Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies
Summary
PCR has emerged as a common technique in many molecular biology laboratories. Provided here is a quick guide to several conventional PCR protocols. Because each reaction is a unique experiment, optimal conditions required to generate a product vary. Understanding the variables in a reaction will greatly enhance troubleshooting efficiency, thereby increasing the chance to obtain the desired result.
Abstract
In the biological sciences there have been technological advances that catapult the discipline into golden ages of discovery. For example, the field of microbiology was transformed with the advent of Anton van Leeuwenhoek's microscope, which allowed scientists to visualize prokaryotes for the first time. The development of the polymerase chain reaction (PCR) is one of those innovations that changed the course of molecular science with its impact spanning countless subdisciplines in biology. The theoretical process was outlined by Keppe and coworkers in 1971; however, it was another 14 years until the complete PCR procedure was described and experimentally applied by Kary Mullis while at Cetus Corporation in 1985. Automation and refinement of this technique progressed with the introduction of a thermal stable DNA polymerase from the bacterium Thermus aquaticus, consequently the name Taq DNA polymerase.
PCR is a powerful amplification technique that can generate an ample supply of a specific segment of DNA (i.e., an amplicon) from only a small amount of starting material (i.e., DNA template or target sequence). While straightforward and generally trouble-free, there are pitfalls that complicate the reaction producing spurious results. When PCR fails it can lead to many non-specific DNA products of varying sizes that appear as a ladder or smear of bands on agarose gels. Sometimes no products form at all. Another potential problem occurs when mutations are unintentionally introduced in the amplicons, resulting in a heterogeneous population of PCR products. PCR failures can become frustrating unless patience and careful troubleshooting are employed to sort out and solve the problem(s). This protocol outlines the basic principles of PCR, provides a methodology that will result in amplification of most target sequences, and presents strategies for optimizing a reaction. By following this PCR guide, students should be able to:
● Set up reactions and thermal cycling conditions for a conventional PCR experiment
● Understand the function of various reaction components and their overall effect on a PCR experiment
● Design and optimize a PCR experiment for any DNA template
● Troubleshoot failed PCR experiments
Protocol
1. Designing Primers
Designing appropriate primers is essential to the successful outcome of a PCR experiment. When designing a set of primers to a specific region of DNA desired for amplification, one primer should anneal to the plus strand, which by convention is oriented in the 5′ → 3′ direction (also known as the sense or nontemplate strand) and the other primer should complement the minus strand, which is oriented in the 3′ → 5′ direction (antisense or template strand). There are a few common problems that arise when designing primers: 1) self-annealing of primers resulting in formation of secondary structures such as hairpin loops (Figure 1a); 2) primer annealing to each other, rather then the DNA template, creating primer dimers (Figure 1b); 3) drastically different melting temperatures (Tm) for each primer, making it difficult to select an annealing temperature that will allow both primers to efficiently bind to their target sequence during themal cycling (Figure 1c) (See the sections CALCULATING MELTING TEMPERATURE (Tm) and MODIFICATIONS TO CYCLING CONDITIONS for more information on Tms).
- Below is a list of characteristics that should be considered when designing primers.
- Primer length should be 15-30 nucleotide residues (bases).
- Optimal G-C content should range between 40-60%.
- The 3′ end of primers should contain a G or C in order to clamp the primer and prevent “breathing” of ends, increasing priming efficiency. DNA “breathing” occurs when ends do not stay annealed but fray or split apart. The three hydrogen bonds in GC pairs help prevent breathing but also increase the melting temperature of the primers.
- The 3′ ends of a primer set, which includes a plus strand primer and a minus strand primer, should not be complementary to each other, nor can the 3′ end of a single primer be complementary to other sequences in the primer. These two scenarios result in formation of primer dimers and hairpin loop structures, respectively.
- Optimal melting temperatures (Tm) for primers range between 52-58 °C, although the range can be expanded to 45-65 °C. The final Tm for both primers should differ by no more than 5 °C.
- Di-nucleotide repeats (e.g., GCGCGCGCGC or ATATATATAT) or single base runs (e.g., AAAAA or CCCCC) should be avoided as they can cause slipping along the primed segment of DNA and or hairpin loop structures to form. If unavoidable due to nature of the DNA template, then only include repeats or single base runs with a maximum of 4 bases.
Notes:
- There are many computer programs designed to aid in designing primer pairs. NCBI Primer design tool http://www.ncbi.nlm.nih.gov/tools/primer-blast/ and Primer3 http://frodo.wi.mit.edu/primer3/ are recommended websites for this purpose.
- In order to avoid amplification of related pseudogenes or homologs it could be useful to run a blast on NCBI to check for the target specificity of the primers.
2. Materials and Reagents
- When setting up a PCR experiment, it is important to be prepared. Wear gloves to avoid contaminating the reaction mixture or reagents. Include a negative control, and if possible a positive control.
- Arrange all reagents needed for the PCR experiment in a freshly filled ice bucket, and let them thaw completely before setting up a reaction (Figure 2). Keep the reagents on ice throughout the experiment.
- Standard PCR reagents include a set of appropriate primers for the desired target gene or DNA segment to be amplified, DNA polymerase, a buffer for the specific DNA polymerase, deoxynucleotides (dNTPs), DNA template, and sterile water.
- Additional reagents may include Magnesium salt Mg2+ (at a final concentration of 0.5 to 5.0 mM), Potassium salt K+ (at a final concentration of 35 to 100 mM), dimethylsulfoxide (DMSO; at a final concentration of 1-10%), formamide (at a final concentration of 1.25-10%), bovine serum albumin (at a final concentration of 10-100 μg/ml), and Betaine (at a final concentration of 0.5 M to 2.5 M). Additives are discussed further in the trouble shooting section.
- Organize laboratory equipment on the workbench.
- Materials include PCR tubes and caps, a PCR tube rack, an ethanol-resistant marker, and a set of micropipettors that dispense between 1 – 10 μl (P10), 2 – 20 μl (P20), 20 – 200 μl (P200) and 200 – 1000 μl (P1000), as well as a thermal cycler.
- When setting up several PCR experiments that all use the same reagents, they can be scaled appropriately and combined together in a master mixture (Master Mix). This step can be done in a sterile 1.8 ml microcentrifuge tube (see Notes).
- To analyze the amplicons resulting from a PCR experiment, reagents and equipment for agarose gel electrophoresis is required. To approximate the size of a PCR product, an appropriate, commercially available molecular weight size standard is needed.
3. Setting up a Reaction Mixture
- Start by making a table of reagents that will be added to the reaction mixture (see Table 1).
- Next, label PCR tube(s) with the ethanol-resistant marker.
- Reaction volumes will vary depending on the concentrations of the stock reagents. The final concentrations (CF) for a typical 50 μl reaction are as follows.
- X buffer (usually supplied by the manufacturer of the DNA polymerase; may contain 15 mM MgCl2). Add 5 μl of 10X buffer per reaction.
- 200 μM dNTPs (50 μM of each of the four nucleotides). Add 1 μl of 10 mM dNTPs per reaction (dATP, dCTP, dTTP and dGTP are at 2.5 mM each).
