Real-time Imaging of Leukotriene B4 Mediated Cell Migration and BLT1 Interactions with β-arrestin
Summary
This paper describes the methodology to determine the chemotactic response of leukocytes to specific ligands and identify interactions between the cell surface receptors and cytosolic proteins using live cell imaging techniques.
Abstract
G-protein coupled receptors (GPCRs) belong to the seven transmembrane protein family and mediate the transduction of extracellular signals to intracellular responses. GPCRs control diverse biological functions such as chemotaxis, intracellular calcium release, gene regulation in a ligand dependent manner via heterotrimeric G-proteins1-2. Ligand binding induces a series of conformational changes leading to activation of heterotrimeric G-proteins that modulate levels of second messengers such as cyclic adenosine monophosphate (cAMP), inositol triphosphate (IP3) and diacyl glycerol (DG). Concomitant with activation of the receptor ligand binding also initiates a series of events to attenuate the receptor signaling via desensitization, sequestration and/or internalization. The desensitization process of GPCRs occurs via receptor phosphorylation by G-protein receptor kinases (GRKs) and subsequent binding of β-arrestins3. β-arrestins are cytosolic proteins and translocate to membrane upon GPCR activation, binding to phosphorylated receptors (most cases) there by facilitating receptor internalization 4-6.
Leukotriene B4 (LTB4) is a pro-inflammatory lipid molecule derived from arachidonic acid pathway and mediates its actions via GPCRs, LTB4 receptor 1 (BLT1; a high affinity receptor) and LTB4 receptor 2 (BLT2; a low affinity receptor)7-9. The LTB4-BLT1 pathway has been shown to be critical in several inflammatory diseases including, asthma, arthritis and atherosclerosis10-17. The current paper describes the methodologies developed to monitor LTB4-induced leukocyte migration and the interactions of BLT1 with β-arrestin and , receptor translocation in live cells using microscopy imaging techniques18-19.
Bone marrow derived dendritic cells from C57BL/6 mice were isolated and cultured as previously described 20-21. These cells were tested in live cell imaging methods to demonstrate LTB4 induced cell migration. The human BLT1 was tagged with red fluorescent protein (BLT1-RFP) at C-terminus and β-arrestin1 tagged with green fluorescent protein (β-arr-GFP) and transfected the both plasmids into Rat Basophilic Leukomia (RBL-2H3) cell lines18-19. The kinetics of interaction between these proteins and localization were monitored using live cell video microscopy. The methodologies in the current paper describe the use of microscopic techniques to investigate the functional responses of G-protein coupled receptors in live cells. The current paper also describes the use of Metamorph software to quantify the fluorescence intensities to determine the kinetics of receptor and cytosolic protein interactions.
Protocol
Methodology
Description of Microscope
Live cell imaging experiments performed using TE-FM Epi-Fluorescence system attached to Nikon Inverted Microscope Eclipse TE300. The microscope equipped with heating stage. A cool snap HQ digital B/W CCD (Roper Scientific) camera and LAMDA 10-2 optical filter changer (Sutter instrument company) is attached to microscope. Excitation and emission wavelengths are controlled with filter wheels and controlled by Lamba 10-2 filter wheel controller, Sutter Instruments Co. Exposure time 500 ms should be enough to view RFP or GFP in live cells. Hardware control and acquisition of images are controlled by Metamorph software. The choice of the filters for the present study are, filter sets S480/20x, S525/40m and S565/25x, S620/60m for GFP and RFP, respectively; EGFP/DsRed dual dichroic, Chroma Technology. This filter wheel can accommodate up to six filter sets. Microscope is attached with Prior Proscan stage controller with Joy stick. All these hardware attachments can be controlled by Metamorph software. The microscope is also attached with Micromanipulators to hold the micropipettes.
