Monitoring the Assembly of a Secreted Bacterial Virulence Factor Using Site-specific Crosslinking
Summary
This article illustrates the use of pulse-chase radio labeling in combination with site-specific photocrosslinking to monitor interactions between a protein of interest and other factors in E. coli. Unlike traditional chemical cross-linking methods, this approach generates high resolution “snapshots” of an ordered assembly pathway in a living cell.
Abstract
This article describes a method to detect and analyze dynamic interactions between a protein of interest and other factors in vivo. Our method is based on the amber suppression technology that was originally developed by Peter Schultz and colleagues1. An amber mutation is first introduced at a specific codon of the gene encoding the protein of interest. The amber mutant is then expressed in E. coli together with genes encoding an amber suppressor tRNA and an amino acyl-tRNA synthetase derived from Methanococcus jannaschii. Using this system, the photo activatable amino acid analog p-benzoylphenylalanine (Bpa) is incorporated at the amber codon. Cells are then irradiated with ultraviolet light to covalently link the Bpa residue to proteins that are located within 3-8 Å. Photocrosslinking is performed in combination with pulse-chase labeling and immunoprecipitation of the protein of interest in order to monitor changes in protein-protein interactions that occur over a time scale of seconds to minutes. We optimized the procedure to study the assembly of a bacterial virulence factor that consists of two independent domains, a domain that is integrated into the outer membrane and a domain that is translocated into the extracellular space, but the method can be used to study many different assembly processes and biological pathways in both prokaryotic and eukaryotic cells. In principle interacting factors and even specific residues of interacting factors that bind to a protein of interest can be identified by mass spectrometry.
Introduction
It is often essential to identify and characterize interactions between a protein of interest and other cellular components to define its role in a biological pathway, to understand its assembly, or to elucidate its mechanism of action. A wide variety of approaches are commonly used to study protein-protein interactions, but all have their limitations. The simplest unbiased method to identify proteins that bind to a protein of interest is copurification (or coimmunoprecipitation) but this approach requires that a protein complex remain intact during the purification procedure. Weak or transient protein interactions can be stabilized through chemical cross-linking, but this method typically requires the use of compounds that link proteins through primary amines that are widely scattered in the protein sequence. Complicated cross-linking patterns that are difficult to interpret can be generated, especially when proteins of interest are components of multiprotein complexes. Furthermore, because the spacer arms of chemical cross-linkers generally exceed 10 Å in length, covalent bonds can be formed with proteins that are located in proximity to but do not interact directly with the protein of interest. Methods such as affinity chromatography and two-hybrid screens are also often useful to identify protein-protein interactions. The former approach, however, requires reproducing conditions that promote physiologically significant interactions and cannot be used to detect weak interactions. The latter approach requires that binding interactions can be replicated in the host organism and are maintained when a protein of interest and its binding partners are placed in the context of a fusion protein. Two-hybrid methods also tend to generate false-positive results2. Once a protein-protein interaction has been established, considerable work is often required to map the site of interaction. Perhaps the most significant drawback of traditional approaches is that they do not provide any information about the temporal sequence of intermolecular interactions or the kinetics of binding.