- 1.5 mM Mg2+. Add only if it is not present in the 10X buffer or as needed for PCR optimization. For example, to obtain the 4.0 mM Mg2+ required for optimal amplicon production of a conserved 566 bp DNA segment found in an uncharacterized Mycobacteriophage add 8 μl of 25 mM MgCl2 to the reaction (Figure 3).
- 20 to 50 pmol of each primer. Add 1 μl of each 20 μM primer.
- Add 104 to 107 molecules (or about 1 to 1000 ng) DNA template. Add 0.5 μl of 2ng/μl genomic Mycobacteriophage DNA.
- Add 0.5 to 2.5 units of DNA polymerase per 50 μl reaction (See manufacturers recommendations) For example, add 0.5 μl of Sigma 0.5 Units/μl Taq DNA polymerase.
- Add Q.S. sterile distilled water to obtain a 50 μl final volume per reaction as pre-determined in the table of reagents (Q.S. is a Latin abbreviation for quantum satis meaning the amount that is needed). Thus, 33 μl per reaction is required to bring the volume up to 50 μl. However, it should be noted that water is added first but requires initially making a table of reagents and determining the volumes of all other reagents added to the reaction.
4. Basic PCR Protocol
- Place a 96 well plate into the ice bucket as a holder for the 0.2 ml thin walled PCR tubes. Allowing PCR reagents to be added into cold 0.2 ml thin walled PCR tubes will help prevent nuclease activity and nonspecific priming.
- Pipette the following PCR reagents in the following order into a 0.2 ml thin walled PCR tube (Figure 4): Sterile Water, 10X PCR buffer, dNTPs, MgCl2, primers, and template DNA (See Table 1). Since experiments should have at least a negative control, and possibly a positive control, it is beneficial to set up a Master Mix in a 1.8 ml microcentrifuge tube (See explanation in Notes).
- In a separate 0.2 ml thin walled PCR tubes (Figure 4) add all the reagents with the exception of template DNA for a negative control (increase the water to compensate for the missing volume). In addition, another reaction (if reagents are available) should contain a positive control using template DNA and or primers previously known to amplify under the same conditions as the experimental PCR tubes.
- Taq DNA polymerase is typically stored in a 50% glycerol solution and for complete dispersal in the reaction mix requires gentle mixing of the PCR reagents by pipetting up and down at least 20 times. The micropipettor should be set to about half the reaction volume of the master mix when mixing, and care should be taken to avoid introducing bubbles.
- Put caps on the 0.2 ml thin walled PCR tubes and place them into the thermal cycler (Figure 5). Once the lid to the thermal cycler is firmly closed start the program (see Table 2).
- When the program has finished, the 0.2 ml thin walled PCR tubes may be removed and stored at 4 °C. PCR products can be detected by loading aliquots of each reaction into wells of an agarose gel then staining DNA that has migrated into the gel following electrophoresis with ethidium bromide. If a PCR product is present, the ethidium bromide will intercalate between the bases of the DNA strands, allowing bands to be visualized with a UV illuminator.
Notes:
- When setting up multiple PCR experiments, it is advantageous to assemble a mixture of reagents common to all reactions (i.e., Master Mix). Usually the cocktail contains a solution of DNA polymerase, dNTPs, reaction buffer, and water assembled into a 1.8 ml microcentrifuge tube. The amount of each reagent added to the Master Mix is equivalent to the total number of reactions plus 10% rounded up to the nearest whole reaction. For instance, if there are 10 x 0.1 = 1 reaction, then (10 + 1) x 5 μl 10X buffer equals 55 μl of 10X buffer for the Master Mix. The reagents in the Master Mix are mixed thoroughly by gently pumping the plunger of a micropipettor up and down about 20 times as described above. Each PCR tube receives an aliquot of the Master Mix to which the DNA template, any required primers, and experiment-specific reagents are then added (see Tables 1 and 7).
- The following website offers a calculator for determining the number of copies of a template DNA (http://www.uri.edu/research/gsc/resources/cndna.html). The total number of copies of double stranded DNA may be calculated using the following equation:
Number of copies of DNA = (DNA amount (ng) x 6.022×1023) / (length of DNA x 1×109 ng/ml x 650 Daltons)
Calculating the number of copies of DNA is used to determine how much template is needed per reaction. - False positives may occur as a consequence of carry-over from another PCR reaction which would be visualized as multiple undesired products on an agarose gel after electrophoresis. Therefore, it is prudent to use proper technique, include a negative control (and positive control when possible).
- While ethidium bromide is the most common stain for nucleic acids there are several safer and less toxic alternatives. The following website describes several of the alternatives including Methylene Blue, Crystal Violet, SYBR Safe, and Gel Red along with descriptions of how to use and detect the final product (http://bitesizebio.com/articles/ethidium-bromide-the-alternatives/).
- While most modern PCR machines use 0.2 ml tubes, some models may require reactions in 0.5 ml tubes. See your thermal cyclers manual to determine the appropriate size tube.
5. Calculating Melting Temperature (Tm)
- Knowing the melting temperature (Tm) of the primers is imperative for a successful PCR experiment. Although there are several Tm calculators available, it is important to note that these calculations are an estimate of the actual Tm due to lack of specific information about a particular reaction and assumptions made in the algorithms for the Tm calculators themselves. However, nearest-neighbor thermodynamic models are preferred over the more conventional calculation: Tm ≈ 4(G-C) + 2(A-T). The former will give more accurate Tm estimation because it takes into account the stacking energy of neighboring base pairs. The latter is used more frequently because the calculations are simple and can be done quickly by hand. See Troubleshooting section for information about how various PCR conditions and additives affect melting temperature.
For calculating the Tm values by nearest-neighbor thermodynamic models, one of the following calculators is recommended:
http://www6.appliedbiosystems.com/support/techtools/calc/
http://www.cnr.berkeley.edu/~zimmer/oligoTMcalc.html
http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/
http://www.finnzymes.com/tm_determination.html
http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::melting
6. Setting Up Thermal Cycling Conditions
- PCR thermal cyclers rapidly heat and cool the reaction mixture, allowing for heat-induced denaturation of duplex DNA (strand separation), annealing of primers to the plus and minus strands of the DNA template, and elongation of the PCR product. Cycling times are calculated based on the size of the template and the GC content of the DNA. The general formula starts with an initial denaturation step at 94 °C to 98 °C depending on the optimal temperature for DNA polymerase activity and G-C content of the template DNA. A typical reaction will start with a one minute denaturation at 94 °C. Any longer than 3 minutes may inactivate the DNA polymerase, destroying its enzymatic activity. One method, known as hot-start PCR, drastically extends the initial denaturation time from 3 minutes up to 9 minutes. With hot-start PCR, the DNA polymerase is added after the initial exaggerated denaturation step is finished. This protocol modification avoids likely inactivation of the DNA polymerase enzyme. Refer to the Troubleshooting section of this protocol for more information about hot start PCR and other alternative methods.
