.
- Apply the following exclusion criteria.
- Do not acquire images if there are no colonoids present in the FOV.
- Do not acquire images if there are only colonoids that are overlapping in the same focal plane present in the FOV (Figure 1Biv).
- Do not acquire images if there is only colonoid debris present in the FOV (Figure 1Bv). Do not include colonoid debris in the analysis.
- Apply the following exclusion criteria.
- Save images in portable network graphic format and export.
- If imaging additional time points, return the 96-well microtiter plate with colonoids to the incubator at 37 °C, 5% CO2.
5. Image analysis
- Open Fiji ImageJ and import the image dataset by dragging and dropping the files onto the ImageJ toolbar or navigating to File | Open and selecting the files. Once open, combine the files into an image stack by clicking Image| Stacks| Images to Stack. Convert the image stack to 8-bit file format by clicking Image| Type| 8 -bit.
- For each image set, click the Freehand Selections tool on the ImageJ toolbar and manually select the region of interest (ROI) on the transmission image using the computer mouse; the ROI is the perimeter of the colonoid. Then, toggle through the image stack to the corresponding GFP channel image.
- Click Analyze| Set Measurements; in the Set Measurements dialog window, tick Mean Grey Value and leave all other boxes unticked. With the GFP image selected, click Analyze | Measure. Repeat this analysis for every colonoid in the image stack. Once the dataset is analyzed, copy all the data in the Results window and paste them into a spreadsheet software application.
6. % max toxicity calculation
- Calculate the mean of the technical replicate Mean Grey Values (MGV) for each condition using equation (1).
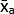 (mean of treatment a) =
(mean of treatment a) = 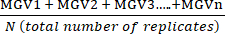 (1)
(1) - Express the mean of each condition as a percentage relative to the mean of the Max Toxicity condition (MT) using equation (2).
%MTa =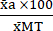 (2)
(2)
7. Calculating the CPI
- Normalize (NORM) the data as follows, using the mean values calculated in step 6.1, subtract the mean of the Untreated condition (UT) from each treatment condition and the Max Toxicity (MT) condition (to remove the background cell death that occurs independent of cytokine treatment). Then, divide each treatment condition by the background-subtracted mean of the Max Toxicity (MT) condition as shown in equation (3).
NORMa =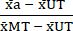 (3)
(3) - Subtract the normalized values calculated in step 7.1 from 1; the resulting values represent cell viability (V) after treatment (as in equation (4) below).
Va = 1 −NORMa (4) - Calculate the coefficient of perturbagen interaction (CPI) using equation (5):
CPI =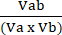 (5)
(5)
Where a denotes the first treatment; b denotes the second treatment; and ab is the combination treatment. CPI values indicate synergistic (<1), additive (=1), or antagonistic (>1) relationships.
Representative Results
Using this protocol, we demonstrated how CD patient colonoids can be used to study the cytotoxic effects of the IBD-relevant cytokines IFN-γ and TNF-α on primary epithelium. We used a commercially available fluorescent cell death dye (SYTOX Green Nucleic Acid Stain), which can only enter cells that have a compromised cell membrane where it is then activated by binding to nucleic acids. We co-treated colonoids with cytokines and the fluorescent cell death dye and performed live cell imaging at 8 h and 24 h with an inverted epifluorescence microscope. Representative transmission/fluorescent overlay images at 8 h indicate that only the IFN-γ + TNF-α-treated colonoids are positive for fluorescent signal; however, there are only a small number of fluorescent cells (Figure 2A). Cell blebbing, a morphological indicator of cell death20, can also be observed in the IFN-γ + TNF-α condition. At 24 h, colonoids treated with IFN-γ + TNF-α display large regions positive for fluorescent signal (Figure 2A). There is also a clear breakdown in the morphology of the colonoid-the central lumen is no longer visible, and the epithelial barrier has been completely disrupted.
To quantify the cell death dye signal, we used open-source image analysis software to calculate the fluorescent intensity of each colonoid. We then normalized the data by expressing the mean of each condition as a percentage of the Max Toxicity treatment. At 8 h, homeostatic or background cell death in the BSA control colonoids was relatively low (7.7% of Max Toxicity) (Figure 2B). There were no statistically significant changes in cell death levels at this time point; however, conditions treated with TNF-α displayed a small increase in cytotoxicity (Figure 2B). After 24 h, cell death levels had increased for all cytokine-treated conditions. However, there was minimal change in cell death for the BSA Control condition between time points (7.5% of Max Toxicity at 24 h). The colonoids treated with IFN-γ + TNF-α had the largest increase in cell death levels compared to BSA control (29.4% of Max Toxicity). The difference in cell death levels between the combined treatment and the single cytokine treatments (IFN-γ, TNF-α) was highly significant. These results suggest the possibility of a cytotoxic synergistic interaction between IFN-γ and TNF-α at 24 h.
