High-throughput Physical Mapping of Chromosomes using Automated in situ Hybridization
Summary
Genome assemblies based on massively parallel DNA sequencing technologies are usually highly fragmented. The development of physical chromosome maps can potentially improve genome assemblies. Here, we demonstrate innovative approaches to chromosome preparation, fluorescent in situ hybridization, and imaging that significantly increase throughput of the physical map development.
Abstract
Projects to obtain whole-genome sequences for 10,000 vertebrate species1 and for 5,000 insect and related arthropod species2 are expected to take place over the next 5 years. For example, the sequencing of the genomes for 15 malaria mosquitospecies is currently being done using an Illumina platform3,4. This Anopheles species cluster includes both vectors and non-vectors of malaria. When the genome assemblies become available, researchers will have the unique opportunity to perform comparative analysis for inferring evolutionary changes relevant to vector ability. However, it has proven difficult to use next-generation sequencing reads to generate high-quality de novo genome assemblies5. Moreover, the existing genome assemblies for Anopheles gambiae, although obtained using the Sanger method, are gapped or fragmented4,6.
Success of comparative genomic analyses will be limited if researchers deal with numerous sequencing contigs, rather than with chromosome-based genome assemblies. Fragmented, unmapped sequences create problems for genomic analyses because: (i) unidentified gaps cause incorrect or incomplete annotation of genomic sequences; (ii) unmapped sequences lead to confusion between paralogous genes and genes from different haplotypes; and (iii) the lack of chromosome assignment and orientation of the sequencing contigs does not allow for reconstructing rearrangement phylogeny and studying chromosome evolution. Developing high-resolution physical maps for species with newly sequenced genomes is a timely and cost-effective investment that will facilitate genome annotation, evolutionary analysis, and re-sequencing of individual genomes from natural populations7,8.
Here, we present innovative approaches to chromosome preparation, fluorescent in situ hybridization (FISH), and imaging that facilitate rapid development of physical maps. Using An. gambiae as an example, we demonstrate that the development of physical chromosome maps can potentially improve genome assemblies and, thus, the quality of genomic analyses. First, we use a high-pressure method to prepare polytene chromosome spreads. This method, originally developed for Drosophila9, allows the user to visualize more details on chromosomes than the regular squashing technique10. Second, a fully automated, front-end system for FISH is used for high-throughput physical genome mapping. The automated slide staining system runs multiple assays simultaneously and dramatically reduces hands-on time11. Third, an automatic fluorescent imaging system, which includes a motorized slide stage, automatically scans and photographs labeled chromosomes after FISH12. This system is especially useful for identifying and visualizing multiple chromosomal plates on the same slide. In addition, the scanning process captures a more uniform FISH result. Overall, the automated high-throughput physical mapping protocol is more efficient than a standard manual protocol.
Protocol
1. High-pressure Polytene Chromosome Preparation from Ovarian Nurse Cells
- Dissect half-gravid Anopheles females at 25 hours post-blood feeding under a dissection microscope and fix ovaries in fresh modified Carnoy’s solution with methanol (100% methanol: glacial acetic acid, 3:1). At this stage the ovaries are at Christophers’ III stage of development when follicles have an oval shape and a transparent area with nurse cells within follicles has a round shape13. Put ovaries from approximately five females into 500 μl of modified Carnoy’s solution in a 1.5 ml Eppendorf tube and keep them at room temperature for 24 hours. Transfer ovaries to -20 °C for a long-term storage.
- Prepare modified Carnoy’s solution with ethanol (100% ethanol: glacial acetic acid, 3:1) and 50% propionic acid just prior to making slides. Place one pair of ovaries on a dust-free slide in one drop of modified Carnoy’s solution. Split ovaries into approximately 6 sections with dissecting needles and place them into drops of 50% propionic acid on clean slides under a dissection MZ6 Leica stereomicroscope. Use a separate slide for each section.
- Separate follicles dissecting using needles and wipe out remaining tissue with filter paper or paper towel under a dissection microscope. Add a new drop of 50% propionic acid to the follicles and allow them to sit for 3-5 minutes at room temperature. Place a coverslip on top of the 50% propionic acid droplet. Let the slide stand for approximately 1 minute.