- The next step is to set the thermal cycler to initiate the first of 25 to 35 rounds of a three-step temperature cycle (Table 2). While increasing the number of cycles above 35 will result in a greater quantity of PCR products, too many rounds often results in the enrichment of undesirable secondary products. The three temperature steps in a single cycle accomplish three tasks: the first step denatures the template (and in later cycles, the amplicons as well), the second step allows optimal annealing of primers, and the third step permits the DNA polymerase to bind to the DNA template and synthesize the PCR product. The duration and temperature of each step within a cycle may be altered to optimize production of the desired amplicon.
The time for the denaturation step is kept as short as possible. Usually 10 to 60 seconds is sufficient for most DNA templates. The denaturation time and temperature may vary depending on the G-C content of the template DNA, as well as the ramp rate, which is the time it takes the thermal cycler to change from one temperature to the next. The temperature for this step is usually the same as that used for the initial denaturation phase (step #1 above; e.g., 94 °C).
A 30 second annealing step follows within the cycle at a temperature set about 5 °C below the apparent Tm of the primers (ideally between 52 °C to 58 °C).
The cycle concludes with an elongation step. The temperature depends on the DNA polymerase selected for the experiment. For example, Taq DNA polymerase has an optimal elongation temperature of 70 °C to 80 °C and requires 1 minute to elongate the first 2 kb, then requires an extra minute for each additional 1 kb amplified. Pfu DNA Polymerase is another thermostable enzyme that has an optimal elongation temperature of 75 °C. Pfu DNA Polymerase is recommended for use in PCR and primer extension reactions that require high fidelity and requires 2 minutes for every 1 kb to be amplified. See manufacturer recommendations for exact elongation temperatures and elongation time indicated for each specific DNA polymerase. - The final phase of thermal cycling incorporates an extended elongation period of 5 minutes or longer. This last step allows synthesis of many uncompleted amplicons to finish and, in the case of Taq DNA polymerase, permits the addition of an adenine residue to the 3′ ends of all PCR products. This modification is mediated by the terminal transferase activity of Taq DNA polymerase and is useful for subsequent molecular cloning procedures that require a 3′-overhang.
- Termination of the reaction is achieved by chilling the mixture to 4 °C and/or by the addition of EDTA to a final concentration of 10 mM.
7. Important Considerations When Troubleshooting PCR
If standard PCR conditions do not yield the desired amplicon, PCR optimization is necessary to attain better results. The stringency of a reaction may be modulated such that the specificity is adjusted by altering variables (e.g., reagent concentrations, cycling conditions) that affect the outcome of the amplicon profile. For example, if the reaction is not stringent enough, many spurious amplicons will be generated with variable lengths. If the reaction is too stringent, no product will be produced. Troubleshooting PCR reactions may be a frustrating endeavor at times. However, careful analysis and a good understanding of the reagents used in a PCR experiment can reduce the amount of time and trials needed to obtain the desired results. Of all the considerations that impact PCR stringency, titration of Mg2+ and/or manipulating annealing temperatures likely will solve most problems. However, before changing anything, be sure that an erroneous result was not due to human error. Start by confirming all reagents were added to a given reaction and that the reagents were not contaminated. Also take note of the erroneous result, and ask the following questions: Are primer dimers visible on the gel after electrophoresis (these run as small bands <100 b near the bottom of the lane)? Are there non-specific products (bands that migrate at a different size than the desired product)? Was there a lack of any product? Is the target DNA on a plasmid or in a genomic DNA extract? Also, it is wise to analyze the G-C content of the desired amplicon.
- First determine if any of the PCR reagents are catastrophic to your reaction. This can be achieved by preparing new reagents (e.g., fresh working stocks, new dilutions), and then systematically adding one new reagent at a time to reaction mixtures. This process will determine which reagent was the culprit for the failed PCR experiment. In the case of very old DNA, which often accumulates inhibitors, it has been demonstrated that addition of bovine serum albumin may help alleviate the problem.
- Primer dimers can form when primers preferentially self anneal or anneal to the other primer in the reaction. If this occurs, a small product of less than 100 bp will appear on the agarose gel. Start by altering the ratio of template to primer; if the primer concentration is in extreme excess over the template concentration, then the primers will be more likely to anneal to themselves or each other over the DNA template. Adding DMSO and or using a hot start thermal cycling method may resolve the problem. In the end it may be necessary to design new primers.
- Non-specific products are produced when PCR stringency is excessively low resulting in non-specific PCR bands with variable lengths. This produces a ladder effect on an agarose gel. It then is advisable to choose PCR conditions that increase stringency. A smear of various sizes may also result from primers designed to highly repetitive sequences when amplifying genomic DNA. However, the same primers may amplify a target sequence on a plasmid without encountering the same problem.
- Lack of PCR products is likely due to reaction conditions that are too stringent. Primer dimers and hairpin loop structures that form with the primers or in the denatured template DNA may also prevent amplification of PCR products because these molecules may no longer base pair with the desired DNA counterpart.
- If the G-C content has not been analyzed, it is time to do so. PCR of G-C rich regions (GC content >60%) pose some of the greatest challenges to PCR. However, there are many additives that have been used to help alleviate the challenges.
8. Manipulating PCR Reagents
Understanding the function of reagents used on conventional PCR is critical when first deciding how best to alter reaction conditions to obtain the desired product. Success simply may rely on changing the concentration of MgCl2, KCl, dNTPs, primers, template DNA, or DNA polymerase. However, the wrong concentration of such reagents may lead to spurious results, decreasing the stringency of the reaction. When troubleshooting PCR, only one reagent should be manipulated at a time. However, it may be prudent to titrate the manipulated reagent.