This paper describes a simple method that overcomes some of the limitations of other approaches that are used to study protein-protein interactions. Initially a single amber mutation is introduced into the gene that encodes a protein of interest. The amber mutant is then coexpressed with an amber suppressor tRNA and an amino acyl-tRNA synthetase derived from Methanococcus jannaschii that have been engineered to incorporate the photoactivatable amino acid analog p-benzoylphenylalanine (Bpa) only at amber codons3. Irradiation of cells with long wavelength ultraviolet (UV) light (~365 nm) then facilitates formation of a covalent bond between Bpa and proteins that are within ~3-8 Å4,5. In some experimental contexts, cross-links can also be formed between activated Bpa and nonprotein components of cells such as lipids and nucleic acids6. Molecules that are terminated at the amber codon should generally be easily distinguishable from the full-length protein on SDS-PAGE and cannot be cross-linked to other proteins. By subjecting cells to pulse-chase radiolabeling prior to cross-linking and then immunoprecipitating or affinity purifying cross-linking products, the sequence and duration of protein-protein interactions can be established. The ability to generate temporal information about intermolecular interactions is especially valuable to study the progression of a protein of interest through any ordered multistep assembly, trafficking or signaling pathway and to identify pathway intermediates. Like chemical cross-linking, site-specific cross-linking detects protein-protein interactions that occur under physiological conditions, but the introduction of a cross-linker at a single position facilitates the analysis of interactions between individual segments of a protein and other factors. This unique feature of site-specific cross-linking is especially advantageous when the protein of interest has multiple segments or domains that have the potential to interact with distinct binding partners. Furthermore, site-specific cross-linking can be used in combination with methods such as mass spectrometry to map the residues that mediate protein-protein interactions with precision. Although this method is optimized for use in E. coli, the incorporation of photactivatable amino acid analogs into proteins by amber suppression has been reported in Mycobacterium, yeast and mammalian cells7-11. Thus, in principle our method can be used in other systems.
We have used this method extensively to identify intermolecular interactions that facilitate the biogenesis of EspP, an E. coli O157:H7 virulence factor. EspP is a member of a superfamily of proteins known as "autotransporters" that contain a large N-terminal extracellular domain ("passenger domain") and aC-terminal domain ("b domain") that folds into a cylindrical b barrel structure and anchors the protein to the outer membrane (OM)12. The mechanism by which the passenger domain is translocated across the OM has been a long-standing mystery. Like all bacterial cell surface proteins, EspP must be transported across the inner membrane, shuttled across the periplasm (the space between the inner and outer membranes), and targeted to the OM in a series of ordered events. Following its translocation across the OM, the EspP passenger domain is also separated from the b domain by a proteolytic cleavage and released from the cell surface. By combining site-specific cross-linking with pulse-chase radiolabeling, we have shown that both domains of the protein initially interact with the molecular chaperone Skp in the periplasm6. Subsequently, discrete segments of the b domain interact in a stereospecific fashion with components of a heterooligomer called the Bam complex, which catalyzes the integration of b barrel proteins into the bacterial OM by an unknown mechanism. Perhaps most interestingly, we have found that the passenger domain is in contact with one of the Bam complex subunits (BamA) as it traverses the OM13. Our results have enabled us to construct a detailed model for autotransporter assembly14 and have implicated the Bam complex in the transport of the passenger domain across the OM.
Protocol
1. Plasmid Construction
- After cloning a gene that encodes a protein of interest into an appropriate expression vector, introduce amber mutations at desired locations using a site-directed mutagenesis kit.
NOTE: Structural information and surface exposure predictions can serve as useful guides to determine the positions of mutations. Because the efficiency of amber suppression and cross-linking can vary considerably, amber mutations should be introduced at multiple positions. Furthermore, because Bpa is an analog of tyrosine and phenylalanine, try to introduce amber mutations at positions that normally encode large hydrophobic amino acids to minimize perturbations of protein structure and function. Finally, when choosing an expression vector keep in mind that overproducing a protein of interest at a very high level might limit the efficiency of Bpa incorporation.
2. Culture Preparation
- Use electroporation to transform an appropriate E. coli strain with a plasmid that encodes the amber suppression system (pDULE-pBpa; see refrence4) and a compatible plasmid that encodes the protein of interest with an amber mutation.
- Plate cells on LB agar containing appropriate antibiotics (use 5 µg/ml tetracycline to select for pDULE-pBpa). Incubate the plates O/N at 37 °C.
- On the following day, start O/N cultures from single colonies in 3-5 ml M9 medium14 containing 0.2% glycerol, all of the L-amino acids except methionine and cysteine (40 µg/ml) and antibiotics. Incubate at 37 °C.