- Wrap the slide with filter paper and plastic. Using a Dremel 200 rotary tool with a Flex-Shaft attachment and soft plastic tip set between 3000-5000 RPM, express the follicles by swirling the tip in circles and lightly pressing the coverslip to evenly spread the nuclei. This step should take approximately 1 minute. Check the spread quality with an Olympus CX41 Phase Microscope using a 20x objective.
- Prepare a sandwich by placing an additional coverslip next to the coverslip covering the chromosomes, and cover them with a second microscope slide. This reduces the chance of crushing the slide in the vise. Wrap the slides with a plastic sheet that come in glass slide containers and filter paper to hold the sandwich and to protect the slide from scratching due to the vise.
- Apply pressure to the slides via mechanical vise. A pressure of 85-120 inch-lbs is sufficient and is achieved by using a torque wrench. This step is necessary for flattening the chromosomes as much as possible.
- Remove the second microscope slide and the additional coverslip. Heat the slide with the coverslip covering the chromosomes to 55 °C on a slide denaturation/hybridization system for 10-15 minutes to further flatten the chromosomes. Dip the slide in liquid nitrogen for at least 15 seconds, and, when bubbling has stopped, proceed to quickly remove the coverslip with a razor blade. Immediately place slides in cold 50% ethanol for 5 minutes. Dehydrate slides in 70%, 90%, 100% ethanol for 5 minutes each. Air dry slides.
2. FISH Using an Automated Slide Staining System
Program is set up with the Xmatrx automated slide staining system to run the following steps, except the probe labeling. The detailed nick-translation labeling protocol is described in the documentation provided with DNA polymerase from Fermentas.
- Before FISH, prepare fluorescent probes by labeling genomic BAC DNA with a fluorochrome using a nick-translation protocol. Mix 1 μg of DNA, 0.05 mM each of unlabeled dATP, dCTP, and dGTP and 0.015 mM dTTP, 1 μl Cy3- or Cy5-dUTP, 0.05 mg/ml BSA, 5 μl of 10x nick-translation buffer, 20 units of DNA polymerase I, 0.0012 units of DNase I, and nuclease-free water to 25 μl. The DNA polymerase I/DNase I ratio is selected empirically to obtain probes with a size range from 300 to 500 bp. Incubate the mix at 15 °C for 1 hour.
- Put slides and reagents into the Xmatrx automated slide staining system and start the program to run the following steps. Apply 800 μl of 1x PBS for 20 min. Blow slides with air. Perform formalin fixation by applying 450 μl of 4% formalin in 1x PBS for 1 min followed by washes with 100% ethanol for 1 sec twice and for 2 min once. Blow slides with air.
- Heat slides at 45 °C for 2 min to avoid bubbling when applying the probes. Apply 20 μl of the DNA probes, add drops of mineral oil to avoid evaporation of a hybridization solution, and place a coverslip on top. Denature chromosomes and DNA probes by heating slides at 90 °C for 10 min.
- For hybridization, incubate slides at 42 °C for 14 hours with coverslips on. The hybridization solution consists of 2x SSC, 100 mM sodium phosphate, 1x Denhardt’s solution, 100 μg/ml of sodium azide, and 10% dextran sulfate in formamide.
- For stringency washes, heat slides at 42 °C for 2 min, remove coverslips, wash slides in 2x SSC for 1 sec 4 times. Blow slides with air. Apply 800 μl of 0.4x SSC at 42 °C for 10 min 2 times. Wash slides in 2x SSC at 25 °C for 10 min.
- Perform chromosome staining by applying 50 μl of 1 μM YOYO-1 in 1x PBS. Apply drops of mineral oil to avoid evaporation of the staining solution, put on coverslips, and incubate at 25 °C for 10 min. Remove coverslips, wash slides in 2x SSC for 1 sec 4 times. Blow slides with air. Apply 15 μl of ProLong Gold antifade reagent. Put on coverslips.