- Magnesium salt Mg2+ (final reaction concentration of 0.5 to 5.0 mM)
Thermostable DNA polymerases require the presence of magnesium to act as a cofactor during the reaction process. Changing the magnesium concentration is one of the easiest reagents to manipulate with perhaps the greatest impact on the stringency of PCR. In general, the PCR product yield will increase with the addition of greater concentrations of Mg2+. However, increased concentrations of Mg2+ will also decrease the specificity and fidelity of the DNA polymerase. Most manufacturers include a solution of Magnesium chloride (MgCl2) along with the DNA polymerase and a 10X PCR buffer solution. The 10 X PCR buffer solutions may contain 15 mM MgCl2, which is enough for a typical PCR reaction, or it may be added separately at a concentration optimized for a particular reaction. Mg2+ is not actually consumed in the reaction, but the reaction cannot proceed without it being present. When there is too much Mg2+, it may prevent complete denaturation of the DNA template by stabilizing the duplex strand. Too much Mg2+ also can stabilize spurious annealing of primers to incorrect template sites and decrease specificity resulting in undesired PCR products. When there is not enough Mg2+, the reaction will not proceed, resulting in no PCR product. - Potassium salt K+ (final reaction concentration of 35 to 100 mM)
Longer PCR products (10 to 40 kb) benefit from reducing potassium salt (KCl) from its normal 50 mM reaction concentration, often in conjunction with the addition of DMSO and/or glycerol. If the desired amplicon is below 1000 bp and long non-specific products are forming, specificity may be improved by titrating KCl, increasing the concentration in 10 mM increments up to 100 mM. Increasing the salt concentration permits shorter DNA molecules to denature preferentially to longer DNA molecules. - Deoxynucleotide 5′-triphosphates (final reaction concentration of 20 and 200 μM each)
Deoxynucleotide 5′-triphosphates (dNTPs) can cause problems for PCR if they are not at the appropriate equivalent concentrations (i.e., [A] = [T] = [C] = [G]) and/ or due to their instability from repeated freezing and thawing. The usual dNTP concentration is 50 μM of EACH of the four dNTPs. However, PCR can tolerate concentrations between 20 and 200 μM each. Lower concentrations of dNTPs may increase both the specificity and fidelity of the reaction while excessive dNTP concentrations can actually inhibit PCR. However, for longer PCR-fragments, a higher dNTP concentration may be required. A large change in the dNTP concentration may necessitate a corresponding change in the concentration of Mg2+. - Thermal stable DNA polymerases
PCR enzymes and buffers associated with those enzymes have come a long way since the initial Taq DNA polymerase was first employed. Thus, choosing an appropriate enzyme can be helpful for obtaining desired amplicon products. For example the use of Taq DNA polymerase may be preferred over Pfu DNA polymerase if processivity and/or the addition of an adenine residue to the 3′ ends of the PCR product is desired. The addition of a 3′ adenine has become a useful strategy for cloning PCR products into TA vectors whit 3′ thymine overhangs. However, if fidelity is more important an enzyme such as Pfu may be a better choice. Several manufactures have an array of specific DNA polymerases designed for specialized needs. Take a look at the reaction conditions and characteristics of the desired amplicon, and then match the PCR experiment with the appropriate DNA polymerase. Most manufactures have tables that aid DNA polymerase selection by listing characteristics such as fidelity, yield, speed, optimal target lengths, and whether it is useful for G-C rich amplification or hot start PCR. - Template DNA
DNA quality and purity will have a substantial effect on the likelihood of a successful PCR experiment. DNA and RNA concentrations can be determined using their optical density measurements at 260 nm (OD260). The mass of purified nucleic acids in solution is calculated at 50 μg/ml of double stranded DNA or 40 μg/ml for either RNA or single stranded DNA at an OD260 =1.0. DNA extraction contaminants are common inhibitors in PCR and should be carefully avoided. Common DNA extraction inhibitors of PCR include protein, RNA, organic solvents, and detergents. Using the maximum absorption of nucleic acids OD260 compared to that of proteins OD280 (OD260/280), it is possible to determine an estimate of the purity of extracted DNA. Ideally, the ratio of OD260/280 is between 1.8 and 2.0. Lower OD260/280 is indicative of protein and/ or solvent contamination which, in all probability, will be problematic for PCR.
In addition to the quality of template DNA, optimization of the quantity of DNA may greatly benefit the outcome of a PCR experiment. Although it is convenient to determine the quantity in ng/μl, which is often the output for modern nanospectrophotometers, the relevant unit for a successful PCR experiment is the number of molecules. That is, how many copies of DNA template contain a sequence complementary to the PCR primers? Optimal target molecules are between 104 to 107 molecules and may be calculated as was described in the notes above.
9. Additive Reagents
Additive reagents may yield results when all else fails. Understanding the reagents and what they are used for is critical in determining which reagents may be most effective in the acquisition of the desired PCR product. Adding reagents to the reaction is complicated by the fact that manipulation of one reagent may impact the usable concentration of another reagent. In addition to the reagents listed below, proprietary commercially available additives are available from many biotechnology companies.
10. Additives That Benefit G-C Rich Templates
- Dimethylsulfoxide (final reaction concentration of 1-10% DMSO)
In PCR experiments in which the template DNA is particularly G-C rich (GC content >60%), adding DMSO may enhance the reaction by disrupting base pairing and effectively lowering the Tm. Some Tm calculators include a variable entry for adding the concentration of DMSO desired in the PCR experiment. However, adding more than 2% DMSO may require adding more DNA polymerase as it has been demonstrated to inhibit Taq DNA polymerase. - Formamide (final reaction concentration of 1.25-10%)
Like DMSO, formamide also disrupts base pairing while increasing the stringency of primer annealing, which results in less non-specific priming and increased amplification efficiency. Formamide also has been shown to be an enhancer for G-C rich templates. - 7-deaza-2′-deoxyguanosine 5′-triphosphate (final reaction concentration of dc7GTP; 3 dc7GTP:1 dGTP 50 μM)
Using 3 parts, or 37.5 μM, of the guanosine base analog dc7GTP in conjunction with 1 part, or 12.5 μM, dGTP will destabilize formation of secondary structures in the product. As the amplicon or template DNA is denatured, it will often form secondary structures such as hairpin loops. Incorporation of dc7GTP into the DNA amplicon will prohibit formation of these aberrant structures.
Note:
dc7GTP attenuates the signal of ethidium bromide staining which is why it is used in a 3:1 ratio with dGTP.
- Betaine (final reaction concentration of 0.5M to 2.5M)
Betaine (N,N,N-trimethylglycine) is a zwitterionic amino acid analog that reduces and may even eliminate the DNA melting temperature dependence on nucleotide composition. It is used as an additive to aid PCR amplification of G-C rich targets. Betaine is often employed in combination with DMSO and can greatly enhance the chances of amplifying target DNA with high G-C content.
11. Additives That Help PCR in the Presence of Inhibitors
- Non ionic detergents function to suppress secondary structure formation and help stabilize the DNA polymerase. Non ionic detergents such as Triton X-100, Tween 20, or NP-40 may be used at reaction concentrations of 0.1 to 1% in order to increase amplicon production. However, concentrations above 1% may be inhibitory to PCR. The presence of non ionic detergents decreases PCR stringency, potentially leading to spurious product formation. However, their use will also neutralize the inhibitory affects of SDS, an occasional contaminant of DNA extraction protocols.
- Addition of specific proteins such as Bovine serum albumin (BSA) used at 400 ng/μl and/ or T4 gene 32 protein at 150 ng/μl aid PCR in the presence of inhibitors such as FeCl3, hemin, fulvic acid, humic acid, tannic acids, or extracts from feces, fresh water, and marine water. However, some PCR inhibitors, including bile salts, bilirubin, EDTA, NaCl, SDS, or Triton X-100, are not alleviated by addition of either BSA or T4 gene 32 protein.
12. Modifications to Cycling Conditions
- Optimizing the annealing temperature will enhance any PCR reaction and should be considered in combination with other additives and/ or along with other modifications to cycling conditions. Thus, in order to calculate the optimal annealing temperature the following equation is employed:
TaOPT = 0.3 TmPrimer + 0.7 TmProduct -14.9
TmPrimer is calculated as the Tm of the less stable pair using the equation:
TmPrimer = ((ΔH/(ΔS+R x ln(c/4)))-273.15 + 16.6 log[K+] Where ΔH is the sum of the nearest neighbor enthalpy changes for hybrids; ΔS is the sum of the nearest neighbor entropy changes; R is the Gas Constant (1.99 cal K-1 mol-1); C is the primer concentration; and [K+] is the potassium concentration.