A. Measuring Leukotriene B4 Directed Dendritic Cell Migration
Using the above described microscope setting, we developed methods for following ligand directed dendritic cell migration in real time. Two micromanipulators (Narishige) are attached to the microscope. The chemotaxis of mouse bone marrow derived dendritic cells (BMDCs) towards LTB4 gradient was recorded. Here, we describe the methods to prepare the micro-pipettes using micro-pipette puller, loading the ligand into micro-pipette and setting up the chemotaxis experiment on the heated stage of the microscope to monitor directional migration of cells. This method can be applied to test the efficacy of distinct chemoattractants in inducing migration as well as effect of inhibitors of chemotaxis in live cells. The isolation of BMDCs21 from mouse has been described in several publications including JoVE22.
1. Preparation of BMDCs
- Plate the BMDCs (after culturing for 10 days) on to the cover slip bottom Fluoro dish and allow them to grow for 16 hr ahead of the experiment.
- Wash the cells with 3 mL of 1X PBS at least 2 times (by adding 3 mL of 1X PBS to the plate and aspirating the buffer out).
- Add 2 mL of 1XPBS to plate. The cells are now ready for the experiment. Keep the cells in 37 °C incubator prior to experiment.
Preparation of ligand loaded micro pipette:
- Fix the glass capillary tubes (Glass standard, 0.75 mm x 0.4 mm; 6″, Cat# 625500, A.M. Systems, INC) on the vertical micropipette puller (Narishige International USA, Inc).
- Keep the temperature at 52 °C and allow the glass to melt and pull apart to form sharp edge tips.
- Fix the pipette onto the micro-pipette holder.
- Smooth the rough edges of micro pipette using Micro forge M-900 (Narishige International USA, Inc) by watching under microscope.
- Prepare at least 10-20 pipettes for each experiment. Note: The micropipettes can also be purchased from World Precision Instruments (μ TIP, T1801TW1F).
- Take desired concentration (500 μL of 100 nM LTB4 in the current experiment) of ligand into 1 mL effendorf tube and keep the micro pipette tip in the solution and allow the solution to enter into the micro pipette (back filling).
- Take 1 mL of ligand and into the syringe (1 cc tuberculin syringe) and slowly fill the ligand using Micro-Fill needle (MF 28G-5, World Precision Instruments) into micro pipette from top.
- Attach the tubing, which fits tightly to the pipette to pipette and tube to needle (21 G 11/2) attached to 1 mL syringe, which is filled with ligand.
- Now, carefully move the ligand filled micro-pipette to the microscope stage and fix onto micro manipulators.
- Apply little pressure from syringe to make sure ligand is slowly releasing from the tip.
- Bring the plated immature BMDCs to the microscope and keep them on the heated plate (37 °C) stage.
- Remove the lid of fluoro dish.
- Focus the cells using oil immersion 60X objective lens.
- Carefully, bring down the ligand loaded pipette using ‘Coarse Manual Manipulator MN-188NE’ to the proximity of the cells.
- Focus the pipette using ‘Hydraulic Fine Micromanipulator MN-188 NE’.
- Apply little pressure from syringe to make sure that ligand is releasing into dish.
- Set the image acquisition parameters to acquire in bright filed (10 ms exposure) in every 15 sec for 2 hrs using Metamorph software.
- Keep monitoring every 30 min for progression of migration towards ligand.
- If cells are crowded at pipette tip, change the location of the tip in the plate to continue the migration of experiment.
B. Measuring GPCR (BLT1) and Cytosolic Protein (β-arrestin) Interactions, Translocation in Live Cells
1. Preparation of Plasmid DNA Constructs
The details of DNA plasmid constructs were described in Jala et al. (2005)18.
2. Transfection of Human BLT1-RFP and β-Arrestin-GFP into RBL-2H3 Cells
- Maintain the Rat Basophilic Leukomia (RBL-2H3) Cells (60 to 70% confluence) at 37 °C in a humidified atmosphere of 95% air, 5% CO2 as monolayer cultures in growth media (DMEM supplemented with 15% FBS, 2 mM L-glutamine, 100 U/ mL penicillin and 100 μg/ mL streptomycin) in T75 flasks.