3. Slide Reading with an Automatic Fluorescent Imaging System
This section details the use of the Duet software, which comes standard with the ACCORD PLUS automated scanning system. Instructions begin after turning on in this order: Olympus U-RFL-T power supply for a halogen bulb, computer Dell precision T3500, microscope Olympus BX61 with a connected camera Olympus U-CMAD3.
- For setting up 10x Pre-Scan, open the Duet software in the ACCORD PLUS automated scanning system. Click “Online” button. Enter new Case ID and assign a Slide ID. Click the dot labeled “BF.” This is the brightfield option. Set a scan choice to “10x Prescan.” Use “2500x circle,” “10000x circle” or “rectangle.” Click “Set&run” for 10x Pre-Scan.
- Click the “OK” button to run 10x Pre-Scan. Follow the prompts to adjust the scanning properly. Click “Finish” to start the scan. Press the “Main” tab to go back to the main screen. Click “Offline” button. Find the Case ID and Slide ID that was assigned and click “Offline Scan.” In the black box at the top left area click on an arrow and select “10x Prescan.” Using the arrows (< || Δ > a.k.a. “back,” “pause,” “play,” “forward” buttons), go through the scanned images.
- After finding an image of interest, double click on the screen the middle of the target region and press “Snap.” This will target an image for capture later on. After selecting all targets, click “Classify.” Select “10x Prescan.” Right click an image and get the chance to classify the images. Select “Polytene.”
- Set up 40x Pre-Scan in the Duet software. Click “Main” to go back to the main menu. Select “Online” again. Select a slide. Change “BF” to “FL.” Change the task name to “Revisit-X40-RG.” Change the section right below the last setting to “Revisit-ALL.” Click “Set&Run.” Press “OK.” Follow the prompts again to set up the automation. Click the “Start matching views” button to match 10x and 40x images. Click “Finish” to start the scan. Once done, click “Classify” and look at the images.
4. Representative Results
Figure 1 graphically depicts the scheme of the high-pressure chromosome preparation. This step involves the process of squashing and flattening chromosomes using a Dremel tool and mechanical vise, as well as chromosome visualization using a phase-contrast microscope. Figure 2 illustrates the hybridization of a fluorescently labeled probe to target DNA on the chromosome slide preparations using an automated slide staining system, the use of an automated scanning microscope for visualizing and mapping the probes after the FISH experiment, and placing and orienting genomic scaffolds on the chromosomes.
Preparations of ovarian nurse cell polytene chromosomes from females of An. gambiae were made using the high-pressure technique. This method does not damage or change most of the chromosome structure (Figure 3). It flattens bended chromosomes and, thus, reveals hidden fine bands that are not seen on regular preparations (Figure 4).
The probes used in this protocol are genomic BAC DNA clones obtained from a ND-TAM BAC library generated from An. gambiae PEST strain DNA. The genomic DNA for this library was extracted from newly hatched first instar larvae of both sexes. Figure 5 shows the results of FISH using BAC clones hybridized to polytene chromosomes of An. gambiae. This procedure was performed using the Xmatrx automated slide staining system. Using a cytogenetic photomap for An. gambiae10, two BAC clones, 102B24 (GenBank: BH372701, BH372694) and BAC 142O19 (GenBank: BH368703, BH368698), were localized in subdivisions 16C and 16D of the 2R arm, respectively. However, the BLAST search against the An. gambiae PEST strain AgamP3 assembly 14 identified the sequences homologous to 102B24 and 142O19 in subdivisions 16B and 17A of the 2R arm, respectively (Table 1). Therefore, the correspondence between the genomic coordinates and the cytogenetic subdivisions can now be adjusted according to our mapping data. The BLAST search against the An. gambiae M form and S form genome assemblies found that the two BAC clones are located in different contigs, but in the same scaffolds within each form (Table 1). The identified contigs can now be associated with specific chromosomal locations. Moreover, the identified scaffolds can now be properly oriented within the cytological subdivisions 16CD. Interestingly, the distances between the 102B24 and 142O19 sequences in the scaffolds are 1,892,981 bp, 1,658,391 bp, and 1,688,426 bp in the PEST strain, M-form, and S-form genome assemblies, respectively. Because, the PEST strain is a hybrid between the M and S forms, this difference is likely due to incorrect assembly of the PEST genome.