- On the next day, centrifuge O/N cultures at 2,500 x g for 5 min at RT to pellet cells. Resuspend the cells in 5 ml fresh M9 medium and repeat the centrifugation.
NOTE: This wash step is critical to maintain plasmids that encode ampicillin resistance because small amounts of b lactamase that are present in the medium from O/N cultures degrade freshly added ampicillin before significant cell growth occurs. - Resuspend the cells in 2 ml fresh medium and measure cell concentration in a spectrophotometer (OD550).
- Add cells to a 250 ml Erlenmeyer flask containing 50 ml M9 medium and antibiotics at OD550=0.03. Incubate the cultures at 37 °C in a shaking water bath.
3. Bpa Incorporation and Preparation for Photocrosslinking
NOTE: The protocol described below is for a sample time course that has three time points (0, 1, and 5 min). Adjust the volumes accordingly if a different number of time points is desired. An outline of the procedure is illustrated in Figure 1.
- While the cultures are growing, prepare a fresh 1 M (1,000x) solution of Bpa in 1 M NaOH (27 mg Bpa in 100 μl 1 M NaOH). Use a dark walled tube to keep the solution in the dark.
- When the cultures reach early to mid-log phase(OD550=0.2), add 50 μl 1 M Bpa to each culture. Add Bpa one drop at a time while swirling the flask to avoid precipitation. Next, add any inducers that are needed to drive expression of the amber mutant. Continue to incubate the cultures at 37 °C for an additional 30 min.
- During the 30 min induction period label six disposable 15 ml centrifuge tubes with the time point (0 min, 1 min, 5 min) and either "+ UV" or "- UV". Place the tubes on ice.
- Fill a second ice bucket with ice and place a 6-well tissue culture plate on top of the ice.
- Thaw high specific activity (1,000 mCi/mmol) 35S-methionine and 35S-cysteine (or a premade mixture of the two compounds) for the pulse labeling and prepare (or thaw) a solution of nonradioactive L-cysteine and L-methionine (100 mM each) for the chase.
- Turn on the UV lamp five min prior to radiolabeling.
NOTE: The lamp should be adjustable so that it can be positioned ~3-4 cm from the samples in the multiwell plate. - Add ice (~2 ml) to each tube labeled "- UV" and to two of the wells in the multiwell plate.
NOTE: Ice should be added to a third well immediately before the 5 min time point. It is essential to place ice directly into the tubes and wells to stop intracellular reactions immediately at each time point and to thereby obtain accurate "snapshots" of a specific biochemical pathway. - At the end of the 30 min induction period transfer 25 ml of the culture to a prewarmed 125 ml disposable Erlenmeyer flask. Place the flask in the 37 °C shaking water bath.
4. Pulse-chase Labeling and UV Irradiation
NOTE: The pulse-chase labeling and UV irradiation protocol has a lot of steps that must be performed in a timely manner and that require considerable organization and advance preparation. All of the reagents and tubes should be readily accessible. A protocol for an experiment that has 0, 1, and 5 min time points is presented below.
- At time 0:00 (min:sec) transfer 25 ml of culture into 125 ml flask.
- At time 0:30 add 30 µCi/ml radioactive amino acids; swirl quickly to mix. Place the flask into the 37 °C shaking water bath.
- At time 1:00 add 250 µl of the 100 mM methionine-cysteine solution; swirl quickly to mix.
- Immediately pipette 4 ml of the culture into one of the wells of the chilled multiwell plate that contains ice. Pipette a second 4 ml aliquot into the 15 ml tube labeled "0 min – UV". Return the flask to the water bath.
- At time 2:00 pipette 4 ml of the culture into the second well of the chilled multiwell plate that contains ice. Pipette another 4 ml aliquot into the 15 ml tube labeled "1 min – UV". Return the flask to the water bath.