- Detach the cells from the dish/flasks by using 6 mL of trypsin-EDTA (0.05% trypsin, 0.53 mM EDTA) and incubating for 5 min at 37 °C in a humidified atmosphere of 95% air, 5% CO2. Gently shake the flasks to monitor the extent of detachment of the cells.
- Add 6 mL of growth media to flask containing 6 mL of Trypsin-EDTA and collect the cells by mixing them by pipetting up and down multiple times. Transfer the cells into 15 mL falcon tubes.
- Count the cells using a hemocytometer and take 4 x 106 cells in 15 mL centrifuge tubes and centrifuge at 480g rpm for 3 min and resuspend in 200 μL of transfection media (DMEM, 20% FBS, 50 mM HEPES). If you plan to perform 3 transfections, prepare the cells and tranfection media accordingly by increasing cell number and the volume of transfection medium.
- Prepare the plasmids to be transfected at the concentrations of at least 1.5 μg/μL. We prepare the DNA plasmid by using QIAGEN Maxi DNA plasmid kit. Add the individual or both plasmid DNAs encoding for human BLT1-RFP (25 μg) and β-arrestin1-GFP (15 μg) to sterile electrophoresis cuvettes, (Bio-Rad # 16552088), which has 0.4 cm electrode gap.
- Take 200 μL of the above resuspended cells and place them in cuvettes containing either hBLT1-RFP or β-arrestin1-GFP or both together and mix them gently with 1 mL sterile pipette.
- Leave the above cuvettes at room temperature for 10 min.
- Electroporate the cells in Gene Pulser II with voltage 250 v and capacity 500 μF.
- After electroporation let the cells stay at room temperature for 10 min.
- Take 1 mL of regular growth medium and mix it with electroporated cells in the cuvettes.
- Distribute 300 μL of this mixture into 35 mm tissue glass bottom culture dishes (Fluoro dish, World Precision Instruments) containing 2 mL of regular growth media and incubate for 1 hr at 37 °C in a humidified atmosphere of 95% air, 5% CO2. Allow the cells to adhere to bottom of the dish.
- Replace the media after 1 h of incubation with regular growth media and allow them grow at 37 °C in a humidified atmosphere of 95% air, 5% CO2 incubator for 18-24 hrs.
3. Live Cell Imaging
Acquisition of images
- After 18-24 hrs, wash the cells 2 or 3 times with 2 mL of warm phenol red free RPMI containing 10 mM HEPES, pH 7.55 (Add 2 mL of warm media directly to plate and aspirate media. Repeat 2 or 3 times). Add 1.8 mL of warm phenol red free RPMI containing 10 mM HEPES.
- Place 30 mm glass coverslip-bottomed culture dishes containing RBL-2H3 cells, transfected with hBLT1-RFP and β-arrestin1-GFP on the heated microscope stage (37 °C).
- Collect the cell images at 600x magnification using 60 X objective Nikon Plan Apo 60X/1.4 numerical aperture oil immersion lens (available in our microscope system).
- Put one oil drop on the 60 x objective lense and focus the cells with regular bright field transmitted light by touching the bottom of the plate.
- To monitor the fluorescence of transfected proteins, switch from bright field (transmitted light) to fluorescence filter (RFP filter, if you want to see receptor or GFP, if you want to see β-arrestin1).
- Choose the bright and healthy cell, which expresses hBLT1-RFP on the cell surface using RFP filter and capture the image.
- Then shift to GFP filter and make sure that β-arrestin-GFP expressed in the cytoplasm and capture the image. Then pseudo color both the images, before capturing time lapsed live cell images to make sure that bleed through of fluorescence is not taking occurring between RFP and GFP.
- Once the choice of your cell made, set the parameters according to your purpose using software (Metamorph Software in this instance).
- In the current demonstration, sixteen bit images are acquired with the camera binning set to 1 X 1 combined with 60 X objective Nikon Plan Apo 60X/1.4 numerical aperture oil immersion lens.
