An Assay to Study the Regulation of Macrophage Phagocytosis by Mesenchymal Stem Cells
Abstract
Source: Evans, J. F., et al. Mesenchymal Stem Cell Regulation of Macrophage Phagocytosis; Quantitation and Imaging. J. Vis. Exp. (2021).
This video demonstrates an assay to study mesenchymal stem cell (MSC) regulation of macrophage phagocytosis in a co-culture. Upon co-culturing MSC with macrophages, non-opsonized zymosan particles conjugated to a pH-sensitive fluorophore are added to be phagocytosed by the macrophages. With the increase in phagocytosis, the fluorescence intensity rises, allowing the determination of MSC-mediated regulation of phagocytic activity.
Protocol
NOTE: All medium preparation and cell culture techniques are carried out under aseptic conditions using a biosafety cabinet with laminar flow. All culture incubation steps described are carried out using an incubator designed to maintain an atmosphere of 37 °C, 5% CO2, and 95% humidity.
1. Cell culture
- Preparation of growth medium
- For MSC (mesenchymal stem cells) and LADMAC (mouse bone marrow-derived macrophages; it is derived from the names of the researchers who established the cell line: Laskin, Dower, and Malech), add 50 mL of FBS and 5 mL of 100x antibiotic/antimycotic mix to 500 mL of high glucose DMEM (Dulbecco's modified Eagle medium).
- For the macrophages (MΦ), add 50 mL of FBS, 100 mL of LADMAC condition medium (prepared following the steps of section 1.2), and 5 mL of 100x antibiotic mix to 500 mL of high glucose DMEM.
NOTE: LADMAC cells produce colony-stimulating factor (CSF-1) necessary to support the growth of the MΦ cells.
- Propagate cells in preparation for co-culture
- To seed MSC and MΦ cells from the frozen stock, equilibrate 9 mL of appropriate growth medium prepared in section 1.1 in each of the four 100 mm cell cultures treated dishes for each cell type and place in the cell culture incubator for 15 min.
- Meanwhile, quickly thaw the aliquots of 106 frozen cells by incubating them in a 37 °C water bath for 2-3 min or until just thawed. Then, add the thawed MSC cells to 9 mL of fresh MSC growth medium and add the thawed MΦ cells to 9 mL of fresh MΦ growth medium in 15 or 50 mL sterile conical tubes and centrifuge at 125 x g in a swinging bucket rotor for 5 min.
- Aspirate or decant the supernatant and resuspend each pellet in 4 mL of the appropriate fresh growth medium by gently pipetting up and down. Add 1 mL of the cell suspension to each of the four 100 mm dishes for each cell type that have been equilibrated in step 1.2.1.
- Return the dishes with cells to the incubator. Change the medium every 2-3 days until 70%-80% confluent.
2. Seed MSC in experimental dishes, Day 1
NOTE: Prior to seeding the MSC, design the experimental 96-well plate layout for fluorescent spectrophotometry and the chamber slide for dynamic imaging. For the 96-well plate, flank experimental wells with wells containing cells but do not use them in the assay. Mark at least four wells to be used for the reagent blank (RBL). See Table 1 for an example 96-well template and Table 2 for an example of a 4-well chamber slide template. Black or white 96-well plates with clear bottoms are recommended. A 1.5 mm borosilicate glass chamber slide is optimal, but the number of chambers can be adjusted depending on the study design. A summary flow chart of the methods that follow is included in Figure 1.
- At 70%-80% confluency, aspirate the growth medium from the cultures in 100 mm dishes and add 5 mL of phosphate-buffered saline (PBS) to isolate the MSC.
- Aspirate the PBS, add 2 mL of 0.05% trypsin/EDTA (Ethylenediaminetetraacetic acid), and place it in the culture incubator for 3-5 min.
- After 3 min, check the cell detachment using an inverted microscope. If the cells are detached, continue to the next step; if not, continue incubation for another 1-2 min. Do not incubate longer than 5-6 min.