The latter equation can be computed using one of the Tm calculators listed at the following website:
http://protein.bio.puc.cl/cardex/servers/melting/sup_mat/servers_list.html
TmProduct is calculated as follows:
TmProduct = 0.41(%G-C) + 16.6 log [K+] – 675/product length
For most PCR reactions the concentration of potassium ([K+]) is going to be 50 mM. - Hot start PCR is a versatile modification in which the initial denaturation time is increased dramatically (Table 4). This modification can be incorporated with or without other modifications to cycling conditions. Moreover, it is often used in conjunction with additives for temperamental amplicon formation. In fact, hot start PCR is increasingly included as a regular aspect of general cycling conditions. Hot start has been demonstrated to increase amplicon yield, while increasing the specificity and fidelity of the reaction. The rationale behind hot start PCR is to eliminate primer-dimer and non-specific priming that may result as a consequence of setting up the reaction below the Tm. Thus, a typical hot start reaction heats the sample to a temperature above the optimal Tm, at least to 60 °C before any amplification is able to occur. In general, the DNA polymerase is withheld from the reaction during the initial, elongated, denaturing time. Although other components of the reaction are sometimes omitted instead of the DNA polymerase, here we will focus on the DNA polymerase. There are several methods which allow the DNA polymerase to remain inactive or physically separated until the initial denaturation period has completed, including the use of a solid wax barrier, anti-DNA polymerase antibodies, and accessory proteins. Alternatively, the DNA polymerase may simply be added to the reaction after the initial denaturation cycle is complete.
- Touchdown PCR (TD-PCR) is intended to take some of the guess work out of the Tm calculation limitations by bracketing the calculated annealing temperatures. The concept is to design two phases of cycling conditions (Table 5). The first phase employs successively lower annealing temperatures every second cycle (traditionally 1.0 °C), starting at 10 °C above and finishing at the calculated Tm or slightly below. Phase two utilizes the standard 3-step conditions with the annealing temperature set at 5 °C below the calculated Tm for another 20 to 25 cycles. The function of the first phase should alleviate mispriming, conferring a 4-fold advantage to the correct product. Thus, after 10 cycles, a 410-fold advantage would yield 4096 copies of the correct product over any spurious priming.
- Stepdown PCR is similar to TD-PCR with fewer increments in the first phase of priming. As an example, the first phase lowers annealing temperatures every second cycle by 3 °C, starting at 10 °C above and finishing at 2 °C below the calculated Tm. Like TD-PCR, phase two utilizes the standard 3-step conditions with the annealing temperature set at 5 °C below the calculated Tm for another 20 to 25 cycles. This would allow the correct product a 256-fold advantage over false priming products.
- Slowdown PCR is simply a modification of TD-PCR and has been successful for amplifying extremely G-C rich (above 83%) sequences (Table 6). The concept takes into account a relatively new feature associated with modern thermal cyclers, which allows adjustment of the ramp speed as well as the cooling rate. The protocol also utilizes dc7GTP to reduce 2 °structure formation that could inhibit the reaction. The ramp speed is lowered to 2.5 °C s-1 with a cooling rate of 1.5 °C s-1 for the annealing cycles. The first phase starts with an annealing temperature of 70 °C and reduces the annealing temperature by 1 °C every 3 rounds until it reaches 58 °C. The second phase then continues with an annealing temperature of 58 °C for an additional 15 cycles.
- Nested PCR is a powerful tool used to eliminate spurious products. The use of nested primers is particularly helpful when there are several paralogous genes in a single genome or when there is low copy number of a target sequence within a heterogeneous population of orthologous sequences. The basic procedure involves two sets of primers that amplify a single region of DNA. The outer primers straddle the segment of interest and are used to generate PCR products that are often non-specific in 20 to 30 cycles. A small aliquot, usually about 5 μl from the first 50 μl reaction, is then used as the template DNA for another 20 to 30 rounds of amplification using the second set of primers that anneal to an internal location relative to the first set.
Other PCR protocols are more specialized and go beyond the scope of this paper. Examples include RACE-PCR, Multiplex-PCR, Vectorette-PCR, Quantitative-PCR, and RT-PCR.
13. Representative Results
Representative PCR results were generated by following the basic PCR protocols described above. The results incorporate several troubleshooting strategies to demonstrate the effect of various reagents and conditions on the reaction. Genes from the budding yeast Saccharomyces cerevisiae and from an uncharacterized Mycobacteriophage were amplified in these experiments. The standard 3-step PCR protocol outlined in Table 2 was employed for all three experiments described below.
Before setting up the PCR experiment, the genomic DNA from both S. cerevisiae and the Mycobacteriophage were quantified and diluted to a concentration that would allow between 104 and 107 molecules of DNA per reaction. The working stocks were prepared as follows. A genomic yeast DNA preparation yielded 104 ng/μl. A dilution to 10 ng/μl was generated by adding 48 μl into 452 μl of TE pH 8.0 buffer. Since the S. cerevisiae genome is about 12.5 Mb, 10 ng contain 7.41 X 105 molecules. The genomic Mycobacteriophage DNA preparation yielded 313 ng/μl. A dilution to 2 ng/μl was generated by adding 6.4 μl into 993.6 μl of TE pH 8.0 buffer. This phage DNA is about 67 Kb. Thus, 1 ng contains 2.73 X 107 molecules, which is at the upper limit of DNA generally used for a PCR. The working stocks were then used to generate the Master Mix solutions outlined in Table 7. Experiments varied cycling conditions as described below.
In Figure 3a, genomic DNA from S. cerevisiae was used as a template to amplify the GAL3 gene, which encodes a protein involved in galactose metabolism. The goal for this experiment was to determine the optimal Mg2+ concentration for this set of reagents. No MgCl2 was present in the original PCR buffer and had to be supplemented at the concentrations indicated with a range tested from 0.0 mM to 5.0 mM. As shown in the figure, a PCR product of the expected size (2098 bp) appears starting at a Mg2+ concentration of 2.5 mM (lane 6) with an optimal concentration at 4.0 mM (lane 9). The recommended concentration provided by the manufacturer was 1.5 mM, which is the amount provided in typical PCR buffers. Perhaps surprisingly, the necessary concentration needed for product formation in this experiment exceeded this amount.
A different DNA template was used for the experiment presented in Figure 3b. Genomic DNA from a Mycobacteriophage was used to amplify a conserved 566 bp DNA segment. Like the previous experiment, the optimal Mg2+ concentration had to be determined. As shown in Figure 3b, amplification of the desired PCR product requires at least 2.0 mM Mg2+ (lane 5). While there was more variability in the amount of product formed at increasing concentrations of MgCl2, the most PCR product was observed at 4 mM Mg2+ (lane 9), the same concentration observed for the yeast GAL3 gene.
Notice that in the experiments presented in Figures 3A and 3B, a discrete band was obtained using the cycling conditions thought to be optimal based on primer annealing temperatures. Specifically, the denaturation temperature was 95 °C with an annealing temperature of 61 °C, and the extension was carried out for 1 minute at 72 °C for 30 cycles. The final 5 minute extension was then done at 72 °C. For the third experiment presented in Figure 3c, three changes were made to the cycling conditions used to amplify the yeast GAL3 gene. First, the annealing temperature was reduced to a sub-optimal temperature of 58 °C. Second, the extension time was extended to 1 minute and 30 seconds. Third, the number of cycles was increased from 30 to 35 times. The purpose was to demonstrate the effects of sub-optimal amplification conditions (i.e., reducing the stringency of the reaction) on a PCR experiment. As shown in Figure 3c, what was a discrete band in Figure 3a, becomes a smear of non-specific products under these sub-optimal cycling conditions. Furthermore, with the overall stringency of the reaction reduced, a lower amount of Mg2+ is required to form an amplicon.