| BAC (accession) |
PEST-strain AgamP3 Coordinates |
M-form contigs Coordinates |
M-form scaffolds Coordinates |
S-form contigs Coordinates |
S-form scaffolds Coordinates |
| 102B24 (BH372694) |
2R:AAAB0100 8844_26 (16B) 43,681,342- 43,681,874 |
ABKP020247 53.1 32,513- 33,045 |
EQ090167.1 1,858,377- 1,858,909 |
ABKQ010123 32.1 4,299- 4,832 |
EQ099711.1 215,891- 216,424 |
| 102B24 (BH372701) |
2R:AAAB0100 8844_28 (16B) 43,788,213- 43,788,969 |
ABKP020247 51.1 24,816- 25,575 |
EQ090167.1, 1,772,683- 1,773,442 |
ABKQ010123 35.1 27,782- 28,540 |
EQ099711.1, 305,514- 306,272 |
| 142O19 (BH368698) |
2R:AAAB0100 8805_5 (17A) 45,428,580- 45,429,264 |
ABKP020247 01.1 2,499- 3,185 |
EQ090167.1 315,806- 316,492 |
ABKQ010123 76.1 49,242- 49,942 |
EQ099711.1 1,791,625- 1,792,325 |
| 142O19 (BH368703) |
2R:AAAB0100 8805_8 (17A) 45,560,099- 45,574,323 |
ABKP020220 17.1 30,971- 32,469 |
EQ090167.1 200,518- 202,016 |
ABKQ010123 79.1 27,760- 28,508 |
EQ099711.1 1,903,569- 1,904,317 |
Table 1. BLAST results of the BAC-end sequences against the genome assemblies of An. gambiae.
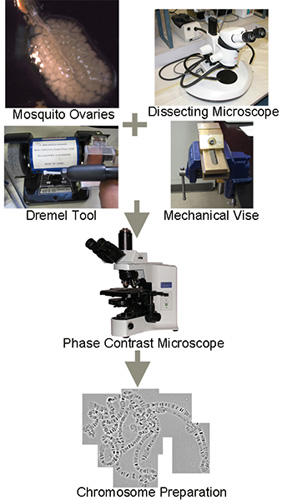
Figure 1. Schematic representation of high-pressure chromosome preparation. Mosquito ovaries are shown at the correct stage of development.
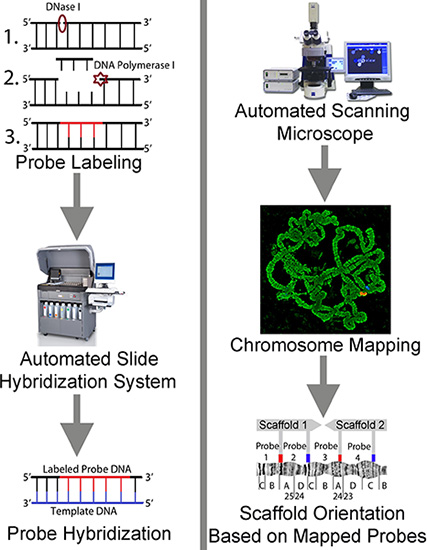
Figure 2. A scheme representing automated FISH, slide scanning, and chromosome mapping of genomic scaffolds.
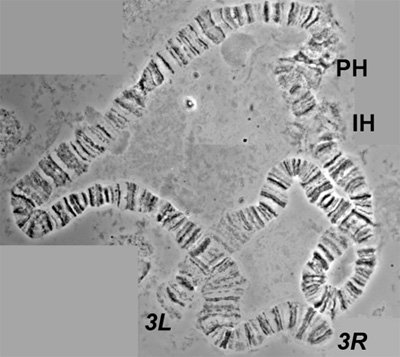
Figure 3. A spread of polytene chromosome 3 of An. gambiae obtained using the high-pressure technique. 3L and 3R mark left and right arms at their telomeres. PH and IH indicate pericentric and intercalary heterochromatin, respectively.