We used the CPI to quantify the cytotoxic interactions between cytokine treatments and to determine if they were synergistic. Interactions between cytokines are considered synergistic when the CPI value is <1, additive when =1, or antagonistic when >1. We calculated CPI values per time point (Figure 2C). At 8 h, the CPI value indicated slight synergism (0.99), with the CPI value decreasing substantially at 24 h (0.83). This analysis confirmed that the interaction between IFN-γ and TNF-α at 24 h was synergistic. Further, it illustrates how in this context synergism between IFN-γ and TNF-α is time-dependent.
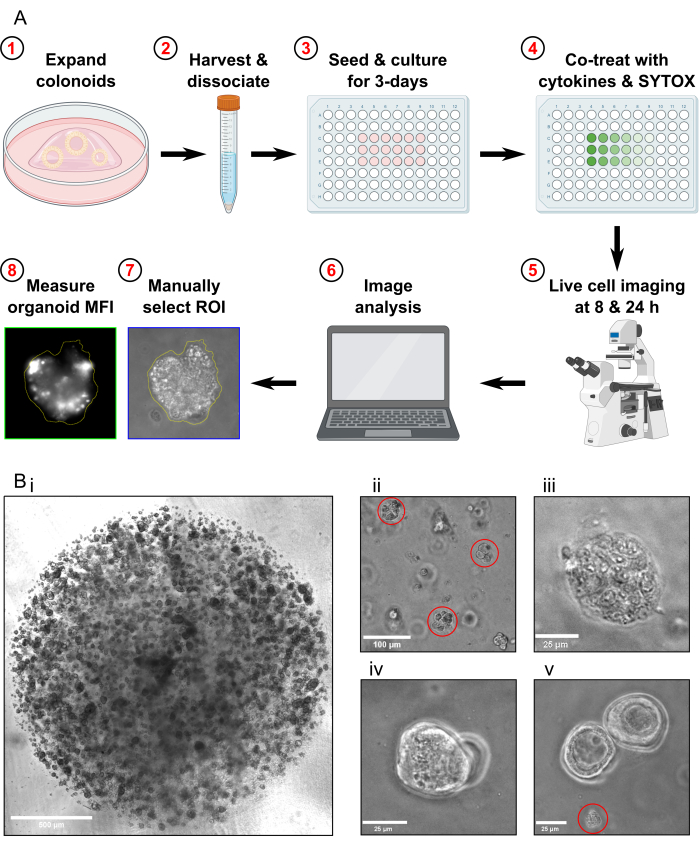
Figure 1: Schematic of experimental workflow and troubleshooting. (A) Schematic overview of protocol. (B) Representative images. (Bi) Light microscopy image illustrating the optimal density of the culture and optimal size of colonoids prior to passaging for an assay. Scale bar = 500 µm. (Bii) Light microscopy image illustrating optimal size of colonoid fragments after dissociation; fragments highlighted in red. Scale bar = 100 µm. (Biii) Light microscopy image of necrotic colonoid morphology after MT treatment (with Triton X-100). Scale bar = 25 µm. (Biv) Light microscopy image of two colonoids overlapping in the same focal plane. Scale bar = 25 µm. (Bv) Light microscopy image of colonoid cell debris present after passaging; debris highlighted in red. Scale bar = 25 µm. Abbreviations: ROI = region of interest; MFI = mean fluorescence intensity; MT = Max Toxicity. Please click here to view a larger version of this figure.