All three experiments illustrate that when Mg2+ concentrations are too low, there is no amplicon production. These results also demonstrate that when both the cycling conditions are correctly designed and the reagents are at optimal concentrations, the PCR experiment produces a discreet amplicon corresponding to the expected size. The results show the importance of performing PCR experiments at a sufficiently high stringency (e.g., discreet bands versus a smear). Moreover, the experiments indicate that changing one parameter can influence another parameter, thus affecting the reaction outcome.
| Reagent | Concentration of stock solutions | Volume | 13X ** Master Mix |
Final Concentration |
| Sterile H2O | Q.S. to 50 μl | Q.S. to 650 μl | ||
| PCR Buffer | 10X | 5 μl | 65 μl | 1X |
| dNTP’s | 10 mM | 1 μl | 13 μl | 200 μM |
| MgCl2 | 25 mM | 3 μl | 39 μl | 1.5 mM |
| Forward Primer | 20 μM = 20 pmol/μl | 1 μl | 13 μl | 20 pmol |
| Reverse Primer | 20 μM = 20 pmol/μl | 1 μl | 13 μl | 20 pmol |
| Template DNA | Variable | Variable | Variable | ~105 Molecules |
| Taq DNA Polymerase | 5 Units/μl* | 0.5 μl | 6.5 μl | 2.5 Units |
| 50 μl/Reaction |
Table 1. PCR reagents in the order they should be added.
*Units may vary between manufacturers
** Add all reagents to the Master Mix excluding any in need of titration or that may be variable to the reaction. The Master Mix depicted in the above table is calculated for 11 reactions plus 2 extra reactions to accommodate pipette transfer loss ensuring there is enough to aliquot to each reaction tube.
| Standard 3-step PCR Cycling | |||
| Cycle step | Temperature | Time | Number of Cycles |
| Initial Denaturation | 94 °C to 98 °C | 1 minute | 1 |
| Denaturation Annealing Extension |
94 °C 5 °C below Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and DNA polymerase dependent |
25-35 |
| Final Extension | 70 °C to 80 °C | 5 minutes | 1 |
| Hold* | 4 °C | ∞ | 1 |
Table 2. Standard 3-step PCR Cycling.
* Most thermal cyclers have the ability to pause at 4°C indefinitely at the end of the cycles.
| 2-step PCR Cycling | |||
| Cycle step | Temperature | Time | Number of Cycles |
| Initial Denaturation | 94 °C to 98 °C | 1 minute | 1 |
| Denaturation Annealing/Extension |
94 °C 70 °C to 80 °C |
10 to 60 seconds Amplicon and DNA polymerase dependent |
25-35 |
| Final Extension | 70 °C to 80 °C | 5 minutes | 1 |
Table 3. 2-step PCR Cycling.
| Hot Start PCR Cycling | |||
| Cycle step | Temperature | Time | Cycles |
| Initial Denaturation | 60 °C to 95 °C | 5 minute then add DNA polymerase | 1 |
| Denaturation Annealing Extension |
94 °C 5 °C below Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and DNA polymerase dependent |
25-35 |
| Final Extension | 70 °C to 80 °C | 5 minutes | 1 |
Table 4. Hot Start PCR Cycling.
| Touchdown PCR Cycling | |||
| Cycle step | Temperature | Time | Cycles |
| Initial Denaturation | 94 °C to 98 °C | 1 minute | 1 |
| Denaturation Annealing Extension |
94 °C X =10 °C above Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and DNA polymerase dependent |
2 |
| Denaturation Annealing Extension |
94 °C X-1 °C reduce 1 °C every other cycle 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and polymerase dependent |
28 |
| Denaturation Annealing Extension |
94 °C 5 °C below Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and DNA polymerase dependent |
20-25 |
| Final Extension | 70 °C to 80 °C | 5 minutes | 1 |
Table 5. Touchdown PCR Cycling.
| Slowdown PCR Cycling | |||
| Cycle step | Temperature | Time | Cycles |
| Initial Denaturation | 94 °C to 98 °C | 1 minute | 1 |
| Denaturation Annealing Extension |
94 °C X °C =10 °C above Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and polymerase dependent |
2 |
| Denaturation Annealing Extension |
94 °C X-1 °C reduce 1 °C every other cycle 70 °C to 80 °C* |
10 to 60 seconds 30 seconds Amplicon and polymerase dependent |
28 |
| Denaturation Annealing Extension |
94 °C 5 °C below Tm 70 °C to 80 °C |
10 to 60 seconds 30 seconds Amplicon and polymerase dependent |
20-25 |
| Final Extension | 70 °C to 80 °C | 5 minutes | 1 |
Table 6. Slowdown PCR Cycling.
*For slowdown PCR, the ramp speed is lowered to 2.5 °C s-1 with a cooling rate of 1.5 °C s-1 for the annealing cycles.
| Stock Solution | Volume added to 50 μl reaction | 13 X Yeast Master Mix | 13 X Phage Master Mix | Final Concentration | |||||||
| Sterile H2O | q.s. to 50 μl = 31 μl or 30.5 | q.s. to 650 μl = 396.5 | q.s. to 520 μl = 403 μl | ||||||||
| PCR Buffer | 10X | 5 μl | 65 μl | 65 μl | 1X | ||||||
| dNTP’s | 10 mM | 1 μl | 13 μl | 13 μl | 200 μM | ||||||
| MgCl2 | Titration | Added to each reaction | Added to each reaction | Added to each reaction | Variable see titration | ||||||
| Forward Primer | 20 μM = 20 pmol/μl | 1 μl | 13 μl | 13 μl | 20 pmol | ||||||
| Reverse Primer | 20 μM = 20 pmol/μl | 1 μl | 13 μl | 13 μl | 20 pmol | ||||||
| Template DNA | 2 ng/μl phage or 10 ng/μl Yeast | 0.5 μl Phage or 1 μl Yeast | 6.5 μl | 13 μl | ~107 Molecules Phage or ~105 Molecules Yeast | ||||||
| Polymerase | 0.5 Units/μl** | 0.5 μl | 6.5 μl | 6.5 μl | 0.5 Units/Reaction | ||||||
| 40 μl + 10(Titration) μl/ Reaction | |||||||||||
| TITRATION | |||||||||||
| [MgCl2] | 0.00 mM | 0.5 mM | 1.0 mM | 1.5mM | 2.0 mM | 2.5 mM | 3.0 mM | 3.5 mM | 4.0 mM | 4.5 mM | 5.0 mM |
| MgCl2 | 0.00 μl | 1.00 μl | 2.00 μl | 3.0 μl | 4.00 μl | 5.00 μl | 6.00 μl | 7.00 μl | 8.00 μl | 9.00 μl | 10.00 μl |
| H2O | 10.00 μl | 9.00 μl | 8.00 μl | 7.00 μl | 6.00 μl | 5.00 μl | 4.00 μl | 3.00 μl | 2.00 μl | 1.00 μl | 0.00 μl |
Table 7. Titration of Mg2+ used in Figure 3.