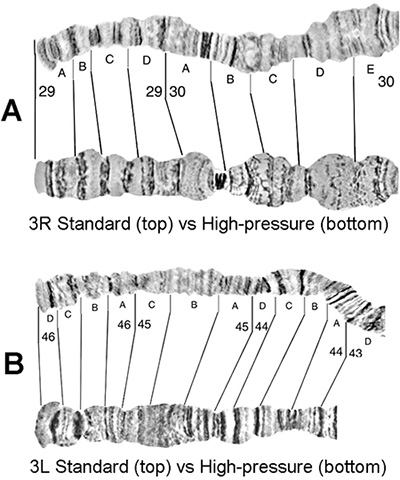
Figure 4. Comparison of An. gambiae polytene chromosomes prepared using the traditional (top) and high-pressure (bottom) technique. The images cover subdivisions 29A-30E of arm 3R (A) and 43D-46D of arm 3L (B).
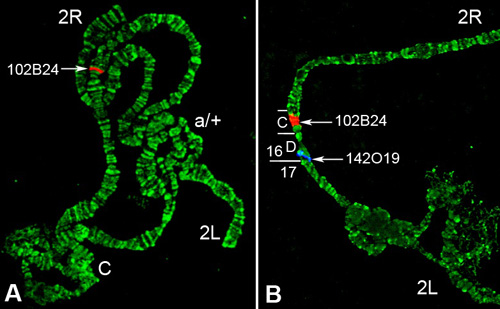
Figure 5. FISH of BAC clones to polytene chromosomes of An. gambiae. A) Hybridization of 102B24 (red signal) with the 2R arm. B) Dual-color FISH of 102B24 (red signal) and 142O19 (blue signal) to subdivisions 16C and 16D of the stretched 2R arm, respectively. Arrows indicate signals of hybridization of BAC clones labeled with Cy3 (red) and Cy5 (blue). C-the centromeric region. a/+ shows the heterozygote 2La inversion. Chromosomes were counterstained with the fluorophore YOYO-1.
Discussion
The most critical step of the high-pressure procedure is the proper squashing of ovarian nurse cells isolated at Christophers’ III stage of ovarian development13. Improper squashing can lead to insufficiently spread chromosomes, which can cause problems when trying to determine probe locations after FISH. If the chromosomes are over-squashed, they can be become broken or elongated to the point where banding patterns are lost. Production of multiple slides should lead to a consistency when attempting to squash the slides using the Dremel tool, which will increase overall slide production efficiency. The high-pressure technique was first developed for freshly isolated salivary glands of D. melanogaster9. However, ovaries of mosquitoes are routinely preserved in modified Carnoy’s fixative solution (3 methanol: 1 glacial acetic acid by volume) before they are used for chromosomal preparations. Therefore, we modified the existing high-pressure protocol to make it suitable for the fixed ovarian nurse cell polytene chromosomes of the malaria mosquito An. gambiae. Because the high pressure is applied using a precision vise possessing a highly parallel work surface of the entire slide, it takes significantly less time to prepare the chromosome squash than using a traditional tapping technique with a pencil’s eraser.
Other important steps include sufficiently soaking follicles in 50% propionic acid and heating the slide. Both of these steps are essential in helping to flatten the chromosomes. If they are neglected, chromosomes can appear shiny after dehydration, which potentially leads to an overabundance of background that can be mistaken for a signal in FISH. The high-pressure squashing technique also works well using the same solutions when making mitotic chromosome preparations. The additional spreading and flattening helps to better express the chromosomes from their nuclei. Adding equal pressure should flatten them accordingly to allow for measurements and potentially better resolution of bands visible from staining. The details about isolating mitotic chromosomes from mosquitoes are given elsewhere15. Although regular squash preparations are sufficient for many purposes including FISH 16 and immunostaining17, the high-pressure method not only lowers potential variance from one slide to the next, but also increases overall chromosome quality, leading to higher detail when mapping chromosomes. This procedure can also be used for preparing polytene chromosome membrane slides for laser-capture microdissection.