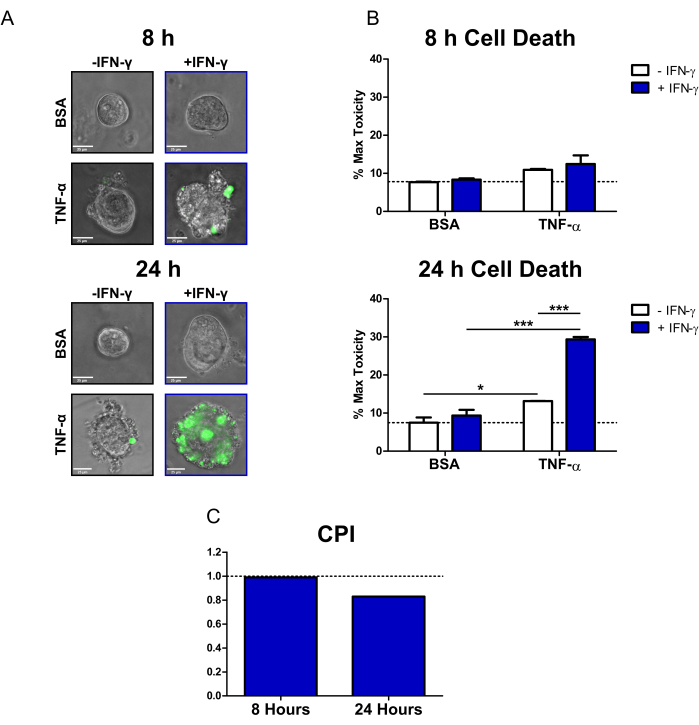
Figure 2: Quantitative analysis of cytokine-induced cell death in human CD colonoids. (A) Representative live microscopy images of CD colonoids treated with SYTOX Green Nucleic Acid Stain (fluorescent cell death dye) and cytokines at 8 h and 24 h; transmission and GFP (green color) channels overlayed. Colonoids were treated as follows: 1) PBS/BSA, 2) 10 ng/mL IFN-γ, 3) 10 ng/mL TNF-α, 4) 10 ng/mL IFN-γ + 10 ng/mL TNF-α. Scale bars = 25 µm. (B) Quantitative analysis of CD colonoids treated with the fluorescent cell death dye and cytokines at 8 and 24 h; data are expressed as a % of the MT condition. N = 2 CD colonoid lines, 11-16 colonoids imaged per condition. (C) CPI calculated per time point using the dataset from B, N = 2 CD colonoid lines. Data are expressed as means ± SE. In B, two-way ANOVA analysis was performed followed by Bonferroni post-tests, *P < 0.05, ***P < 0.001 as indicated. Abbreviations: CD = Crohn's disease; GFP = green fluorescent protein; CPI = coefficient of perturbagen interaction; PBS = phosphate-buffered saline; BSA = bovine serum albumin; TNF-α = tumor necrosis factor-alfa; IFN-γ = interferon-gamma; MT = Max Toxicity. Please click here to view a larger version of this figure.
Table 1: Composition of culture media for protocol. To prepare Organoid Proliferation Media, combine L-WRN conditioned media and Serum-free Media 1:1, then add supplements. Organoid Proliferation Media should be used within 2 weeks of preparation. Please note all complete media should be stored at 4 °C. Please click here to download this Table.
Table 2: Experimental 96-well plate layout and cytokine treatments. Please click here to download this Table.
Discussion
Several methods have been developed for quantitative analyses of cell death in intestinal organoids. Examining the disruption of intestinal organoid morphology by light microscopy is a straightforward approach to quantifying the effects of cytotoxic substances11. However, morphological changes are not a direct measurement of cell death, and the method is only semiquantitative. Another method is to evaluate organoid metabolic activity using an MTT or ATP assay10,11. It is important to note that these assays can only determine changes in cell viability and must be validated with a cell death assay. Other fluorometric cell death assays using DNA binding dyes have been reported12,13. A non-imaging approach using a fluorescent microplate reader is possible and allows for high throughput12. However, this method measures the average signal of an entire well, making it unsuitable for heterogeneous populations. It also requires the use of a microplate reader with Z-height adjustment. Fluorescent imaging-based techniques can be used for single-organoid analysis and capture cellular/subcellular and morphological data. Automated confocal high-content imaging (HCI) systems can generate large amounts of data at a high throughput13. Unfortunately, confocal HCI needs specialized equipment, uses complex protocols, typically requires commercial image analysis software, and is expensive.