- At time 6:00 pipette 4 ml of the culture into the third well of the chilled multiwell plate that contains ice. Pipette another 4 ml aliquot into the 15 ml tube labeled "5 min – UV".
- Place the ice bucket with the multiwell plate under the preheated UV lamp and irradiate each sample individually for 4 min.
NOTE: In experiments in which time points are spaced at least 4 min apart each sample should be irradiated as soon as it is pipetted into the multiwell plate. - Transfer the cells into the chilled 15 ml tubes labeled "0 min +UV", 1 min +UV", etc.
- Centrifuge cells at 2,500 x g for 10 min at 4 °C.
- Resuspend each sample in 300 µl M9 medium or PBS and add 33 µl 100% cold trichloroacetic acid (TCA) to precipitate proteins. Centrifuge the TCA precipitates in a microfuge at top speed for 10 min and remove the supernatant. At this point samples can be stored at – 20 °C.
5. Immunoprecipitation and Crosslinking Product Detection
- Add 50 µl solubilization buffer (15% glycerol, 200 mM Trisbase, 15 mM EDTA, 4% SDS, 2 mM PMSF) to each sample and heat with agitation at 95 °C for 5 min to solubilize precipitated protein.
- Add 1 ml radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1.0% Igepal CA-630, 0.5% deoxycholate, 0.1% SDS) and perform standard immunoprecipitations15 using one-fifth to one-half of each sample.
NOTE: Igepal CA-630 (octylphenyl-polyethylene glycol) is used in place of NP-40, which is no longer commercially available. The two compounds are chemically indistinguishable. - Resolve immunoprecipitated proteins by SDS-PAGE. Stain, destain, and dry gels.
- Expose gels to a phosphor screen O/N and detect radioactive proteins (including crosslinking products) with a phosphorimager.
Representative Results
We used site-specific photocrosslinking to identify proteins that interact with EspP during its journey to the OM. In the experiments that are shown here, phenylalanine codons at residues 1113 and 1214 were replaced with amber codons. Both positions are located in the bdomain, which integrates into the OM and serves as a membrane anchor. The crystal structure of the EspPβ domain16 shows that the two residues are on the periplasmic side of the b barrel but are separated by ~120° (see structure in Figures 2 and 3). E. coli strain AD20217was transformed with pDULE-pBpa and a plasmid (pRI22) encoding one of the two EspP amber mutants13. After IPTG was added to induce EspP synthesis, pulse-chase labeling was conducted and samples were collected at 0, 1, and 5 min time points. Half of each aliquot was UV irradiated while the other half was used as a nonirradiated control. Immunoprecipitations were then conducted using antisera generated against an EspP C-terminal peptide and against other proteins as described below. Immunoprecipitated proteins were resolved by SDS-PAGE.
Two different polypeptides were immunoprecipitated from both irradiated and control cells by the C-terminal anti-EspPantiserum (Figures 2 and 3, lanes 2-4 and 9-11). Initially a ~135 kD precursor form of the protein that contains covalently linked passenger and b domains (proEspP) predominated. At later time points the level of proEspP declined and the free b domain began to predominate as the passenger domain was translocated across the OM and separated from the b domain by a proteolytic cleavage. The UV-irradiated samples, however, clearly had higher molecular weight bands that were not present in the control samples (Figures 2 and 3,lanes 9-11; see also references6,14). These bands result from the crosslinking of proEspP to interacting proteins. Based on the mobility of these polypeptides we estimated the size of the interacting proteins and then determined whether they might correspond to known OM protein assembly factors. To test our predictions we performed additional immunoprecipitations using antisera generated against the putative interacting partners. We found that a ~150 kD polypeptide that was observed when Bpa was incorporated at both residues 1113 and 1214 could be immunoprecipitated with an antiserum against the 17 kD periplasmic chaperone Skp (Figures 2 and 3, lane 8). Furthermore, we found that larger polypeptides that were observed when Bpa was incorporated at only one of the two positions could be immunoprecipitated with antisera against the Bam complex subunits BamB and BamD, respectively (Figures 2 and 3, lanes 12-14).