- Collect the fluorescence images for RFP and GFP fluorochromes simultaneously at 30 sec time interval (if the translocation/interactions are too fast, one can reduce the time intervals) of these using filter wheels controlled by Metamorph software.
- The Camera exposure times set to 1000 msec for RFP and GFP. (Set the specific exposure times according to the fluorescent intensity of RFP and GFP).
- First collect the images for 1 min without adding ligand, which serves as 0 time point.
- Add the 200 μL of ligand (from stock of 10 μM to a final concentration of 1 μM) after 1 min without disturbing the plate/or cell position.
- The fluorescence images are collected for 60 min after adding the ligand and stored as TIFF images with increasing order of the file names.
- All these files can be made as stack files with individual/combined fluorescence images with time stamps using the software.
Process of images and quantification of fluorescence of proteins and localization
- Acquire the images with plain media (no cells) with RFP and GFP fluorescent filters and save them as background images. Use these images to subtract from actual data images obtained above.
- Prepare the stack of images individually for RFP and GFP images.
- Select/define the various regions (surface vs cytosolic) to measure the fluorescence intensity of RFP and GFP (BLT1 and β-arrestin translocation)
- Plot the amount fluorescence intensities as the function of time. This graph will provide the information about kinetics of translocation of given molecule.
Representative Results
A. Measuring Leukotriene B4 Directed Dendritic Cell Migration
The method described in this protocol was used to determine the dendritic cell migration towards leukotriene B4 (Figure 1 and attached video 1)21. We can apply similar methodology to determine the chemotactic capacity of cells to a particular ligand in live cells.
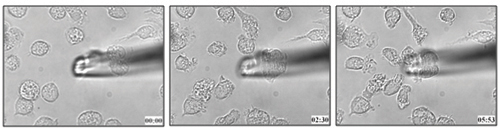
Figure 1. Mouse bone marrow derived dendritic cell migration towards 100 nM LTB4.
Video 1. Live cell migration of BMDCs towards LTB4. Click here to see video.
B. Measuring GPCR (BLT1) and Cytosolic Protein (β-arrestin) Interactions, Translocation in Live Cells
Upon completion of this procedure, one can obtain the following information in GPCR signaling events.
- The localization of GPCR and cytosolic proteins and nucleus (Figure 2).
- Ligand induced interaction of surface GPCR (BLT1 in this case) with cytosolic protein (β-arrestin in this case) (Fig 3).
- Kinetics of receptor internalization and β-arrestin translocation to membrane followed by internalization along with receptors (Figure 4) (Jala et al. 2005)18.
- One can determine the ligand dependent receptor activation status in live cells (attached video 2) and determine critical motifs or processes involved in the receptor activation (Jala et al. 2005)18.
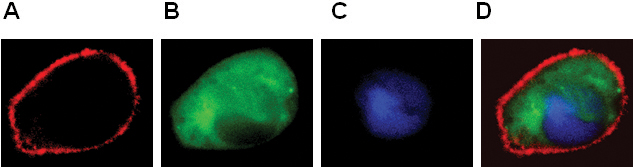
Figure 2. Expression of BLT1-RFP (A), β-arrestin-GFP (B), pNuc-CFP (C) into RBL-2H3 cells. Color combined image shown in panel D.
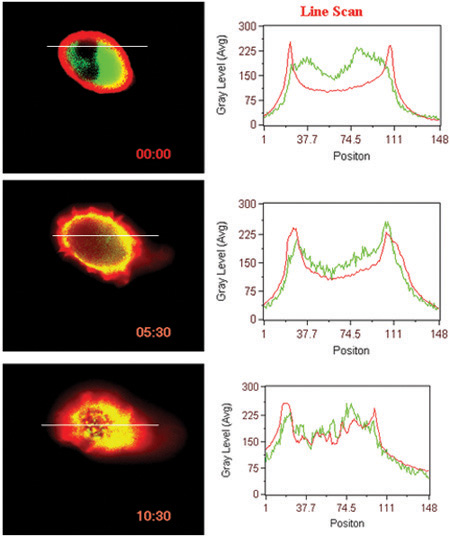
Figure 3. Ligand induced translocation of receptor and β-arrestin. Line scan of fluorescent intensities in the cells are shown.