Our protocol for quantitative analysis of colonoid cell death at multiple time points is straightforward, simple, and inexpensive. However, compared to automated HCI and plate reader systems, it is time-consuming and has reduced throughput. Another limitation of our method is the use of widefield as opposed to confocal microscopy. Confocal microscopy is more suitable for imaging thick 3D samples such as organoids as it reduces out-of-focus signal and can acquire serial optical sections (Z-stacks). However, confocal imaging typically requires longer acquisition times and high-intensity lasers that increase phototoxicity/photobleaching. It is critical to note that fluorescent cell death dyes like SYTOX Green are only suitable for measuring forms of cell death where there is loss of cell membrane integrity such as necrosis, late apoptosis-associated secondary necrosis, necroptosis, and pyroptosis21. There are some forms of regulated cell death where the cell membrane remains impermeable at least during the early phases of cell death, such as caspase-dependent apoptosis. However, this protocol could be easily modified to also incorporate imaging of a caspase 3/7 activity fluorescent reporter22. This would provide additional data to help characterize the specific cell death modality.
We used our protocol to demonstrate the cytotoxic synergistic interaction between the cytokines IFN-γ and TNF-α (Figure 2C), which we have previously reported in CD patient-derived organoids9,10. The physiological relevance of this form of synergism has also been demonstrated in murine models of hemophagocytic lymphohistiocytosis and sepsis23. Several mathematical reference models and approaches have been implemented for quantifying synergy between combinations of biological agents24,25. They differ in terms of their complexity, the number of factors they consider, and the threshold for considering an interaction to be synergistic24,25. Some models need prior knowledge of the biological agents tested, make certain assumptions about the activity of agents, and can require comprehensive dose-response curves for each single and combination treatment25. The method we selected to measure synergy is a modification of the coefficient of drug interaction (CDI) model, which has previously been used to measure the inhibitory effects of chemotherapy drug combinations on cancer cell line proliferation26. The CDI is a Bliss independence model; when calculating the predicted combined effect of two perturbagens Bliss independence assumes that they target separate pathways and have independent mechanisms of action27. For an interaction between perturbagens to be synergistic the actual combined effect must be greater than the predicted effect. This model is appropriate for our experimental setup as IFN-γ and TNF-α are known to have different receptors and downstream signaling components. Further, Bliss independence allows for the calculation of a coefficient of interaction to quantify synergism and does not require dose-response datasets.
There are a few key factors that must be considered to ensure optimal results for this protocol. It is important that colonoids are propagated to a high density (Figure 1Bi), that they are approximately 25-50 µm in diameter, and are actively proliferating before attempting to seed cells. The use of suboptimal cultures of colonoids for assays may result in insufficient cell numbers, low colonoid recovery, and inconsistent experiments. For reproducible results, it is also important to seed the density of colonoids consistently between experiments. It has previously been demonstrated that the in vitro response to inflammatory cytokines can be influenced by cell seeding density28,29. Another common issue is the formation of air bubbles in the BME dome, which can affect imaging. This can be prevented by using the reverse pipetting technique. This technique also results in more consistent seeding.
Further, if imaging multiple time points, prepare a Max Toxicity condition for each time point. Triton-X 100, a nonionic surfactant, is commonly used as a positive control (Max Toxicity condition) for cytotoxicity assays. The addition of Triton-X 100 lyses and kills the colonoids, allowing the fluorescent cell death dye to enter the cells. Using a Max Toxicity condition from an earlier time point will result in inaccurate and inconsistent normalization of data due to the fluorescent signal decaying over time.
A final point to consider is the choice of BME used for colonoid culture. There are several commercial producers of BME; however, for our protocol, we have only tested the brand included in the Table of Materials. A recent study using patient-derived pancreatic cancer organoids found that the commercial source of BME altered cell proliferation rates but had no significant effect on response to chemotherapy drugs or gene expression30. With this in mind, we expect the trend of results to be similar between BME brands for our protocol, but we recommend using the same brand consistently.
We demonstrated how this protocol can be used for the analysis of IFN-γ and TNF-α induced cell death using CD patient-derived colonoids. Patient-derived intestinal organoids are a powerful tool to study CD as they retain many characteristics of the disease, including increased sensitivity to the cytotoxic effects of TNF-α31. However, the protocol could be easily modified to investigate cytotoxic effects of perturbagens other than cytokines or disease states other than IBD such as colorectal cancer (we have successfully tested the protocol using non-IBD colonoids). We believe this method is useful for any research area concerned with cell death mechanisms, epithelial barrier function, or intestinal mucosal immunology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank the patients for their informed consent and participation in the research study, and the clinical personnel for their excellent assistance. Figure 1A was created with BioRender.com. This work was supported by grants from Science Foundation Ireland-namely a career development award (CDA) to K.N. (SFI-13/CDA/2171), a research centre grant (SFI-12/RC/2273), and a research centre spoke award (SFI-14/SP/2710) to APC Microbiome Ireland. P.F. also received funding from SFI/20/RP/9007.