These experiments provide both temporal and spatial information about EspP interactions during its assembly. The observation that interactions between proEspP and Bam complex proteins peaked later and persisted longer than interactions with Skp suggests that proEspP interacts with Skp first and then binds to the Bam complex. Moreover, the cross-linking of two residues located on opposite sides of the b domain to discrete Bam complex subunits suggests that the b barrel interacts with the Bam complex in a stereospecific fashion during its integration into the OM. This observation has important implications for understanding the mechanism by which the Bam complex catalyzes the membrane integration of b barrel proteins.
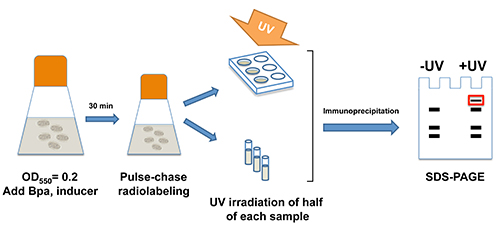
Figure 1. Outline of pulse-chase labeling and site-specific photocrosslinking protocol. Bpa and any inducers needed to drive the expression of the amber mutant are added when E. coli cultures are in log phase (OD550=0.2). Pulse-chase radiolabeling is conducted 30 min later. At appropriate time points aliquots are removed, placed in a multiwell plate and subjected to UV irradiation individually. An additional aliquot is removed at each time point but is not irradiated. The protein of interest is immunoprecipitated and run on SDS-PAGE. Bands that are observed in irradiated (“+UV”) but not control (“-UV”) samples arise from the cross-linking of the protein of interest to an interacting protein.
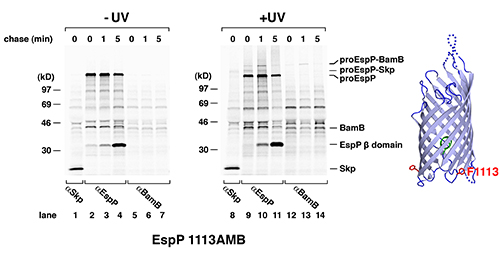
Figure 2. Interaction of EspP residue 1113 with Skp and BamB. E. coli transformed with pDULE-pBpa and a plasmid encoding the autotransporter EspP with an amber mutation at residue 1113 were grown in M9 medium and subjected to pulse-chase radiolabeling. An aliquot of the culture was UV irradiated at each time point, and an equal aliquot was left untreated. Immunoprecipitations were then conducted with the indicated antisera. The position of residue 1113 in the EspPβ domain is shown on the right.
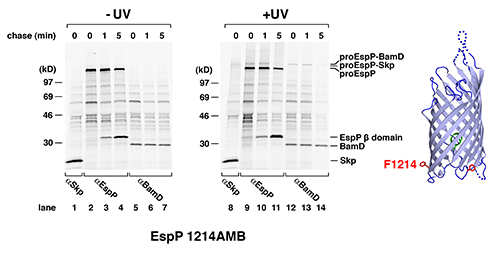
Figure 3. Interaction of EspP residue 1214 with Skp and BamD. The experiment described in Figure 2 was repeated except that the cells were transformed with pDULE-pBpa and a plasmid encoding the autotransporter EspP with an amber mutation at residue 1214. As shown on the right, residue 1214 is located ~120° away from residue 1113.