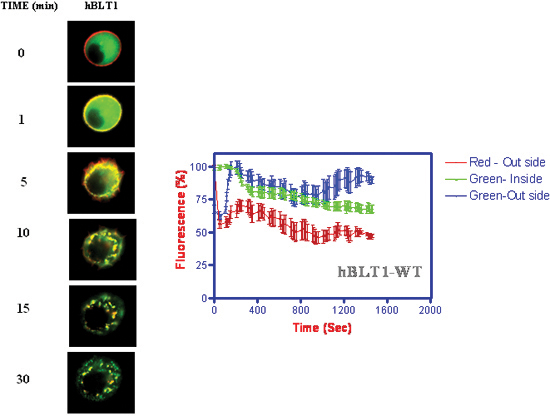
Figure 4. Kinetics of receptor internalization and β-arrestin translocation upon addition of 1 μM LTB4. The fluorescence intensities were measured as a function of time at membrane and cytosolic locations of the cell.
Video 2 Live imaging of cells expressing BLT1-RFP and β-arrestin-GFP upon the addition of 1 μM LTB4. Click here to watch video
Discussion
Live cell imaging is a powerful tool to demonstrate the function and interactions of specific proteins as they occur in real-time. The methods described in this manuscript clearly show that LTB4 can induce rapid migration of dendritic cells. These methods not only expand the aspects of LTB4 function to diverse cell types, they allow similar methods to be applied to a variety of other chemokines and testing their efficacy as chemotactic agents on different leukocyte sub populations. Fluorescence imaging in this system allows for monitoring signaling events in real-time. Furthermore, these methods may also be extended to examine the tight association of molecules in vivo by following fluorescence resonance energy transfer (FRET) using appropriate flurochromes tagged molecules.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The research is supported by National Institutes of Health grants AI-52381, CA138623 and Kentucky Lung Cancer Research Board and institutional support from James Graham Brown Cancer Center.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Cell lines: | ||||
| Rat Basophilic Leukomia Cell line (RBL-2H3) or HEK293 cells. | ATCC | CRL-2256 | ||
| Media: | ||||
| Delbecco’s modified Eagle’s Medium (DMEM) | Invitrogen | 11995 | ||
| Phenol red free RPMI or DMEM | Invitrogen | 11835-030 | ||
| Fetal Bovine Serum | Invitrogen | 16000-044 | ||
| L-Glutamine (200 mM) | Invitrogen | 25030 | ||
| Penicillin-streptomycin (10000 U/mL) | Invitrogen | 15140 | ||
| Trypsin, 0.05% (1X) with EDTA 4Na, liquid | Invitrogen | 25300 | ||
| HEPES (1M) | Invitrogen | 15630 | ||
| Others: | ||||
| 35 mm sterile glass coverslip-bottomed Fluoro dishes (0.17 mm thick) (WillCo-dish) | WPI | FD35-100 | ||
| Sterile Gene Pulser Cuvette (0.4 cm electrode gap) (Bio-Rad) | Bio-Rad | 16552088 | ||
| Instruments/software: | ||||
| Gene Pulser II electroporater | Bio-Rad | |||
| TE-FM Epi-Fluorescence system attached to Nikon Inverted Microscope Eclipse TE300 | Nikon | |||
| Metamorph Software | Universal Imaging | |||
| Vertical Micro-pipette puller | Narishige International | |||
| Micro-Forge M-900 | Narishige International | |||
| Hadraulic Micromanipulator MO-188NE | Narishige International | |||
| Coarse Manual Manipulator, MN-188NE | Narishige International | |||
| cDNA constructs: | ||||
| cDNA of G-Protein coupled receptor tagged with red fluorescence protein at C-terminus (hBLT1-RFP) | Jala et al 2005 | |||
| cDNA of cytosolic protein tagged with GFP (β-arrestin1-GFP in present study). | Jala et al 2005 |
References
- Wess, J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 11, 346-354 (1997).
