Fluorescence Anisotropy-Based Detection of Protein-Protein Interactions
Abstract
Source: Gijsbers, A., et al. Fluorescence Anisotropy as a Tool to Study Protein-protein Interactions. J. Vis. Exp. (2016).
In this video, we describe the fluorescence anisotropy technique to study the interactions between the fluorophore-tagged Shwachman-diamond syndrome (SBDS) protein and the elongation factor-like 1 GTPase (EFL1). On incubating SBDS proteins with gradually increasing concentrations of EFL1, a steady increase in anisotropy is observed, indicating a successful interaction between the two proteins.
Protocol
1. Labeling of SBDS-FlAsH with the FlAsH Fluorescent Dye 4',5'-Bis(1,3,2 dithioarsolan-2-yl) Fluorescein
- Mix 3 nmol of the SBDS-FlAsH protein (the FlAsH-tag corresponds to the tetracysteine motif Cys-Cys-Pro-Gly-Cys-Cys genetically engineered in the C-terminus of the recombinant SBDS protein) with 3 nmol of the 4',5'-bis(1,3,2 dithioarsolan-2-yl) fluorescein dye in 5 µl volume of Anisotropy buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 5 mM β-mercaptoethanol).
- Let the reaction proceed for 8 hr at 4 °C. Dialyze the sample against Anisotropy buffer overnight to remove the free dye.
- Use the Lambert-Beer law to quantify the % of labeled protein. Measure the absorbance at 280 nm and 508 nm in a spectrophotometer using a quartz cuvette of appropriate volume. NOTE: Consider the following molar absorption coefficients (M-1 cm-1):
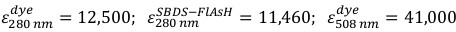 Equation 1
Equation 1 - Calculate the concentration of labeled SBDS-FlAsH protein using Equation 2.
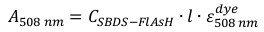 Equation 2
Equation 2 - Calculate the concentration of total SBDS protein using Equation 3 by substituting the calculated CSBDS-FlAsH from the previous step.
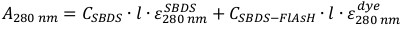 Equation 3
Equation 3 - Calculate the percentage of labeled protein using Equation 4.
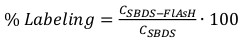 Equation 4
Equation 4
2. Fluorescence Anisotropy Experiments
NOTE: Anisotropy experiments were done in a spectrofluorometer equipped with a polarization toolbox and data collection was performed using the anisotropy program provided in the software of the equipment. The excitation wavelength was set at 494 nm with a spectral bandwidth of 8 nm and the emission was recorded using a band-pass filter of 530±25 nm. Measurements were done at 25 °C in a 200 µl cuvette with a 5 mm path length.
- In a fluorescence cuvette, place 200 µl of 30 nM SBDS-FlAsH in an anisotropy buffer and titrate 2 µl of 30 µM EFL1. Mix thoroughly and let the reaction stand for 3 min before measuring the anisotropy value.
- Repeat step 2.1 until a total volume of 40 µl of EFL1 has been added.
3. Data Analysis
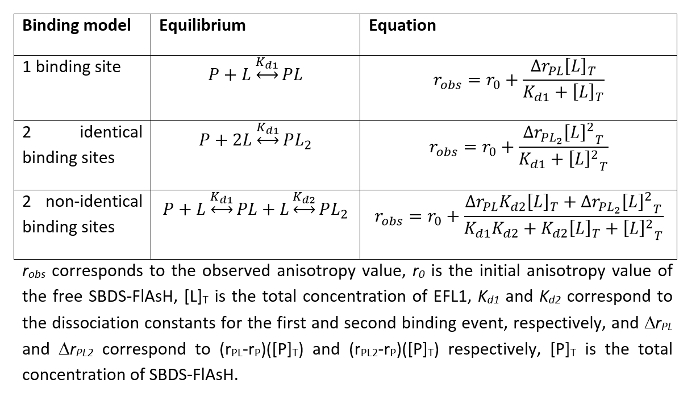
- Fit the data to the appropriate binding model using a nonlinear least squares regression algorithm. Equations for the most common binding models are presented in Table 1.
- Evaluate the best model that describes the interaction between the proteins by inspecting the residuals of the fit. Support the chosen model with additional experiments.
Table 1. Common protein-protein interaction binding models and the mathematical equations that describe them.
Disclosures
The authors have nothing to disclose.
Materials
| Tris Base | Formedium | TRIS01 | Ultra pure |
| dye 4’,5’-bis(1,3,2 dithioarsolan-2- yl) fluorescein | ThermoFischer Scientific | LC6090 | This kit contains the dye to label a FlAsH tag |
| Fluorescence cell | Hellma Analytics | 111-057-40 | |
| Spectrophotometer | Agilent Technologies | G6860AA | Cary 60 UV-Vis |
| Spectrofluorometer | Olis | No applicable | Olis DM 45 with Polarization Toolbox |
| 2-Mercaptoethanol | Sigma-Aldrich | M6250-100ML | |
| NaCl | Formedium | NAC02 | Sodium Chloride |
| MgCl2 | Merck Millipore Corporation | 1725711000 | Magnesium Chloride |
| Glycerol | Sigma-Aldrich | G5516 | |
| Glycerol | Tecsiquim, S.A. de C.V. | GT1980-6 |