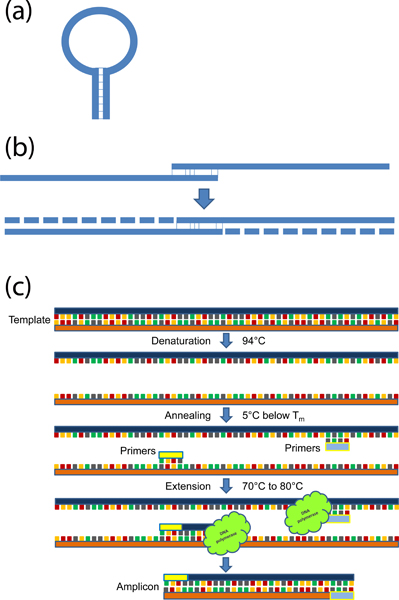
Figure 1. Common problems that arise with primers and 3-step PCR amplification of target DNA. (a) Self-annealing of primers resulting in formation of secondary hairpin loop structure. Note that primers do not always anneal at the extreme ends and may form smaller loop structures. (b) Primer annealing to each other, rather than the DNA template, creating primer dimers. Once the primers anneal to each other they will elongate to the primer ends. (c) PCR cycles generating a specific amplicon. Standard 3-step PCR cycling include denaturation of the template DNA, annealing of primers, and extension of the target DNA (amplicon) by DNA polymerase.

Figure 2. Ice bucket with reagents, pipettes, and racks required for a PCR. (1.) P-200 pipette, (2.) P-1000 pipette, (3.) P-20 pipette, (4.) P-10 pipette, (5.) 96 well plate and 0.2 ml thin walled PCR tubes, (6.) Reagents including Taq polymerase, 10X PCR buffer, MgCl2, sterile water, dNTPs, primers, and template DNA, (7.) 1.8 ml tubes and rack.

Figure 3. Example of a Mg2+ titrations used to optimize a PCR experiment using a standard 3-step PCR protocol. (a) S. cerevisiae Yeast genomic DNA was used as a template to amplify a 2098 bp GAL3 gene. In lanes 1 – 6, where the Mg2+ concentration is too low, there either is no product formed (lanes 1-5) or very little product formed (lane 6). Lanes 7 – 11 represent optimal concentrations of Mg2+ for this PCR experiment as indicated by the presence of the 2098 bp amplicon product. (b) An uncharacterized mycobacteriophage genomic DNA template was used to amplify a 566 bp amplicon. Lanes 1 – 4, the Mg2+ concentration is too low, as indicated by the absence of product. Lanes 5 – 11 represent optimal concentrations of Mg2+ for this PCR as indicated by the presence of the 566 kb amplicon product. (c) . S. cerevisiae Yeast genomic DNA was used as a template to amplify a 2098 bp GAL3 gene as indicated in panel a. However, the annealing temperature was reduced from 61 °C to 58 °C, resulting in a non-specific PCR bands with variable lengths producing a smearing effect on the agarose gel. Lanes 1 – 4, where the Mg2+ concentration is too low, there is no product formed. Lanes 5 – 8 represent optimal concentrations of Mg2+ for this PCR as seen by the presence of a smear and band around the 2098 kb amplicon product size. Lanes 9 – 11 are indicative of excessively stringent conditions with no product formed. (a-c) Lanes 12 is a negative control that did not contain any template DNA. Lane M (marker) was loaded with NEB 1kb Ladder.
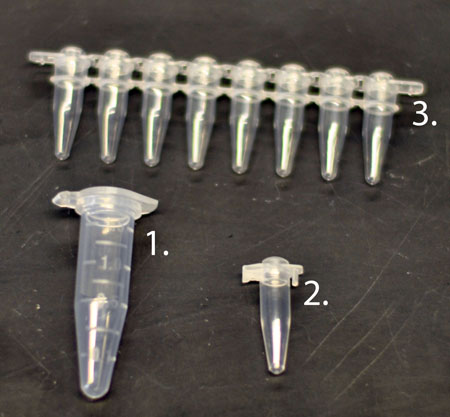
Figure 4. Sterile tubes used for PCR. (1.) 1.8 ml tube (2.) 0.2 ml individual thin walled PCR tube, (3.) 0.2 ml strip thin walled PCR tubes and caps.
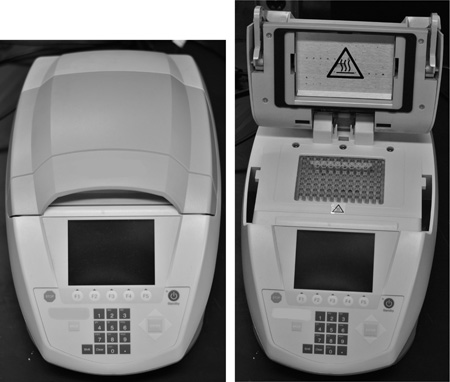
Figure 5. Thermal cycler. Closed thermal cycler left image. Right image contains 0.2 ml thin walled PCR tubes placed in the heating block of an open thermal cycler.
Discussion
PCR has become an indispensible tool in the biological science arsenal. PCR has altered the course of science allowing biologists to yield power over genomes, and make hybrid genes with novel functions, allowing specific and accurate clinical testing, gaining insights into genomes and diversity, as well as simply cloning genes for further biochemical analysis. PCR application is limited only by the imagination of the scientist that wields its power. There are many books and papers that describe new specialized uses of PCR, and many more will be developed over the next generation of biological science. However, regardless of the anticipated approaches, the fundamental framework has remained the same. PCR, in all its grandeur, is an in vitro application to generate large quantities of a specific segment of DNA.
Designing a PCR experiment requires thought and patience. The results shown in Figure 3 exemplify one of the major challenges when designing an optimization strategy for PCR. That is, as one parameter of PCR is changed, it may impact another. As an example, if the initial PCR was carried out at the sub-optimal annealing temperature (58°C) with an optimal Mg2+ concentration of 2.0 mM, then the result would produce a smear as seen in Figure 3c. An attempt to resolve the smear might involve setting up PCR conditions with reactions containing 2.0 mM MgCl2 and adjusting the annealing temperature to 61°C. However, as seen in Figure 3a, this would not yield any product. Consequently, it is advisable to titrate reagents, rather than adding one concentration to a single reaction, when troubleshooting spurious results. Also, the most common adjustments that are required for optimizing a PCR experiment are to change the Mg2+ concentration and to correct the annealing temperatures. However, if these changes do not minimize or abrogate aberrant results, titration of additives and /or changing the cycling condition protocols as described in Tables 2-6 may alleviate the problem. If all else fails, redesign the primers and try, try again.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Special thanks to Kris Reddi at UCLA for setting up reagents and pouring gels and to Erin Sanders at UCLA for inspiration, guidance, and support and proofreading the manuscript. I would also like to thanks Giancarlo Costaguta and Gregory S. Payne for supplying the yeast genomic DNA and primers to amplify the GAL3 gene. I would also like to thank Bhairav Shah for taking pictures of the lab equipment and reagents used to make figures 2 – 4. Funding for this project was provided by HHMI (HHMI Grant No. 52006944).