- Add 5 mL of fresh growth medium to the dish with the detached cells. Rinse the dish using the trypsin/medium mixture and collect cells into a clean, sterile 50 mL conical tube using a sterile serological pipette.
- Centrifuge for 5 min at 125 x g. Aspirate the supernatant and resuspend the cell pellet in 10 mL of fresh growth medium by gently pipetting up and down.
- To count the cells, add 10 µL of the cell suspension to 30 µL of 0.4% trypan blue solution in a 1.5 mL microcentrifuge tube. Then, pipette 10 µL of the mixture under the coverslip of a hemocytometer counting chamber.
- Using an inverted or bright field microscope, with the 10x objective, count the unstained (viable) cells in four of the 1 mm2 squares. Calculate the cell number using the formula: cells/mL = cell count/# 1 mm2 squares x dilution factor x 104.
NOTE: For this example, the number of 1 mm2 squares counted is four, and the dilution factor is 4. - Use the equation C1V1 = C2V2 to calculate for and prepare a 1 x 105 cell/mL suspension. Prepare 1 mL of cell/mL suspension for every column of the 96-well plate to be filled and 0.75 mL for every chamber of the 4-well chamber slide to be filled according to the plate design.
NOTE: C1 = cell count, V1 = what is being calculated, C2 = 1 x 105, V2 = 5.50 mL. - Determine V1, and subtract it from the desired 5.50 mL of the total volume to determine the volume of the fresh medium needed to prepare the suspension. Add the volume of cells (V1) from the original suspension to the volume of fresh medium for a total of 5.50 mL with 1 x 105 cells/mL.
- Add 100 µL of 1 x 105 cell/mL suspension to each well of the 96-well plate according to the plate design using a multichannel micro pipettor and 530 µL to each of the wells of the chamber slide using a micropipette according to the plate design. Incubate overnight.
NOTE: For this experiment, MSCs are plated in columns 1, 2, 3, and 6 of the 96-well plate (Table 1) and wells 1 and 2 of the 4-well chamber slide (Table 2). Therefore, at least 5.50 mL of a 1 x 105 cell/mL suspension is prepared. MΦ at 80% confluence is treated with inflammatory mediators the same day the MSCs are seeded in the experimental plates.
3. Isolate the MΦ and prepare the co-cultures, Day 2
- Remove the activation medium from the MΦ cells and replace it with 5 mL of fresh growth medium. Scrape gently with a cell lifter and collect the cells into a 50 mL conical tube.
- Count the cells using trypan blue exclusion and hemocytometry as described for MSC in, steps 2.6-2.7.
- Using the equation in steps 2.8-2.9, prepare two separate suspensions of 2 x 105 cells/mL, one with cells from the control and one with cells from the treated MΦ cultures.
NOTE: Prepare 1 mL of cell/mL suspension for every column of the 96-well plate to be filled and 0.75 mL for every chamber of the 4-well chamber slide to be filled according to the plate design. - Gently aspirate the medium from the experimental wells of the 96-well plate containing MSC. Using a multichannel micropipette, add 100 µL of the treated and the control MΦ cell suspensions in the appropriate experimental wells with and without MSC according to the plate design. Incubate overnight.
- Gently aspirate the medium from the 4-well chamber slide containing MSC, and, using a micropipette, add 530 µL of MΦ cell suspension to the appropriate wells according to the design. Incubate overnight.
4. Phagocytosis assay, kinetic fluorescent 96-well plate read, Day 3
- Set the assay parameters
- On the day of the assay, turn on the fluorescent plate reader and computer-set the temperature on the system to 37 °C.
- Open the System Pro software and check the instrument icon in the upper-left corner to determine whether the instrument is connected to the computer. If the red circle with a line is visible, click on the instrument icon and choose the instrument in the pop-up window to make the connection.
- Select New Experiment to access the Plate Set-Up Helper pop-up window. From the Plate Set-Up Helper menu, select Configure Your Acquisition Settings.