Materials
| Advanced DMEM/F12 | Gibco | 12634010 | |
| Amphotericin B Solution | Sigma-Merck | A2942 | |
| A-83-01 | Sigma-Merck | SML0788 | |
| BioRender | Science Suite Inc. | N/A | Scientific illustration software |
| Bovine Serum Albumin | Sigma-Merck | A2058 | Essentially IgG-free, low endotoxin |
| B27 Supplement | Invitrogen | 17504-044 | |
| CHIR-99021 | Sigma-Merck | SML1046 | |
| Costar 48-well Clear TC-treated Multiple Well Plates, Individually Wrapped, Sterile | Corning | 3548 | |
| Cultrex Basement Membrane Extract, Type 2, Pathclear | R&D Systems | 3532-010-02 | Basement membrane extract |
| Dimethyl sulfoxide | Sigma-Merck | D2650 | |
| Dulbecco′s Phosphate Buffered Saline | Sigma-Merck | D8537 | |
| EVOS FL Digital Inverted Fluorescence Microscope | Invitrogen | AMF4300 | Digital inverted epifluorescence microscope |
| EVOS 40x Objective, fluorite, LWD, phase-contrast | ThermoFisher Scientific | AMEP4683 | Long working distance 40x fluorescence objective |
| Fiji/ImageJ (Windows version) | Open-source software | N/A | Image analysis software |
| Foetal Bovine Serum | Sigma-Merck | F9665 | |
| Gentamicin Solution | Sigma-Merck | G1397 | |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | 100-0485 | Enzyme-free cell dissociation reagent |
| GlutaMAX-1 | Gibco | 35050061 | L-alanyl-L-glutamine dipeptide supplement |
| GraphPad Prism 5 (Windows version) | Dotmatics | N/A | Data graphics and statistics software |
| Greiner 15 mL Polypropylene Centrifuge Tube, Sterile with conical bottom & Screw Cap | Cruinn | 188261CI | |
| HEPES 1 M | Gibco | 15630080 | |
| Human recombinant EGF (animal free) | Peprotech | AF-100-15 | |
| N-Acetylcysteine | Sigma-Merck | A9165 | |
| Nicotinamide | Sigma-Merck | N0636 | |
| Normocin | InvivoGen | ant-nr-05 | Broad range antimicrobial reagent |
| Nunc Edge 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplate | ThermoFisher Scientific | 15543115 | |
| N2 supplement | Invitrogen | 17502-048 | |
| Recombinant Human IFN-gamma Protein | R&D Systems | 285-IF | Resuspend in sterile filtered 0.1% PBS/BSA |
| Recombinant Human TNF-alpha Protein | R&D Systems | 210-TA | Resuspend in sterile filtered 0.1% PBS/BSA |
| SB202190 | Sigma-Merck | S7067 | |
| Snap Cap Low Retention Microcentrifuge Tubes | ThermoFisher Scientific | 3451 | |
| SYTOX Green Nucleic Acid Stain – 5 mM Solution in DMSO | Invitrogen | S7020 | Fluorescent cell death dye, protect from light |
| Triton X-100 | Sigma-Merck | 93420 | |
| Trypan Blue solution | Sigma-Merck | T8154 | |
| Tryple Express | Gibco | 12604013 | Enzymatic dissociation reagent |
| Y-27632 | MedChemExpress | HY-10071 | Inhibitor of ROCK-I and ROCK-II |
References
- Patankar, J. V., Becker, C. Cell death in the gut epithelium and implications for chronic inflammation. Nat Rev Gastroenterol Hepatol. 17 (9), 543-556 (2020).
- Zeissig, S., et al. Downregulation of epithelial apoptosis and barrier repair in active Crohn’s disease by tumour necrosis factor alpha antibody treatment. Gut. 53 (9), 1295-1302 (2004).
- Bartee, E., Mcfadden, G. Cytokine synergy: An underappreciated contributor to innate anti-viral immunity. Cytokine. 63 (3), 237-240 (2013).
- Fish, S. M., Proujansky, R., Reenstra, W. W. Synergistic effects of interferon γ and tumour necrosis factor α on T84 cell function. Gut. 45 (2), 191-198 (1999).