Discussion
This paper describes a method that combines site-specific photocrosslinking with pulse-chase labeling to examine the dynamics of interactions between a protein of interest and other components of a living cell. The method is especially useful to study multistep processes in which the protein interacts sequentially with other factors. Unlike traditional chemical cross-linking approaches, site-specific photocrosslinking provides detailed information about the status of individual segments or even individual residues of a protein of interest. This information is especially valuable in the analysis of proteins that have multiple independent domains or modules or, as illustrated here, that have relatively closely spaced residues that come in contact with distinct subunits of an oligomeric complex. Perhaps the greatest advantage of the method is that it can provide information about the sequence of events in a multistep pathway and the kinetics of protein-protein interactions. Furthermore, the incorporation of the crosslinking group into a protein of interest during its synthesis eliminates the need to perform time-consuming chemical reactions and to treat cells with chemical reagents that might alter the native environment in unpredictable ways. In principle, the use of long wavelength UV light to activate Bpa should not significantly damage cell components.
The key technical factors that influence the success of this method are the efficiency of amber suppression and the efficiency of UV-induced crosslinking. The efficiency of amber suppression can be estimated by determining the ratio of the full-length protein to the truncated protein that is produced when translation terminates at the amber codon. Amber suppression is clearly affected by the position (i.e. the translational context) of the amber mutation, but may also be affected by the overall rate of translation or other physiological parameters. We have found that the efficiency of amber suppression often exceeds 50% when we use laboratory strains of E. coli that harbor pDULE-pBpa and that are grown under the conditions described here. If an amber mutation is only weakly suppressed, we recommend altering the position of the mutation. It may also be useful to alter growth conditions or the host cell if possible. Of course the efficiency of crosslinking is strongly affected by the proximity of the benzophenone group of Bpa to reactive groups in interacting proteins. Because crosslinking efficiency is often difficult to predict, we recommend introducing Bpa at multiple positions in a protein of interest. It should also be noted that the strength of the UV light source and the distance of the lamp from the samples are critical. We have found that the tetracycline used to select for pDULE-pBpa fortuitously turns a pale yellow color that is visible after clear culture media (such as M9 medium) are UV irradiated for several minutes under a high intensity lamp. If the medium does not turn color, we recommend repeating the irradiation with a different bulb or light source.
The success of this method is also limited by the stability and dynamics of the protein-protein interactions under investigation. Very weak or transient interactions may be difficult to detect, but such interactions are also difficult to detect using other methods as well. Likewise, it may be difficult to use this method to determine the sequence of events in a pathway if those events are not well resolved temporally. From a practical point of view, the method is most useful for the analysis of transitions that occur over a time scale of minutes rather than seconds. Nevertheless, we have found that relatively transient interactions can sometimes be captured by lowering the incubation temperature of the culture and thereby slowing reaction rates12,14. In principle, the method should also be useful to detect protein-protein interactions that are ordinarily short-lived if cells that harbor mutations that slow down reactions or trap reaction intermediates can be obtained.
In some cases, the identification of the crosslinking partner may be challenging. As illustrated here, it is generally helpful to be able to generate hypotheses about potential crosslinking partners based on knowledge of the location, function and/or assembly status of the protein of interest. The identity of crosslinking partners can be tested not only by performing immunoprecipitations with appropriate antisera, but also by using knock-out strains or depletion strains that do not make the putative crosslinking partner. Knock-out strains can also be useful to provide an additional control when the crosslinking products are weak or immunoprecipitation results are ambiguous. In principle crosslinking partners can be identified in the absence of other information if they can be purified and subjected to mass spectrometry or if they have a unique chemical "signature" that can be determined using methods such as protease mapping or nonenzymatic digestion (e.g. cyanogen bromide digestion). Analysis of the crosslinking product by mass spectrometry, however, generally requires the use of larger cultures than those described here and the development of an effective purification protocol. We have found that when the protein of interest is abundant and the crosslinking reaction is highly efficient (i.e. in an "ideal situation"), the crosslinking product obtained after irradiating a ~500 ml culture of E. coli is sufficient for mass spectrometry analysis. Of course by analyzing a crosslinking product by mass spectrometry it becomes theoretically possible to identify a specific peptide in the target protein that interacts with a protein of interest and to thereby map the protein-protein interaction with great precision.