- Select Monochromator from the settings menu for the optical configuration. Select FL (fluorescence) for the read mode and Kinetic for the read type.
- In the same configurations window, under Category, click on Wavelengths, and then set bandwidths to 9 nm for excitation and 15 nm for emission.
- Set the Number of Wavelength Pairs to 1. Set the wavelengths LM1 to 510 nm for excitation and to 540 nm for emission.
- Continue through the categories and select Plate Type next. Select the option that matches the plate type being used.
NOTE: It is important that the selection matches the plate type. The read heights are set by the system according to the selection. - Next, select Read Area and highlight the area of the 96-well plate included in the experimental design.
- Select PMT and Optics, set PMT Gain to High and Flashes per read to 6. Check the box next to Read from Bottom if using a clear-bottom plate.
- Select Timing and set for 10-minute intervals over a 70-minute period.
- Select Shake, check the Before First Read box, and set it for 5 seconds. Check the Between Reads box and set it for 3 seconds. Set shake intensity for both to Low and Linear.
- Close the window. When the Plate Set-Up Helper window pops up, select Configure Your Plate Layout. Highlight the BL wells of the plate design and click on Plate Blank.
- Highlight each of the experimental rows, click on Add, name the group, and then select a color.
- Under Assignment Options below the plate design, select Series, and then define the series in the window and click on OK. Repeat for each group.
- Prepare the zymosan suspension
- While waiting for the system temperature to reach 37 °C, resuspend 1 mg of zymosan particles in 5 mL of the live-cell imaging medium.
- Add 1 mL of live-cell imaging medium to the vial containing the particles and collect them into a glass culture tube. Rinse the vial with an additional 1 mL of imaging solution and transfer it to the same glass tube to ensure that all the particles are transferred. Add 3 mL of additional imaging solution to achieve a 0.2 mg/mL of particle suspension.
- Vortex with quick pulses for 30-60 s. Then, use a probe sonicator to sonicate with 60 quick pulses.
NOTE: It is very important to create a homogenous suspension of particles and not let the suspension sit too long before adding to the experimental wells. Clumping recurs rapidly. The particle per cell density may need to be optimized depending on the source, treatment, and density of MΦ used. In the studies presented, conjugated zymosan particles were used. However, conjugated E. coli and S. aureus are also available.
- While waiting for the system temperature to reach 37 °C, resuspend 1 mg of zymosan particles in 5 mL of the live-cell imaging medium.
- Add zymosan and read the plate
- Aspirate the medium from the experimental wells and rinse 1x with 100 µL of the live-cell imaging solution. Rinse the RBL wells with the live-cell imaging solution as well.
- Aspirate the live-cell imaging solution from experimental and RBL wells and replace it with 100 µL of the zymosan particle suspension.
- Open the plate tray of the fluorescent reader using the touchpad interface. Set the plate, without the lid, in the tray with the A1 well in the upper-left corner. Close the tray using the touchpad and click on the green Read button in the top menu.
- When the read is complete, save the file to the appropriate folder and export the data in a spreadsheet format by clicking on File and selecting Export.
- Calculate the mean ± SEM relative fluorescence for each replicate group at each time point (10 min, 20 min, 30 min, etc.) in the spreadsheet (Supplementary File 1) and transfer the data to graphing software.
- Use a line graph format to present the data and apply two-way ANOVA (Time x Treatment/MSC as factors) followed by Tukey's multiple comparisons test to determine differences between individual groups.
NOTE: While the 96-well plate read is in progress, prepare the dynamic imaging plate for time-lapse image acquisition. Ensure that the dynamic imaging proceeds with the analysis of one experimental well at a time.