- Wakisaka, Y., Sugimoto, S., Sato, T. Organoid medicine for Inflammatory Bowel Disease. Stem Cells. 40 (2), 123-132 (2022).
- Flood, P., Hanrahan, N., Nally, K., Melgar, S. Human intestinal organoids: Modeling gastrointestinal physiology and immunopathology – current applications and limitations. Eur J Immunol. 54 (2), e2250248 (2024).
- Matsuzawa-Ishimoto, Y., et al. An intestinal organoid-based platform that recreates susceptibility to t-cell-mediated tissue injury. Blood. 135 (26), 2388-2401 (2020).
- Lee, C., et al. Intestinal epithelial responses to IL-17 in adult stem cell-derived human intestinal organoids. J Crohns Colitis. 16 (12), 1911-1923 (2022).
- Woznicki, J. A., et al. TNF-α synergises with IFN-γ to induce caspase-8-JAK1/2-STAT1-dependent death of intestinal epithelial cells. Cell Death Dis. 12 (10), 864 (2021).
- Flood, P., et al. DNA sensor-associated type I interferon signaling is increased in ulcerative colitis and induces jak-dependent inflammatory cell death in colonic organoids. Am J Physiol Gastrointest Liver Physiol. 323 (5), G439-G460 (2022).
- Grabinger, T., et al. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis. 5 (5), e1228 (2014).
- Bode, K. J., Mueller, S., Schweinlin, M., Metzger, M., Brunner, T. A fast and simple fluorometric method to detect cell death in 3D intestinal organoids. Biotechniques. 67 (1), 23-28 (2019).
- Mertens, S., et al. Drug-repurposing screen on patient-derived organoids identifies therapy-induced vulnerability in KRAS-mutant colon cancer. Cell Rep. 42 (4), 112324 (2023).
- Vandussen, K. L., et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 64 (6), 911-920 (2015).
- Miyoshi, H., Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 8 (12), 2471-2482 (2013).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Woznicki, J. A., et al. Human BCL-G regulates secretion of inflammatory chemokines but is dispensable for induction of apoptosis by IFN-γ and TNF-α in intestinal epithelial cells. Cell Death Dis. 11 (1), 68 (2020).
- Edgar, R. D., et al. Culture-associated DNA methylation changes impact on cellular function of human intestinal organoids. Cell Mol Gastroenterol Hepatol. 14 (6), 1295-1310 (2022).
- Mubaid, F., Brown, C. M. Less is more: Longer exposure times with low light intensity is less photo-toxic. Microscopy Today. 25 (6), 26-35 (2017).
- Ziegler, U., Groscurth, P. Morphological features of cell death. News Physiol Sci. 19 (3), 124-128 (2004).
- Zhang, Y., Chen, X., Gueydan, C., Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 28 (1), 9-21 (2018).
- Tamura, H., et al. Evaluation of anticancer agents using patient-derived tumor organoids characteristically similar to source tissues. Oncol Rep. 40 (2), 635-646 (2018).
- Karki, R., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 184 (1), 149-168.e17 (2021).
- Geary, N. Understanding synergy. Am J Physiol Endocrinol Metab. 304 (3), E237-E253 (2013).
- Duarte, D., Vale, N. Evaluation of synergism in drug combinations and reference models for future orientations in oncology. Curr Res Pharmacol Drug Discov. 3, 100110 (2022).
- Wong, F. C., Woo, C. C., Hsu, A., Tan, B. K. The anti-cancer activities of Vernonia amygdalina extract in human breast cancer cell lines are mediated through caspase-dependent and p53-independent pathways. PLoS One. 8 (10), e78021 (2013).
- Ryall, K. A., Tan, A. C. Systems biology approaches for advancing the discovery of effective drug combinations. J Cheminform. 7, 7 (2015).
- Sukho, P., et al. Effect of cell seeding density and inflammatory cytokines on adipose tissue-derived stem cells: An in vitro study. Stem Cell Rev Rep. 13 (2), 267-277 (2017).
- Vaughan-Jackson, A., et al. Density dependent regulation of inflammatory responses in macrophages. Front Immunol. 13, 895488 (2022).
- Lumibao, J. C., et al. The impact of extracellular matrix on the precision medicine utility of pancreatic cancer patient-derived organoids. bioRxiv. , (2023).
- Lee, C., et al. TNFα induces LGR5+ stem cell dysfunction in patients with Crohn’s disease. Cell Mol Gastroenterol Hepatol. 13 (3), 789-808 (2022).

.