The protocol described here can also be modified in different ways and adapted to different conditions. For example, in cases where it is not possible or desirable to grow cells in a defined methionine and cysteine-free medium, a "pulse-chase" might be performed by inducing synthesis of a protein of interest for a brief period of time, adding an inhibitor of protein synthesis and then monitoring the subsequent fate of the newly synthesized protein by immunoprecipitation or Western blot. In addition, in cases where antibodies are not available to a protein of interest it should be possible to tag the protein (e.g. with a His tag) and to use other affinity methods (e.g. nickel chromatography) to isolate crosslinking products.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Materials
| QuikChange II Site-Directed Mutagenesis Kit | Agilent | 200521 | |
| BPA (H-p-Bz-Phe-OH) | Bachem | F-2800 | |
| TRAN35S-LABEL, Metabolic Labeling Reagent (35S-L-methionine and 35S-L-cysteine, >1,000 Ci/mmol) | MP Biomedicals | 51006 | |
| Spectroline SB-100P Super-High-Intensity UV Lamp, 365 nm | Spectronics Corporation | SB-110P | |
| Spectroline Replacement Bulb 100S | Spectronics Corporation | 11-992-15 | |
| Falcon Tissue Culture Plate, 6-well | Becton Dickinson Labware | 353046 | |
| Disposable 125 ml Erlenmeyer flask | Corning | 430421 | |
| Innova 3100 shaking water bath | New Brunswick Scientific | n.a. |
References
- Wang, L., Xie, J., Schultz, P. G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35, 225-249 (2006).
- von Mering, C., et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 417, 399-403 (2002).
- Chin, J. W., Martin, A. B., King, D. S., Wang, L., Schultz, P. G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. PNAS. 99, 11020-11024 (2002).
- Farrell, I. S., Toroney, R., Hazen, J. L., Mehr, R. A., Chin, J. W. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods. 2 (5), 377-384 (2005).
- Wittelsberger, A., Mierke, D. F., Rosenblatt, M. Mapping ligand-receptor interfaces: approaching the resolution limit of benzophenone-based photaffinity scanning. Chem. Biol. Drug Des. 71, 380-383 (2008).
- Ieva, R., Tian, P., Peterson, J. H., Bernstein, H. D. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. PNAS. 108, 383-391 (2011).
- Wang, F., Robbins, S., Guo, J., Shen, W., Schultz, P. G. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One. 5, (2010).
- Deiters, A., et al. Adding amino acids with novel reactivity to the genetic code of Saccharyomycescerevisiae. J. Am. Chem. Soc. 125, 11782-11783 (2003).
- Young, T. S., Ahmad, I., Brock, A., Schultz, P. G. Expanding the genetic repertoire of the methylotrophic yeast Pichiapastoris. 생화학. 48 (12), 2643-2653 (2009).
- Hino, N., et al. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat. Methods. 2 (3), 201-206 (2005).
- Liu, W., Brock, A., Chen, S., Chen, S., Schultz, P. G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 4 (3), 239-244 (2007).
- Leyton, D. L., Rossiter, A. E., Henderson, I. R. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 10 (3), 213-225 (2012).
- Ieva, R., Bernstein, H. D. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. PNAS. 106, 19120-19125 (2009).
- Pavlova, O., Peterson, J. H., Ieva, R., Bernstein, H. D. Mechanistic link between barrel assembly and the initiation of autotransporter secretion. PNAS. 110, (2013).
- Harlow, D., Lane, D. . Using antibodies: a laboratory manual. , (1999).
- Barnard, T. J., Dautin, N., Lukacik, N., Bernstein, P., Buchanan, S. K. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat. Struct. Mol. Biol. 14 (12), 1214-1220 (2007).
- Akiyama, Y., Ito, K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem. Biophys. Res. Commun. 167 (2), 711-715 (1990).