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Taq Polymeras | Sigma | D4545-25UN | |
| dNTPs | Qiagen | 201225 | |
| 0.2 ml Thin Wall Tubes PCR tubes | Bio Rad | 223-9469 | |
| 0.2 ml Thin Wall Tube caps | Bio Rad | 223-9472 | |
| TE Buffer: | |||
| EDTA disodium salt dihydrate | Sigma | E5134 | |
| Trizma-HCl | Sigma | T-3253 | |
| Custom Phage Primer | Invitrogen | 25441F_RL6_Contig2_68k TACTGCAACGCGATGTTGCG | |
| Custom Phage Primer | Invitrogen | 26007R_RL6_ Contig2_68k TCACCATCAATCGTGGCGGT | |
| Custom Yeast Primer | Integrated DNA Technologies | SacI-Gal3(-256) ACCTGGAGCTCCTATTT TAACATGTGGATTTCTTGAAAGAATGAAATCG | |
| Custom Yeast Primer | Integrated DNA Technologies | Gal3(1848)-SacI CGGATGGAGCTCGCGT GAAGGTAATTTTTTTTTATGTCTCCG | |
| Thermal Cycler | Bio Rad | 170-9703 | My Cycler |
| Agarose | ISC BioExpress | E-3120-500 | |
| Gel electrophoresis | Hoeffer Scientific Instruments | Model HE33 | |
| 1 kb DNA Ladder | New England BioLabs | N3232S | |
| Bio Rad Power Pack | Bio Rad | 164-5050 | PowerPac Basic |
| Gel Doc system | Fotodyne Incorporated | FOTO/Analyst Investigator FX Workstation | |
References
- Albert, J., Fenyo, E. M. Simple sensitive, and specific detection of human immunodeficiency virus type 1 in clinical specimens by polymerase chain reaction with nested primers. J. Clin. Microbiol. 28, 1560-1564 (1990).
- Baskaran, N., et al. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 6, 633-638 (1996).
- Blanchard, M. M., Taillon-Miller, P., Nowotny, P., Nowotny, V. PCR buffer optimization with uniform temperature regimen to facilitate automation. PCR Methods Appl. 2, 234-240 (1993).
- Borer, P. N., Dengler, B., Tinoco, I., Uhlenbeck, O. C. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 86, 843-853 (1974).
- Breslauer, K. J., Frank, R., Blocker, H., Marky, L. A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. U.S.A. 83, 3746-3750 (1986).
- Chou, Q., Russell, M., Birch, D. E., Raymond, J., Bloch, W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 20, 1717-1723 (1992).
- Coleman, W. B., Tsongalis, G. J. . Diagnostics for the clinical laboration. , (2005).
- D’Aquila, R. T., et al. Maximizing sensitivity and specificity of PCR by pre-amplification heating. Nucleic Acids Res. 19, 3749 (1991).
- Dieffenbach, C. W., Lowe, T. M., Dveksler, G. S. General concepts for PCR primer design. PCR Methods Appl. 3, S30-S37 (1993).
- Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K., Mattick, J. S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991).
- Erlich, H. A., Gelfand, D., Sninsky, J. J. Recent advances in the polymerase chain reaction. Science. 252, 1643-1651 (1991).
- Frey, U. H., Bachmann, H. S., Peters, J., Siffert, W. PCR-amplification of GC-rich regions: ‘slowdown PCR. Nat. Protoc. 3, 1312-1317 (2008).
- Haqqi, T. M., Sarkar, G., David, C. S., Sommer, S. S. Specific amplification with PCR of a refractory segment of genomic DNA. Nucleic Acids Res. 16, 11844 (1988).
- Henke, W., Herdel, K., Jung, K., Schnorr, D., Loening, S. A. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25, 3957-3958 (1997).
- Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. . PCR Protocols: A guide to methods and applications. , (1990).
- King, N. . Methods in Molecular Biology. 630, (2010).
- Kleppe, K., Ohtsuka, E., Kleppe, R., Molineux, I., Khorana, H. G. Studies on polynucleotides. XCVI. Repair replications of short synthetic DNA’s as catalyzed by DNA polymerases. J. Mol. Biol. 56, 341-361 (1971).
- Korbie, D. J., Mattick, J. S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 3, 1452-1456 (2008).
- Kramer, M. F., Coen, D. M. Enzymatic amplification of DNA by PCR: standard procedures and optimization. Curr. Protoc. Toxicol. Appendix 3, 1-14 (2001).
- Kramer, M. F., Coen, D. M. The polymerase chain reaction. Curr. Protoc. Protein Sci. Appendix 4, Appendix 4J (2002).
- Kreader, C. A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62, 1102-1106 (1996).
- Kwok, S., et al. Identification of human immunodeficiency virus sequences by using in vitro enzymatic amplification and oligomer cleavage detection. J. Virol. 61, 1690-1694 (1987).
- McConlogue, L., Brow, M. A., Innis, M. A. Structure-independent DNA amplification by PCR using 7-deaza-2′-deoxyguanosine. Nucleic Acids Res. 16, 9869 (1988).
- Mullis, K. B. The unusual origin of the polymerase chain reaction. Sci. Am. 262, 56-65 (1990).
- Mullis, K. B. Target amplification for DNA analysis by the polymerase chain reaction. Ann. Biol. Clin. (Paris). 48, 579-582 (1990).
- Mullis, K. B. The polymerase chain reaction in an anemic mode: how to avoid cold oligodeoxyribonuclear fusion). PCR Methods Appl. 1, 1-4 (1991).
- Mullis, K. B., Faloona, F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155, 335-350 (1987).
- Paabo, S., Gifford, J. A., Wilson, A. C. Mitochondrial DNA sequences from a 7000-year old brain. Nucleic Acids Res. 16, 9775-9787 (1988).
- Rees, W. A., Yager, T. D., Korte, J., von Hippel, P. H. Betaine can eliminate the base pair composition dependence of DNA melting. 생화학. 32, 137-144 (1993).
- Roux, K. H., Hecker, K. H. One-step optimization using touchdown and stepdown PCR. Methods Mol. Biol. 67, 39-45 (1997).
- Rychlik, W., Spencer, W. J., Rhoads, R. E. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 18, 6409-6412 (1990).
- Saiki, R. K., et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 239, 487-491 (1988).
- Saiki, R. K., et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 230, 1350-1354 (1985).
- Sambrook, J. A. R., David, W. . Molecular Cloning A Laboratory Manual. 2, (2001).
- Sarkar, G., Kapelner, S., Sommer, S. S. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 18, 7465 (1990).
- Smit, V. T., et al. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 16, 7773-7782 (1988).
- Wilson, I. G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63, 3741-3751 (1997).
- Wu, R. . Methods in Enzymology. 218, (1993).