- Gether, U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 21, 90-113 (2000).
- Pierce, K. L., Premont, R. T., Lefkowitz, R. J. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 3, 639-650 (2002).
- Lefkowitz, R. J. G. protein-coupled receptors. III. New roles for receptor kinases and beta-arrestins in receptor signaling and desensitization. J Biol Chem. 273, 18677-18680 (1998).
- Shenoy, S. K., Lefkowitz, R. J. Multifaceted roles of beta-arrestins in the regulation of seven-membrane-spanning receptor trafficking and signalling. Biochem J. 375, 503-515 (2003).
- . Beta-arrest or. Nature. 383, 447-450 (1996).
- Serhan, C. N., Haeggstrom, J. Z., Leslie, C. C. Lipid mediator networks in cell signaling: update and impact of cytokines. Faseb J. 10, 1147-1158 (1996).
- Tager, A. M., Luster, A. D. BLT1 and BLT2: the leukotriene B(4) receptors. Prostaglandins Leukot Essent Fatty Acids. 69, 123-134 (2003).
- Toda, A., Yokomizo, T., Shimizu, T. Leukotriene B4 receptors. Prostaglandins Other Lipid Mediat. 68-69, 575-585 (2002).
- Haribabu, B. Targeted disruption of the leukotriene B(4) receptor in mice reveals its role in inflammation and platelet-activating factor-induced anaphylaxis. J Exp Med. 192, 433-438 (2000).
- Subbarao, K. Role of leukotriene B4 receptors in the development of atherosclerosis: potential mechanisms. Arterioscler Thromb Vasc Biol. 24, 369-375 (2004).
- Jala, V. R., Haribabu, B. Leukotrienes and atherosclerosis: new roles for old mediators. Trends Immunol. 25, 315-322 (2004).
- Heller, E. A. Inhibition of atherogenesis in BLT1-deficient mice reveals a role for LTB4 and BLT1 in smooth muscle cell recruitment. Circulation. 112, 578-586 (2005).
- Miyahara, N. Requirement for leukotriene B4 receptor 1 in allergen-induced airway hyperresponsiveness. Am J Respir Crit Care Med. 172, 161-167 (2005).
- Terawaki, K. Absence of leukotriene B4 receptor 1 confers resistance to airway hyperresponsiveness and Th2-type immune responses. J Immunol. 175, 4217-4225 (2005).
- Shao, W. H., Del Prete, A., Bock, C. B., Haribabu, B. Targeted disruption of leukotriene B4 receptors BLT1 and BLT2: a critical role for BLT1 in collagen-induced arthritis in mice. J Immunol. 176, 6254-6261 (2006).
- Kim, N. D., Chou, R. C., Seung, E., Tager, A. M., Luster, A. D. A unique requirement for the leukotriene B4 receptor BLT1 for neutrophil recruitment in inflammatory arthritis. J Exp Med. 203, 829-835 (2006).
- Jala, V. R., Shao, W. H., Haribabu, B. Phosphorylation-independent beta-arrestin translocation and internalization of leukotriene B4 receptors. J Biol Chem. 280, 4880-4887 (2005).
- Jala, V. R., Haribabu, B. Real-time analysis of G protein-coupled receptor signaling in live cells. Methods Mol Biol. 332, 159-165 (2006).
- Del Prete, A., A, . Regulation of dendritic cell migration and adaptive immune response by leukotriene B4 receptors: a role for LTB4 in up-regulation of CCR7 expression and function. Blood. 109, 626-631 (2007).
- Salogni, L. Activin A induces dendritic cell migration through the polarized release of CXC chemokine ligands 12 and 14. Blood. 113, 5848-5856 (2009).
- Boudreau, J., Koshy, S., Cummings, D., Wan, Y. Culture of myeloid dendritic cells from bone marrow precursors. J Vis Exp. , (2008).