Limitations to the high-pressure method include slide breakage and overstretching of the chromosomes. Slide breakage caused by placing too much stress on the slide via the vise is possible, but is limited by using the pressures denoted in the article. For overstretching of the chromosomes, if the Dremel tool is applied to the slide for an extended period of time, there is the possibility that the chromosomes can become too stretched out and lose resolution. This can be remedied by applying the tool for a brief period of time, checking under the microscope, and applying more time with the Dremel tool if the chromosomes are insufficiently spread.
A traditional FISH protocol includes a number of washing and incubation steps, which are usually 5-20 min long and require almost full attention of a researcher for the whole duration of the experiment. Moreover, the number of slides that can be handled manually is usually limited to a few slides in a given experiment. In contrast, an automated slide staining system performs all steps (including washing, incubation, probe application, denaturation, hybridization, coverslip application and removal) automatically. This frees up to 6-8 working hours for a researcher in a FISH experiment. Moreover, an automated FISH system can significantly increase the throughput. For example, the Xmatrx system is capable of processing up to 40 assays on single preparation slides and up to 80 assays on dual preparation slides simultaneously. Among limitations of this system is that it is not efficient for a small number of FISH experiments as it requires preparing large volumes of solutions. In addition, the FISH protocol programmed in the system may require modifications and adjustments for new applications.
A slide scanning system is an automated version of a fluorescent microscope. Automated stage moving and a simple microscope control panel free a researcher’s time and make operating the microscope extremely simple. For instance, the software in the ACCORD PLUS scanning system allows for easy capturing of multiple channels of fluorescence, and easy to manage image acquisition. An example of this is the inclusion of z-stack capturing, rather than taking a single image; the software captures a configurable z-stack of images to ensure that at least one image remains in focus. Although this system is made more for multiple image acquisition (a lot of cells on a single slide), it still makes finding chromosomes on the slide much easier than navigating through the slide manually.
Both the automated slide staining system and the scanning system are routinely used for FISH diagnostics of chromosomal abnormalities in human cells by cytogenetics clinics. In this study, we developed a protocol that utilizes these automated systems for a basic research application. The automated high-throughput physical mapping protocol can facilitate rapid development of physical chromosome maps for any species of interest. Although, in this report, we used polytene chromosomes for physical mapping, mitotic chromosomes can also be utilized with these systems. It would be useful for the protocol to be adjusted to mitotic chromosomes because the majority of organisms do not develop readable polytene chromosomes. However, the polytene chromosomes, when available, can provide useful information about the correspondence of functional genome domains with the chromosome structure at the highest resolution. The organizational principles of polytene chromosomes, such as patterns of bands and interbands, have recently been likened to that of regular nonpolytene (interphase) chromosomes18. Therefore, the detailed physical mapping performed on high-pressure chromosome preparations has the potential to link DNA sequences to specific chromosomal structures such as bands, interbands, puffs, centromeres, telomeres, and heterochromatin; thus, creating chromosome-based genome assemblies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the grant from National Institutes of Health 1R21AI094289 to Igor V. Sharakhov. The Xmatrx Automated Slide Staining System and ACCORD PLUS Automated Scanning System were purchased with the help of the Equipment Trust Fund program, Fralin Life Science Institute, Department of Entomology and Department of Biochemistry of Virginia Tech.