Table 1: Example of a 96-well plate design. CBL – Cell blank; MSC – seeded day 1; MΦ+ treated and MΦ untreated seeded day 2; RBL reagent blank added on assay day 3.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| A | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| B | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| C | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| D | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| E | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| F | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| G | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
| H | CBL | MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ | CBL | RBL |
Table 2: Example of a 4-well chamber slide design. MSC – plated day 1; MΦ+ treated and MΦ untreated plated day 2.
| 1 | 2 | 3 | 4 |
| MSC/MΦ | MSC/MΦ+ | MΦ | MΦ+ |
Representative Results
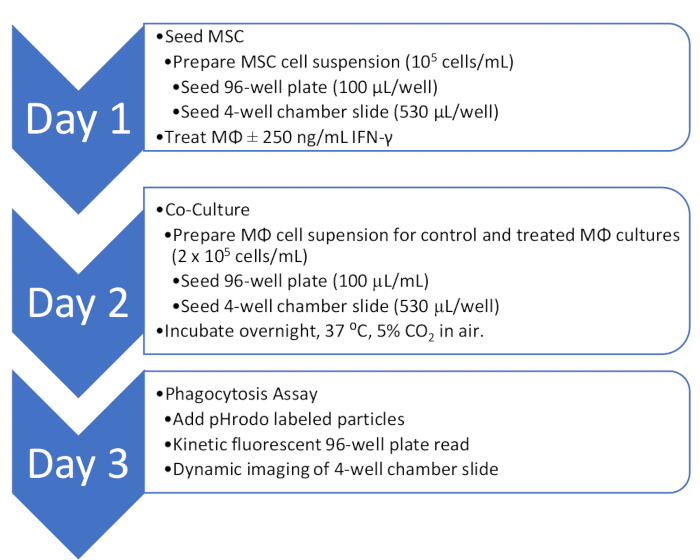
Figure 1: Overview of the co-culture and phagocytosis assay workflow. An outline of the workflow for quantitative and qualitative analysis of MΦ phagocytosis activity in co-culture with MSC.
Supplementary File 1: A representative spreadsheet file of raw kinetic data from an optimal experiment. Please click here to download this File.
Disclosures
The authors have nothing to disclose.
Materials
| 96-well Black Polystyrene Microplate | MilliporeSigma | CLS3603-48EA | |
| 0.4% trypan blue solution | MilliporeSigma | T8154-20ML | |
| 15 mL and 50 mL Conical Sterile Polypropylene Centrifuge Tubes | ThermoFisher | 339653 | |
| Antibiotic-Antimycotic (100X) Gibco | ThermoFisher | 15240096 | |
| Cell lifter | MilliporeSigma | CLS3008-100EA | |
| Culture flasks, tissue culture treated, surface area 75 cm2, canted neck, with cap, filtered | MilliporeSigma | C7231-120EA | |
| D1 ORL UVA [D1] | ATCC | CRL-12424 | Mouse MSC Cell Line |
| DMEM, High Glucose | ThermoFisher | 11965092 | |
| Fetal Bovine Serum, qualified, heat-inactivated | ThermoFisher | 16140071 | |
| Hemocytometer | FisherScientific | 02-671-51B | |
| I-11.15 | ATCC | CRL-2470 | Mouse MΦ Cell Line |
| LADMAC Cell Line | ATCC | CRL-2420 | LADMAC cells secrete the growth factor colony stimulating factor 1 (CSF-1). |
| Live-Cell Imaging solution | ThermoFisher | A14291DJ | |
| PBS, pH 7.4 | ThermoFisher | 10010031 | |
| pH Rhodo Green Zymosan Bioparticles Conjugate | ThermoFisher | P35365 | |
| Spectramax i3X | Molecular Devices | ||
| Sterile Single-Use Vacuum Filter Units, 250 mL, 0.2 µm | ThermoFisher | 568-0020 | |
| Sterile syringe filters, 0.2 micrometer | ThermoFisher | 723-2520 | |
| Tissue-culture-treated culture dishes, 100 mm x 20 mm | MilliporeSigma | CLS430167-100EA | |
| Trypsin-EDTA (0.05%), phenol red | ThermoFisher | 25300054 |