Materials
| Name of the reagent/Equipment | Company | Catalogue number | Comments |
| 22×22 mm microscope coverslips | Fisher | 12-544-10 | |
| 18×18 mm microscope coverslips | Fisher | 12-553-402 | |
| Acetic acid | Fisher | A491-212 | |
| Methanol | Fisher | A412-4 | |
| Propionic acid | Sigma-Aldrich | 402907 | |
| Dremel 200 rotary tool with a Flex-Shaft attachment | Rand | 3″ Multi-purpose Mini Bench Grinder (with rate limiter) | |
| Specially designed 200 μl plastic tips (beaded plastic edges) for Dremel tool | Pipetman | F171300 | Cut and heat fat end of pipet tips |
| Tin coated rapid vise | Avenger Gold Toolmaker | MTC-200-1 | Precision ground square and parallel to within 0.00025 |
| Torque wrench | Craftsman | 44593 | |
| ThermoBrite Slide Denaturation/Hybridization System | Abbott Molecular | 30-144110 | |
| 25 mm barrier slides | Abbott Molecular | XT108-SL | |
| 25 mm coverslips | Abbott Molecular | XT122-90X | |
| Xmatrx Automated Slide Staining System | Abbott Molecular | 08L46-001 | |
| MZ6 Leica stereomicroscope | Leica | VA-OM-E194-354 | A different stereomicroscope can be used |
| Olympus CX41 Phase Microscope | Olympus | CX41RF-5 | A different phase microscope can be used |
| ACCORD PLUS Automated Scanning System | BioView (USA) | BV-5000-ACCP | |
| 10x PBS | Invitrogen | P5493 | |
| 10% NBF (neutral buffered formalin) | Sigma-Aldrich | HT501128 | |
| 99% formamide | Fisher Scientific | BP227500 | |
| Dextran sulfate sodium salt | Sigma | D8906 | |
| 20x SSC buffer | Invitrogen | AM9765 | |
| Sodium phosphate | Sigma-Aldrich | S3264 | |
| 50x Denhardt’s solution | Sigma-Aldrich | D2532 | |
| Sodium azide | Sigma-Aldrich | S2002 | |
| 1 mM YOYO-1 iodide (491/509) solution in DMSO | Invitrogen | Y3601 | |
| ProLong Gold antifade reagent | Invitrogen | P36930 | |
| dATP, dCTP, dGTP, dTTP | Fermentas | R0141, R0151, R0161, R0171 | |
| Cy3-dUTP, Cy5-dUTP | GE Healthcare | PA53022, PA55022 | |
| BSA | Sigma | A3294 | |
| DNA polymerase I | Fermentas | EP0041 | |
| DNase I | Fermentas | EN0521 |
References
- . Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J. Hered. 100, 659-674 (2009).
- Robinson, G. E., et al. Creating a Buzz About Insect Genomes. Science. 331, 1386-1386 (2011).
- Besansky, N. Genome Analysis Of Vectorial Capacity In Major Anopheles Vectors Of Malaria Parasites. White Paper. , 1-15 (2008).
- Alkan, C., Sajjadian, S., Eichler, E. E. Limitations of next-generation genome sequence assembly. Nature. 8, 61-65 (2011).
- Sharakhova, M. V., et al. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 8, R5 (2007).
- Lewin, H. A., Larkin, D. M., Pontius, J., O’Brien, S. J. Every genome sequence needs a good map. Genome Res. 19, 1925-1928 (2009).
- Bhutkar, A., et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. 유전학. 179, 1657-1680 (2008).
- Novikov, D., Kireev, I., Belmont, A. S. High-pressure treatment of polytene chromosomes improves structural resolution. Nature. 4, 483-485 (2007).
- George, P., Sharakhova, M. V., Sharakhov, I. V. High-resolution cytogenetic map for the African malaria vector Anopheles gambiae. Insect Mol. Biol. 19, 675-682 (2010).
- Clements, A. N. . The Biology of Mosquitoes: Development, Nutrition and Reproduction. , (1992).
- Sharakhova, M. V., et al. Imaginal discs–a new source of chromosomes for genome mapping of the yellow fever mosquito Aedes aegypti. PLoS Negl. Trop. Dis. 5, e10 (2011).
- Scriven, P. N., Kirby, T. L., Ogilvie, C. M. FISH for Pre-implantation Genetic Diagnosis. J. Vis. Exp. (48), e2570 (2011).
- Cai, W., Jin, Y., Girton, J., Johansen, J., Johansen, K. M. Preparation of Drosophila Polytene Chromosome Squashes for Antibody Labeling. J. Vis. Exp. (36), e1748 (2010).
- Vatolina, T. Y., et al. Correction: Identical Functional Organization of Nonpolytene and Polytene Chromosomes in Drosophila melanogaster. PLoS ONE. 6, (2011).

